Abstract
Differentiation from a haploid round spermatid to a highly streamlined, motile sperm requires temporal and spatial regulation of the expression of numerous proteins. One form of regulation is the storage of translationally repressed mRNAs. In Drosophila spermatocytes, the transcription of many of these translationally delayed mRNAs during spermiogenesis is in turn directly or indirectly regulated by testis-specific homologs of TATA-box-binding-protein-associated factors (tTAFs). Here we present evidence that expression of Mst77F, which is a specialized linker histone-like component of sperm chromatin, and of protamine B (ProtB), which contributes to formation of condensed sperm chromatin, is regulated at three levels. Transcription of Mst77F is guided by a short, promoter-proximal region, while expression of the Mst77F protein is regulated at two levels, early by translational repression via sequences mainly in the 5′ part of the ORF and later by either protein stabilization or translational activation, dependent on sequences in the ORF. The protB gene is a direct target of tTAFs, with very short upstream regulatory regions of protB (−105 to +94 bp) sufficient for both cell-type-specific transcription and repression of translation in spermatocytes. In addition, efficient accumulation of the ProtB protein in late elongating spermatids depends on sequences in the ORF. We present evidence that spermatocytes provide the transacting mechanisms for translational repression of these mRNAs, while spermatids contain the machinery to activate or stabilize protamine accumulation for sperm chromatin components. Thus, the proper spatiotemporal expression pattern of major sperm chromatin components depends on cell-type-specific mechanisms of transcriptional and translational control.
Keywords: tTAF target genes, Protamine, Mst77F, Transcriptional regulation, Translational repression, Translational activation
Introduction
The development of functional sperm requires stage- and cell-type-specific activation of numerous genes involved in the regulation of meiotic divisions, formation of the spermatid acrosome and flagellum, and extreme nuclear condensation. Developing male germ cells in both Drosophila and mammals store many translationally repressed mRNAs for later spermatid morphogenesis (Renkawitz-Pohl et al., 2005; Sassone-Corsi, 2002). In Drosophila, only a few genes are transcribed after the meiotic divisions (Barreau et al., 2008; Vibranovski et al., 2010), and therefore spermatid morphogenesis largely depends on stored, often translationally repressed mRNAs transcribed in spermatocytes (Fuller, 1993; Jayaramaiah Raja and Renkawitz-Pohl, 2005) though it cannot be excluded that stable proteins persist or mRNAs are translated before and after meiosis such as β2-tubulin (Michiels et al., 1993). In-situ hybridization experiments on adult testes indicate that at least 2100 genes are specifically activated in spermatocytes, and the majority of these transcripts persists during spermiogenesis (Zhao et al., 2010).
A striking example of such stored transcripts arises from a common feature of mammalian and Drosophila spermatid differentiation—the replacement of histones by protamines, which results in highly compact chromatin. The protamine-like proteins Protamine A (Mst35Ba, ProtA) and Protamine B (Mst35Bb, ProtB) and the linker histone-like protein Mst77F are major chromatin components of mature sperm in Drosophila. The corresponding mRNAs are transcribed in young spermatocytes but are translated only several days later, during late spermatid elongation stages (Jayaramaiah Raja and Renkawitz-Pohl, 2005; Rathke et al., 2007). Live imaging of testes and single cysts from flies carrying a protB-eGFP transgene revealed that accumulation of ProtB takes place 50–60 h after meiosis. Thus, protB mRNAs are stored in an untranslated state up to 6 days (Awe and Renkawitz-Pohl, 2010). Likewise, the sperm chromatin protein Mst77F is expressed in late spermatids from mRNAs transcribed in spermatocytes and stored. Mst77F genetically interacts with β2-tubulin, which indicates a further function of Mst77F (Fuller et al., 1989; Jayaramaiah Raja and Renkawitz-Pohl, 2005; Tweedie et al., 2009). Indeed, the Mst77F protein colocalizes with microtubules during nuclear shaping, for which it is essential (Jayaramaiah Raja and Renkawitz-Pohl, 2005; Rathke et al., 2010).
In Drosophila and in mammals, “non-prototypical” homologs of subunits of core promoter recognition complexes have been shown to guide cell-type-specific transcription (Goodrich and Tjian, 2010). In Drosophila spermatocytes, testis-specific homologs of TATA-box-binding-protein-associated factors (tTAFs) regulate the expression of many spermatid differentiation genes. Often these genes are transcribed solely in spermatocytes. Five such tTAFs have been characterized and proposed to act in a complex: Cannonball (Can; dTAF5 homolog), No hitter (Nht; dTAF4 homolog), Meiotic arrest (Mia; dTAF6 homolog), Spermatocyte arrest (Sa; dTAF8 homolog), and Ryan express (Rye; dTAF12 homolog) (Hiller et al., 2004, 2001). Germ cells from male flies mutant for any of these tTAF genes do not enter meiotic division and spermiogenesis. Although the cells mature spermatocytes, they fail to express the high levels of many spermiogenesis-relevant genes of normal spermatocytes (Hempel et al., 2006; Hiller et al., 2004; White-Cooper et al., 1998), which suggests that tTAFs directly and/or indirectly control robust transcription of many genes required for spermatid morphogenesis. Of these spermiogenesis-relevant, tTAF-dependent genes, so far only the promoters of fzo, Mst87F, and dj are known to be bound by tTAFs (Chen et al., 2005, 2011). The transcripts of each of these genes are spatiotemporally translated in a distinct, controlled manner. fzo encodes a GTPase relevant for mitochondrial morphogenesis (Hales and Fuller, 1997), Mst87F encodes an outer-dense fiber protein component of the sperm tail (Kuhn et al., 1988), and dj encodes a histone-like protein that is both a transient nuclear component and a persisting component of mitochondrial derivatives of the sperm tail (Santel et al., 1998). Whether other genes are under direct control of tTAFs remains to be investigated, especially as it has been proposed that the function of tTAFs partly counteracts Polycomb to allow transcription of spermiogenesis-relevant genes (Chen et al., 2005, 2011).
Here, we identified the sperm chromatin component ProtB as a direct transcriptional target of the tTAFs. We provide evidence that ProtB and Mst77F expression is regulated at three levels: transcription by tTAFs, translational repression, and selective activation of translation and/or protein accumulation in late spermatids dependent on the specific ORF.
Materials and methods
Fly strains and culture
Drosophila melanogaster strains were maintained on standard medium at 25 °C. w1 (Klemenz et al., 1987; Michiels et al., 1993) was used as wild-type unless otherwise specified. can12 mutants (Hiller et al., 2001) were used for chromatin immunoprecipitation (ChIP) and in-situ hybridization assays.
Cloning of promoter lacZ reporter constructs
Promoter lacZ reporter constructs were generated by PCR using genomic DNA from wild-type flies and appropriate primers with linked EcoRI and BamHI restriction sites. The PCR products were inserted into the transformation vector pChabΔsal, which supplies the 3′ UTR from SV40 (Thummel et al., 1988). Transgenic Drosophila lines were established by injection of DNA as described by Michiels et al. (1993). For protB, three promoter lacZ constructs were generated: pc-protB1 (−329 to +135, and the first intron in addition), pc-protB2ΔInt1 (−105 to +94), and pc-protB2ΔInt1+ ΔEx1 (−105 to +4). For Mst77F, the promoter lacZ constructs pc-77F1 (−278 to +172), pc-77F3 (−89 to +172), and pc-77F3Δ5′UTR (−89 to +38) were generated.
Cloning and monitoring expression of ProtamineB-eGFP under the control of the UAS–GAL4 system and the β2-tubulin (Tub85B) control region
For expression under the control of the UAS–GAL4 system, the complete ORF of protB fused in frame C-terminal to eGFP was cloned into the transformation vector pUASt (Brand and Perrimon, 1993), which contains the basic promoter and the 5′ UTR of the hsp70 gene and the 3′ UTR of SV40 with a polyadenylation signal. Transgenic fly lines were established in a w1 background. The expression of UAS-ProtB-eGFP in male germ cells was driven by bam-GAL4 (Chen and McKearin, 2003).
To promote the expression of ProtB under the control of β2-tubulin (β2t), a construct containing the regulatory region of β2t (from 677 bp upstream of the ATG start codon) and the ORF of protB (all but the stop codon) with a C-terminal eGFP was cloned into the transformation vector pChabΔsalΔlacZ, which provides the SV40 3′ UTR. The presence of the fusion gene in transgenic lines was verified by PCR using the primers EcoRI-β2t-Prsen (5′ GATGAATTCTCATTGTAGGAGCCAGAG 3′) and ProtB-BamHI-as (5′ GATGGATCCCTTGCAAATCCG 3′). As a control, the same β2t regulatory region was fused to eGFP and cloned into pChabΔsalΔlacZ, which provides the SV40 3′ UTR. At least two independent lines for protB constructs were analyzed and revealed the same expression pattern.
Adult testes were dissected, and then the eGFP signals were examined using a Zeiss microscope (AxioPlan2) equipped with appropriate fluorescence filters. Images were individually recorded and figures arranged with Adobe Photoshop CS2/CS5. Schemes designed with Adobe Illustrator CS5.
Cloning and monitoring expression of Mst77F-eGFP or Mst77F-mCherry and C-terminal truncated and Mst77F full-length under the control of the β2-tubulin (Tub85B) regulatory region
For expression of Mst77F under the control of the β2-tubulin (β2t) promoter and the 5′ UTR (Santel et al., 2000) a construct containing the regulatory region of β2t (from 677 bp upstream of the ATG) and the ORF of Mst77F (all but the stop codon) was cloned into the transformation vector pPWG (see The Drosophila Gateway (TM) Vector Collection), which resulted in a C-terminal in-frame fusion to eGFP. The 3′ UTR and polyadenylation signal were from KC10.
To generate C-terminal truncated Mst77F constructs, sequences from −448 to +712 (Mst77FΔ60C-eGFP) or −450 to +394 (Mst77FΔ141C-mCherry) were cloned in frame C-terminal to eGFP or mCherry in pChabΔsalΔlacZ containing the SV40 3′ UTR and polyadenylation signal.
Transgenic fly lines were established in a w1118 background. As pChabΔsalΔlacZ integrates randomly into the genome, three to five independent transgenic lines were analyzed for each construct, and all lines of each construct had the same expression pattern.
Antibodies and immunofluorescence staining
Hoechst staining was used to visualize chromatin. All antibodies were used in immunofluorescence stainings of squashed testis carried out essentially as described in Hime et al. (1996) and Rathke et al. (2007). To follow the fate of the histones, an anti-histone antibody from Chemicon, Millipore (MAB052; 1:1200) was used. This antibody recognizes H1 and all core histones.
Cy5-conjugated anti-mouse (Dianova; 1:100) was used as secondary antibody. Immunofluorescence, eGFP and Hoechst signals were examined using a Zeiss microscope (AxioPlan2) equipped with appropriate fluorescence filters. Images were individually recorded and figures arranged with Adobe Photoshop CS2/CS5. Schemes designed with Adobe Illustrator CS5.
In-situ hybridization and β-Galactosidase assay
Whole-mounts of adult testes were hybridized in-situ according to Morris et al. (2009) with minor modifications such as pre-hybridization, hybridization and washes in hybridization buffer at 55 °C instead of 65 °C. DIG-labeled RNA probes were generated using 500–800 bp fragments of the corresponding ORFs amplified by PCR from genomic DNA and cloned into the pCR®II-TOPO® vector (Invitrogen). β-Galactosidase activity was visualized by a histochemical reaction using the chromogenic substrate X-Gal. Adult testes were fixed in 0.7% glutaraldehyde at room temperature for 15 min, and larval testes were fixed for 5 min. The enzymatic staining reaction was incubated at 37 °C for 10 min (adult testis), 30 min or overnight (ovn, larval testis). After a 30-min time interval the w1 or w1118 host strains show no background activity.
Chromatin immunoprecipitation and quantitative PCR analyses
Chromatin immunoprecipitation (ChIP) and quantitative PCR analyses were performed as previously described in Chen et al. (2005) using anti-Sa antibody. Each ChIP assay contained chromatin of 50 pairs of testes dissected from either w1118 or homozygous can12-mutant flies. Input DNA and DNA immunoprecipitated with specific antibodies (ChIP-DNA) were analyzed by real-time PCR using the gene-specific primers as listed below and TaqMan probes with the Universal PCR Master Mix (Applied Biosystems Inc. #58003365-01) in an ABI 7300 Real-time PCR system. Each PCR reaction was performed in duplicate, and the Ct numbers for each reaction were collected and averaged. The amount of PCR product was quantified using the absolute quantification method with a standard curve. At least three independent ChIP reactions, each from an independent biological replicate, were performed for each data point. CycA was used as an internal control (Chen et al., 2011). The ChIP DNA was first normalized with the input DNA amount (Input %), and then the raw input % data were converted to data expressing the fold change over the constitutively expressed CycA gene from the same sample. The relative enrichment normalized to CycA was then averaged, and the two different genotypes, wt and can12, were compared. The gene-specific primers used for PCR were as follows: protB: Forward, CAAAACCTACGCCAACTATATGGAATAA; Reverse, CGTAACCACCTAACAGAAGGATGT; and FAM, TATCCGCC;GGCGTCTATCCGCCGGCGTC; Mst77F: Forward, AGCTAGTCGGATTGCAAACAGAATAT; Reverse, TTTCTTGATCAGTATCAGTCGAGCTG; and FAM, ACGGACGGACGAATAT; Mst87F: Forward. GTCAAACCGATATACCTGTGCGTAA; Reverse, ATGTGTTCAGGCCGAAAGGA; and FAM, CCAGATTTTGTATCATTATTATTTG; and CycA: Forward, CAACAGCAAGAAGGCAACGA; Reverse, GAGTCCGATTATGCTCTGCTCTT; and FAM, CCCTTCCTTCTCTCTTTCTC.
Results
Transcription of protB and Mst77F in primary spermatocytes requires tTAFs
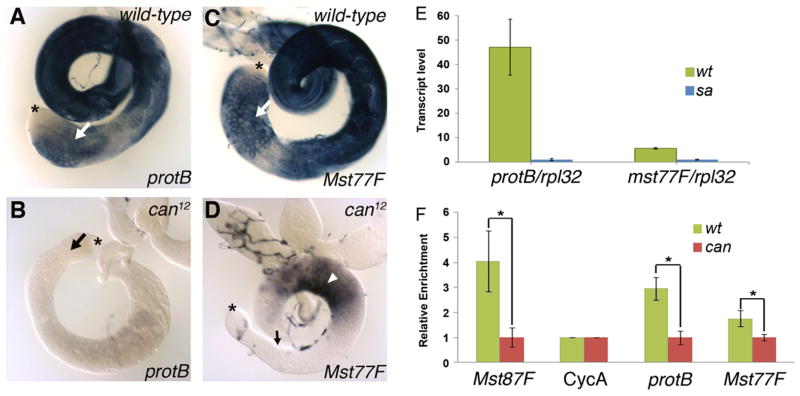
tTAF function was required for normal expression of protB and Mst77F transcripts in spermatocytes. In-situ hybridization to wild-type testes using antisense RNA probes for protB and Mst77F detected transcripts in spermatocytes (Fig. 1A, C, arrows) and in elongating spermatid stages, consistent with data of Jayaramaiah Raja and Renkawitz-Pohl (2005). In-situ hybridization to can12-mutant testis showed dramatic reduction in protB transcripts at all spermatocyte stages (Fig. 1B) in comparison to wild-type testis (Fig. 1A). Also the level of transcripts hybridizing with the Mst77F probe was dramatically reduced in the spermatocyte region of the can12 mutant testis (Fig. 1D, arrow), although some signal cross hybridizing with the Mst77F probe was still detected (Fig. 1D, arrowhead), which contains late-stage arrested spermatocytes and/or dying cysts in the mutant testis. Consistent with the in-situ hybridization data, a comparison of transcript levels in wild-type versus tTAF (sa−/−) mutant testes from microarray data of Chen et al. (2011) indicated that the level of protB transcripts was 12-fold lower and the level of Mst77F transcripts was over 4-fold lower in the tTAF (sa−/−) mutant than in wild-type testes (Fig. 1E).
Fig. 1.
Transcription of protB and Mst77F depends on tTAFs. (A–D) In-situ hybridization of whole-mount wild-type (A, C) and can12 (B, D) testes using protB-specific (A, B) and Mst77F-specific (C, D) probes. The staining reactions were incubated for the same length of time. Arrows point toward the spermatocytes; the arrowhead in D points toward the basal region; and asterisks indicate the tips of the testis, where germ-line stem cells reside. (E) Transcriptome analysis of transcript levels of protB and Mst77F in wild-type (wt) and tTAF (sa−/−) (sa) mutant testes. The results are from three independent experiments; error bars indicate the sd. (F) Quantitative real-time PCR analysis of anti-Sa ChIP using wild-type and can12 testes showing the enrichment of Sa at different targets. The results are from three independent experiments and are normalized to 1; error bars indicate the sd. All ChIP results were normalized to CycA within the same genotype, and then the results of the two genotypes were compared (*: P value <0.05 using two-sample t-test).
The reduction in Mst77F transcript levels in tTAF mutants compared to the wild-type observed by in-situ hybridization and transcriptome analysis using microarray data may underrepresent the actual effects of the tTAF (sa−/−) mutation on Mst77F transcription. Both the in-situ probe for Mst77F and the microarray assay likely detected signal from transcripts of the 18 Mst77F pseudogenes (Mst77Y-1 to Mst77Y-18) found on the Y chromosome in Drosophila melanogaster, in addition to the signal from transcripts of the autosomal Mst77F for which all our reporter constructs and fusion genes were designed. Compared to Mst77F, these Y-linked, possibly functional Mst77 copies have an additional 12 bp within their 5′ UTRs (Krsticevic et al., 2010; Russell and Kaiser, 1993). According to Krsticevic et al. (2010), 72% of the Mst77F-like mRNA expressed in wild-type testes arises from the autosomal Mst77F gene, while the remaining 28% arises from the Mst77Y genes. If this significant contribution of Mst77Y transcripts is independent of TAFs, it may account for both the only modest 4-fold apparent reduction (25% remaining transcripts) in Mst77F transcripts observed in the tTAF-mutant testes by microarray (Fig. 1E) as well as the transcripts detected by in-situ hybridization in the basal region of tTAF-mutant testes (Fig. 1D).
Chromatin immunoprecipitation (ChIP) assays indicated that protB might be a direct target of the tTAF Sa (Fig. 1F). Since the level of Sa protein does not change in can12-mutant testes (Chen et al., 2005), the occupancy of the tTAF Sa at promoters of target genes in testes from flies with a tTAF can mutation can be analyzed by ChIP using antibodies against Sa. In ChIP assays with anti-Sa, the genomic region containing the protB promoter in wild-type testes was enriched ~3-fold more than in can12-mutant testes (Fig. 1F). This compares favorably with a positive control tested in the same experiment, the Mst87F gene promoter (Chen et al., 2005), which was enriched 4.1-fold more in wild-type than in can12 mutant testes. The Mst77F locus in wild-type testes was enriched only modestly, i.e., ~1.8-fold more than in can12 mutant testes (Fig. 1F) in the same experiment.
Short upstream sequences regulate cell-type-specific expression of ProtB and Mst77F
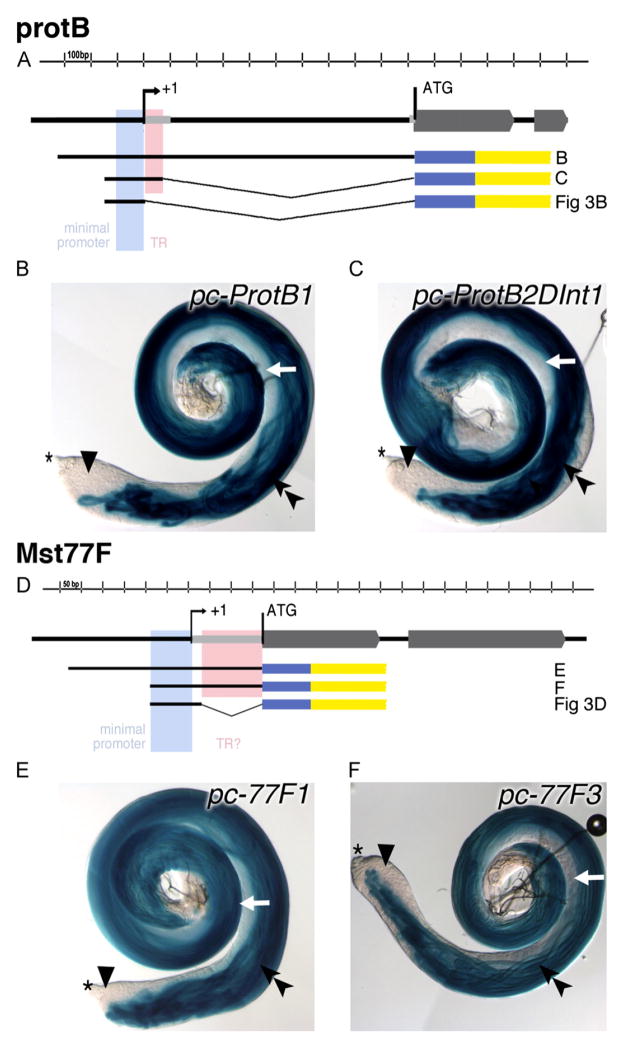
Deletion analyses of the region proximal to the transcription start site of protB and of Mst77F demonstrated that a very short upstream region was sufficient to ensure transcription of each gene in male germ cells. Our previous studies with protB-eGFP gene fusions indicated that a genomic fragment from 1683 bp upstream to the ATG encoding the translational start, i.e., a fragment containing the 5′ UTR and 655 bp upstream of the predicted transcriptional start site, was sufficient for a high level of cell-type- and stage-specific ProtB-eGFP expression (Jayaramaiah Raja and Renkawitz-Pohl, 2005). Similarly, studies with Mst77F-eGFP gene fusions indicated that a genomic fragment from 450 bp upstream to the ATG, i.e., a fragment containing the 5′ UTR plus 278 bp upstream of the predicted transcriptional start site, was sufficient (Jayaramaiah Raja and Renkawitz-Pohl, 2005). Further deletion analysis using transgenes in which regulatory regions drove expression of β-Galactosidase (summarized in Table 1) revealed that a genomic fragment containing sequences from 105 bp upstream of the predicted transcriptional start site of protB plus the first 94 bp of the 5′ UTR region (pc-protB2ΔInt1) was sufficient for β-Galactosidase expression in male germ cells (Fig. 2A, C). β-Galactosidase activity appeared only in elongated spermatid bundles (Fig. 2C; double arrowhead), whereas early haploid spermatids and younger stages revealed no β-Galactosidase activity (Fig. 2C; white arrow and arrowhead). The level of β-Galactosidase expression from pc-protB2ΔInt was comparable to that from the full-length promoter construct pc-protB1 (Fig. 2B, C; testes were incubated in parallel under the same conditions). Likewise, deletion analyses of similar reporter constructs for Mst77F (Fig. 2D) revealed that a region including from 89 bp upstream of the predicted transcriptional start site plus the 172 bp 5′ UTR was sufficient to drive strong expression of β-Galactosidase from a reporter transgene (pc-77F3) in spermatids, similar to a full-length promoter-lacZ construct (pc-77F1; Fig. 2E, F).
Table 1.
Summary of results of reporter gene assays and gene fusion expression of Mst77F and protB.
| Construct name | Promoter | 5′ UTR | ORF | 3′ UTR including polyadenylation signal | Reporter expressed in larval testes (0.5h staining, β-Gal or eGFP life) | Reporter expressed in larval testes (ovn staining β-Gal) | Reporter expressed in spermatocytes in adult testes (10 min staining this work β-Gal or life eGFP) | Reporter expressed in spermatids in adult testes (10 min staining, β-Gal or life eGFP) |
|---|---|---|---|---|---|---|---|---|
| protB-eGFP | protB | protB | protB-eGFP | SV40 | − | − | − | +late nulear |
| pc-ProtB1 | protB | protB | lacZ | SV40 | − | + | − | + |
| pc-ProtB2ΔInt1 | protB | Partial 5′ UTR | lacZ | SV40 | − | + | − | + |
| pc-ProtB2ΔInt1+ΔEx1 | protB | − | lacZ | SV40 | ++ | +++ | Likely | + |
| β2t | β2t | lacZ | SV40 | + | + | ? | +++ Michels et al. (1989) Santel et al. (2000) | |
| β2t-eGFP | β2t | β2t | eGFP | SV40 | Not analyzed | Not analyzed | ++ | +++ |
| β2t-protB-eGFP | β2t | β2t | protB-eGFP | SV40 | Not analyzed | Not analyzed | − | ++late nulear |
| UAS-ProtB-eGFP | UAS-driven by bam-GAL4 | Hsp70 | protB-eGFP | SV40 | Not analyzed | Not analyzed | eGFP in spermatogonia and fades in early spermatocyte stages | -(eGFP mRNA does not persist until late spermatids) |
| Mst77F-eGFP | Mst77F | Mst77F | Mst77F-eGFP | SV40 | − | − | − | ++ late nulear or close to nuclei |
| pc-77F1 | Mst77F | Mst77F | lacZ | SV40 | + | ++ | − | +++ |
| pc-77F3 | Mst77F | Mst77F | lacZ | SV40 | ++ | +++ | − | +++ |
| pc-77F3Δ5′UTR | Mst77F | − | lacZ | SV40 | ++ | +++ | − | +++ |
| β2t-Mst77F-eGFP | β2t | β2t | Mst77F-eGFP | KC10 | Not analyzed | Not analyzed | − | ++ late nulear or close to nuclei |
| Mst77FΔ141C-mCherry | Mst77F | Mst77F | Mst77FΔ141-C-mCherry | SV40 | − | − | − | flagella of late spermatids |
| Mst77FΔ60C-eGFP | Mst77F | Mst77F | Mst77FΔ60C-eGFP | SV40 | Not analyzed | Not analyzed | − | flagella of late spermatids |
| UAS-Mst77F-eGFP | UAS-driven by bam-GAL4 | Hsp70 | Mst77F-eGFP | SV40 | Not analyzed | Not analyzed | eGFP in spermatogonia and fades in early spermatocyte stages | − |
Fig. 2.
Expression of ProtB and Mst77F is controlled by short upstream regions. (A, D) Schematic drawings of the genomic regions and the generated promoter-lacZ constructs of protB and Mst77F. Exons are depicted as gray block arrows, the minimal promoters are in light blue, and the regions tested for contributing to translational control (TR) are in pink. Black thick lines illustrate the regions contained within each promoter-lacZ construct; thin gray lines indicate the deleted regions. The reporter gene lacZ is indicated in blue; the SV40 3′ UTR with a polyadenylation signal is indicated in yellow. (B, C, E, F) Analyses of β-Galactosidase activity in the testis of transgenic flies bearing different protB (B, C) or Mst77F (E, F) promoter lacZ constructs after 10 min of staining reaction. Asterisk indicate the tip of the testis. Double arrowheads, elongated spermatid bundles; arrowheads, spermatocytes; and early spermatids, white arrows.
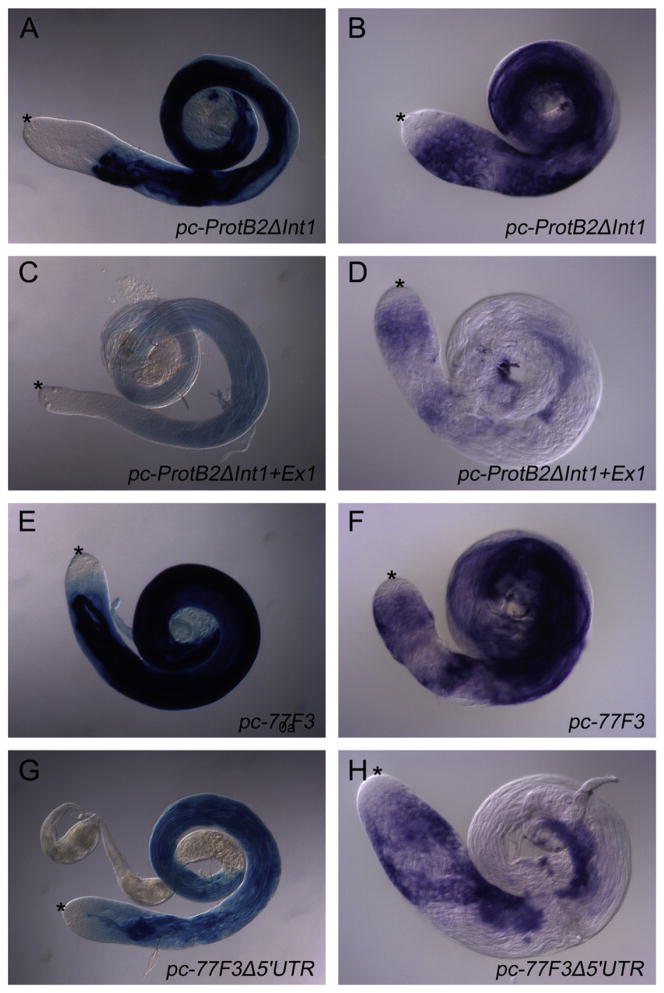
Further deletion of sequences encoding parts of the 5′ UTR of protB and Mst77F (bottom constructs in Fig. 2A, D) substantially reduced the level of reporter expression in spermatids (Fig. 3 respectively). Although the pc-protB2ΔInt1+ΔEx1 and pc-77F3Δ5′UTR reporters had the same regulatory regions upstream of the predicted transcriptional start sites as pc-protB2ΔInt1 and pc-77F3, respectively, the testes from flies bearing the pc-protB2ΔInt1+ΔEx1 and pc-77F3Δ5′UTR constructs (Fig. 3C, G) showed much less β-Galactosidase activity in spermatids than testes from flies bearing the larger reporter constructs incubated in parallel under the same conditions (Fig. 3A, E). It is possible that the further 5′ UTR deletions might have removed or damaged part of the core promoter, an enhancer, or a transcript-stability-mediating element. Alternatively the efficiency of the translation might be reduced. In-situ hybridzation with a lacZ probe (same probe, same time of staining reaction in parallel) revealed that the constructs with the complete 5′ UTR, having a high β-Galactosidase activity (Fig. 3A, E) also had high transcript level (Fig. 3A, F). The constructs from which a substantial segment of the 5′ UTR had been deleted on the other hand had a low β-Galactosidase activity (Fig. 3C, G) and a low lacZ transcript levels (Fig. 3D, H). These data argue against a role of the 5′ UTR in translational efficiency and instead favor a role of these sequences in the regulation of the transcript levels of both Mst77F and protB. In the presence of the 5′ UTR the lacZ transcripts accumulate in spermatocyte stages (Fig. 3B, F), which is apparently not the case if substantial segments of the 5′ UTR are deleted (Fig. 3D, H). The transcript level in spermatocytes appears less effected in Mst77F constructs than in Protamine constructs. This suggests that the 5′ UTR of Mst77F mRNA mainly affects transcript stability while the 5′ UTR of protamines mRNA effects transcript level in spermatocytes and transcript stability in spermatids.
Fig. 3.
The 5′ UTR is essential for high expression of ProtB and Mst77F. (A, C, E, G) Adult testes of reporter lines were stained for β-Galactosidase activity in parallel for 0.5 h. pc-protB2ΔInt (A) and pc-77F3 (E) include the 5′ UTR, whereas pc-protB2ΔInt1ΔEx1 (C) and pc-77F3Δ5′UTR (G) lack most of the 5′ UTR (see Fig. 2A, D). (B, D, F, H) Adult testes from these reporter lines were analyzed by in-situ hybridization with an antisense probe to lacZ RNA.
The 5′ UTR of protB mRNA contains sequences that mediate efficient translational repression in spermatocytes
Comparison of transcripts detected by in-situ hybridization, which were visible in both spermatocytes and spermatids (Fig. 1A, C), with the expression of β-Galactosidase from the reporter constructs, visible only in elongating spermatid bundles (Fig. 2B, C, E, F; double arrowheads), raised the possibility that translational control elements (TCEs) or translational repression elements (TREs) in the UTRs might limit protein expression in spermatocytes. For all the reporter constructs of protB and Mst77F (Fig. 2A, D) and of fusion genes (Jayaramaiah Raja and Renkawitz-Pohl, 2005) tested so far, the 3′ UTR (including the polyadenylation site) was derived from SV40, making the 5′ UTR rather than the 3′ UTR of protB and Mst77F a likely candidate for the localization of TCEs active in spermatocytes. Because in adult testes cytoplasmic β-Galactosidase activity from the reporters in elongating spermatids was already very strong after 10 min of reaction (Fig. 3A, D) and spermatid flagella extend apically far into the spermatocyte region, it was difficult to evaluate whether spermatocytes contain some β-Galactosidase activity. To focus on levels of reporter expression in spermatocytes, we analyzed larval testes lacking spermatids to address translational regulation.
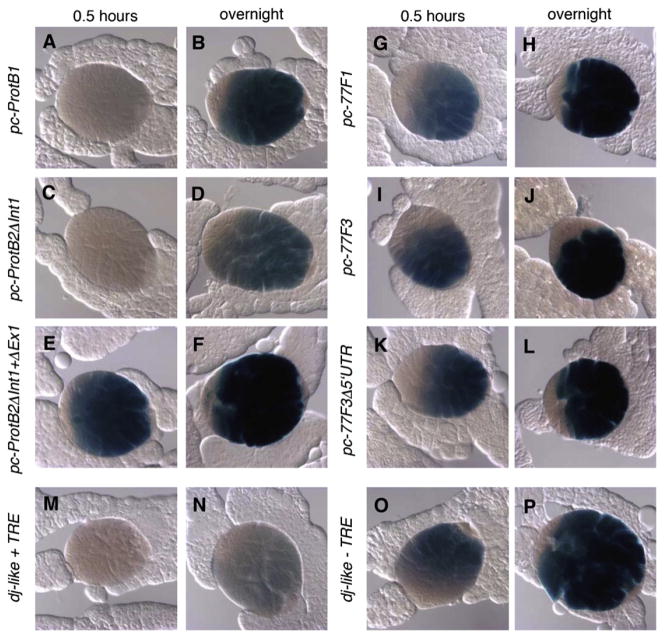
Deletion analysis revealed that sequences within the first 94 bases of the 5′ UTR of the protB locus were required for efficient translational repression of reporter constructs in spermatocytes (Fig. 4). To assay for premature expression of the reporter, testes of third-instar larvae, which contain stages of germ cell differentiation up to onset of the meiotic divisions but normally lack spermatids, were analyzed to focus on expression in spermatocytes. β-Galactosidase reactions were incubated for either 0.5 h or ovn in a single experiment (Fig. 4) to allow different levels to be compared within the linear range of the assay. In reporter constructs containing the intact protB 5′ UTR (Fig. 4A, C), β-Galactosidase activity was not detected in the spermatocytes in larval testes after 0.5 h of staining, similar to larval testes carrying the positive control, a reporter construct dj-like+TRE (Fig. 4M), which we had previously shown mediates translational repression in spermatocytes (Hempel et al., 2006). In contrast we found larval testes incubated from flies bearing the construct pc-protB2ΔInt1+ΔEx1, i.e., having a 90-bp deletion within the 5′ UTR, stained for 0.5 h of incubation were positive for β-Galactosidase activity in primary spermatocytes (Fig. 4E, F). Together these findings indicate that a region of the 5′ UTR of protB is required for effective translational repression of the mRNA in spermatocytes. We note that significant β-Galactosidase activity was detected in reporter constructs containing the intact protB 5′ UTR (Fig. 4B, D) after prolonged incubation of the staining reaction ovn, unlike for dj-like+TRE (Fig. 4N), suggesting that the protB 5′ UTR is not sufficient for full translational repression. However, stronger β-Galactosidase activity was detected after ovn incubation of testes from flies carrying pc-protB2ΔInt1+ΔEx1 (Fig. 4F), which lack the 5′ UTR, than for testes from flies carrying pc-protB2ΔInt1 (Fig. 4D), which contain the 5′ UTR, consistent with sequences within the protB 5′ UTR contributing to strong translational repression in spermatocytes.
Fig. 4.
The 5′ UTR is not sufficient to block translational repression of β-Galactosidase in protB-lacZ and Mst77F-lacZ reporter gene assays. Larval testes of protB (A–F) and Mst77F (G–L) transgenic lines and dj-like reporter lines (M–P) were stained for β-Galactosidase activity in primary spermatocytes in parallel either for 0.5 h or ovn as indicated. (A–D, G–J, M, N) 5′ UTR is present; (E, F, K, L, O, P) most of the 5′ UTR is lacking.
In contrast, the 5′ UTR of Mst77F did not appear to contain sequences sufficient to repress translation of Mst77F-lacZ fusions in spermatocytes (Fig. 4). Larval testes showed significant β-Galactosidase activity in spermatocytes already after only 0.5 h of staining (Fig. 4G, I) and enhanced activity after staining ovn (Fig. 4H, J), similar to constructs from dj-like from which the well-characterized, translational repression element (TRE) (Hempel et al., 2006) had been removed (Fig. 4O, P). These observations suggested that the 5′ UTR of Mst77F was not sufficient for translational repression of Mst77F mRNA in spermatocytes, at least in larval testes (see Fig. 2D TR?). We detected no significant differences between reporter constructs that retained the Mst77F 5′ UTR (Fig. 4G–J) and those from which the bulk of the Mst77F 5′ UTR had been deleted (Fig. 4K, L) after the respective incubation times. Therefore we addressed the question whether the ORF is responsible for translational repression (see below).
Sequences within the ORF regulate accumulation of the Mst77F and ProtB proteins
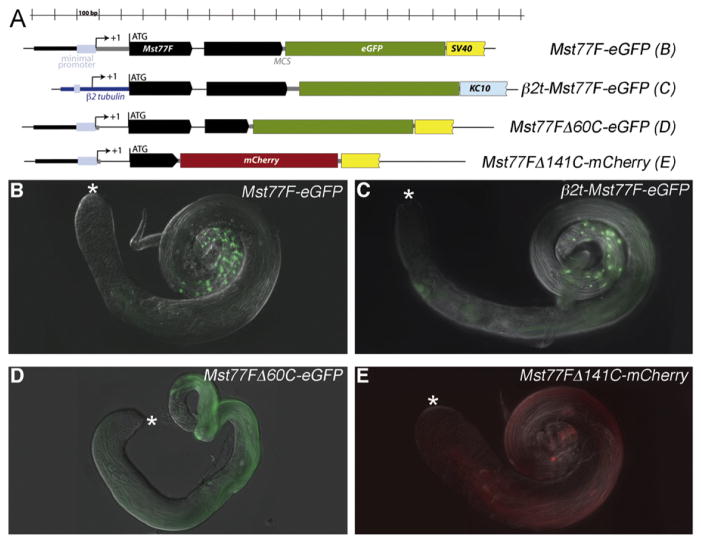
Comparison of reporter transgenes that contained different amounts of the Mst77F protein-coding region fused in frame to eGFP indicated that the accumulation of the Mst77F protein in elongating spermatids was regulated either at the level of translational repression or by protein turnover by sequences in the ORF. We detected no expression of eGFP in larval testes (data not shown) carrying reporter transgenes with the full-length Mst77F protein-coding region up to but not including the stop codon fused in frame to eGFP (Jayaramaiah Raja and Renkawitz-Pohl, 2005). The construct included the 62-bp intron, and the stop codon at the end of the GFP was followed by a 3′ UTR and polyadenylation signal from SV40 (Mst77F-eGFP in Fig. 5A). In adult testes, the Mst77F-eGFP fusion protein was only detected in condensing nuclei in late-stage elongated spermatids (Fig. 5B). Similar results were obtained when an Mst77F-eGFP fusion protein was expressed under control of the promoter and 5′ UTR of the testis-specific tubulin gene β2t (β2t-Mst77F-eGFP, Fig. 5C). Deletion analysis identified a region of the ORF required for localization of Mst77F to the nucleus in elongated spermatids. The NLS (nuclear localization signal) of Mst77F is predicted to lie within the 60 C-terminal amino acids (Nakai and Kanehisa, 1992). Consistent with this prediction, a reporter transgene with a truncation that removed the 3′ most 180 bp (60 codons) of the Mst77F ORF (Mst77FΔ60C-eGFP, Fig. 5A) showed Mst77F-eGFP in flagella of elongated spermatids but not in spermatid nuclei (Fig. 5D). No GFP signal was detected in spermatocytes. Similarly, when portions of the Mst77F ORF carrying only the first 222 bp (thus lacking the last 141 amino acids and the 62-bp intron) was fused in frame to mCherry (Mst77FΔ141C-mCherry, Fig. 5A), the mCherry localized to the flagella of late elongating spermatids, not to the nuclei. Furthermore expression of the Mst77F-mCherry fusion protein was detected solely in late spermatids (Fig. 5E), indicating that sequences located in the first 222 bp of the Mst77FΔ141C-mCherry ORF restrict accumulation of Mst77F protein in spermatocytes and early spermatids, either by specifying translational repression or protein turnover.
Fig. 5.
Translational repression and activation of Mst77F mRNA depends on the Mst77F ORF. (A) Schematic drawings of the constructs used to test the Mst77F ORF for its regulatory capabilities. (B–E) eGFP fluorescence in adult testes of transgenic males carrying (B) Mst77F-eGFP, (C) β2t-Mst77F-eGFP, (D) Mst77FΔ60C-eGFP, and (E) Mst77FΔ141C-mCherry. Asterisk indicate the tips of the testis, where germ-line stem cells reside.
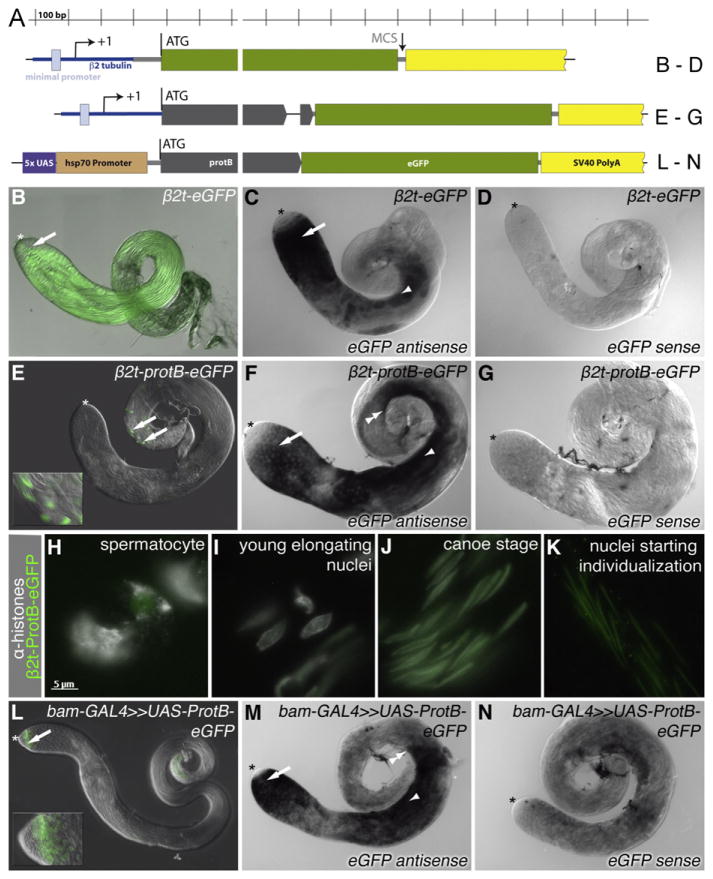
Similar reporter analysis indicated that the protB ORF also contains sequences that block protein expression or accumulation in spermatocytes and early elongating spermatids. For reporter constructs in which eGFP was expressed under the control of the promoter and 5′ UTR of the testis-specific tubulin gene β2t, with the 3′ UTR and polyadenylation signal from SV40 (Figs. 6A, β2t-eGFP), eGFP was detected in adult testes in male germ cells at all stages from spermatocytes to elongated spermatids (Fig. 6B), reflecting the expression of the transcript detected by in-situ hybridization with a probe specific for the eGFP component of the mRNA (Fig. 6C). However, when sequences containing the protein-coding protB ORF (all but the stop codon) and including the 49 bp long second intron were introduced into the reporter construct (β2t-protB-eGFP, Fig. 6A), the ProtB-eGFP fusion protein was mainly only detected in late spermatids (Fig. 6E), in a pattern reminiscent of ProtB-eGFP, expressed under the control of the endogenous protB promoter. In contrast, the transcript for protB-eGFP was detected from early spermatocytes to late elongating spermatids (Fig. 6F), as for the β2t-eGFP reporter transcript (Fig. 6C). While in whole-mount preparations, the ProtB-eGFP fusion protein was detected solely in the nuclei of elongated spermatids (Fig. 6E, arrows and insert), closer inspection of squashed preparations of testes from β2t-protB-eGFP transgenic flies showed a very weak eGFP signal in spermatocytes (Fig. 6H). However, strong fluorescence of eGFP in the squashed preparations appeared only in elongating spermatids, where it localized to nuclei at the canoe-stage, the time when histones were gradually vanishing (Fig. 6J). The β2t-ProtB-eGFP expressing male flies were fertile. Similar results were obtained with the corresponding Mst77F constructs (β2t-Mst77F-eGFP, Fig. 5C). Thus, for both protB and Mst77F, the genomic region containing the ORF plus a small intervening intron was sufficient for the proper spatiotemporal accumulation of the protein. Both the translationally repressed protB mRNA and Mst77F mRNA seemed mainly localized in the cytoplasm (Jayaramaiah Raja and Renkawitz-Pohl, 2005). Splicing mainly takes place in the nucleus. Testing whether these translationally repressed mRNAs might be stored unspliced, we performed RT-PCR with primers flanking the introns of protB and Mst77F using purified mRNA from testes. We found that the vast majority of the stored endogenous mRNAs were spliced (Fig. S1); thus, participation of the intron in translational control in the cytoplasm is unlikely.
Fig. 6.
The ORF of protB confers translation in late spermatids. (A) Diagram of the constructs. (B, E, H–L) eGFP fluorescence; (C, D, F, G, M, N) in-situ hybridization using an eGFP-specific probe. (B, C, D) eGFP under the control of the β2-tubulin promoter and 5′ UTR (β2t-eGFP). (B) Whole mount of adult testis; arrow, late spermatids. (C) Hybridization with an eGFP antisense probe; arrow, early spermatocytes; arrowhead, early spermatids. (D) Hybridization with an eGFP sense probe. (E, F, G) protB-eGFP under the control of the β2-tubulin upstream control region and 5′ UTR (β2t-proB-eGFP). (E) Whole mount of adult testis; arrows, late spermatids; inset, higher magnification. (F) Hybridization with an eGFP antisense probe; arrow, early spermatocytes, arrowhead, early spermatids; double arrowhead, indicates late signal in spermatids. (G) Hybridization with an eGFP sense probe. (H, I, J, K) Squash preparations of β2-protB-eGFP testes, GFP autofluorescence and α-histone, (H) Spermatocytes, (I) Young elongating nuclei, (J) Canoe-stage nuclei. (K) Nuclei starting individualization (L, M, N) bam-GAL4-driven UAS-ProtB-eGFP. (L) Whole mount of adult testis; Arrow, spermatogonia; inset, higher magnification of the tip of this testis. (M) Hybridization with an eGFP antisense probe; arrow, spermatogonia; arrowhead, spermatid stage; double arrow, eGFP transcripts hardly detectable distally to this point. (N) Hybridization with an eGFP sense probe; double arrow, spermatids. In (B–G, L–M), the asterisk marks the hub region of the testis.
Strikingly, when protB-eGFP was expressed in testes under the control of the binary UAS–GAL4 system (Brand et al., 1994) using a heterologous promoter and 5′ UTR from hsp70, and the 3′ UTR and polyadenylation signal of SV40 (UAS-ProtB-eGFP, Fig. 6A), and the bam-GAL4 driver, the ProtB-eGFP fusion protein was strongly detected in late spermatogonia, most likely because bam-GAL4 drives transcription earlier than either the β2t-protB or Mst77F promoters. However, expression of the ProtB-eGFP fusion protein dropped abruptly as cells became early spermatocytes (Fig. 6L), despite the detection of the corresponding transcripts during the spermatocyte growth phase and up to the early spermatid stages (Fig. 6M), consistent with a cell-type-specific mechanism present in early spermatocytes but not in late spermatogonia that either blocks translation or ensures rapid turnover of ProtB protein. The same was observed with UAS-Mst77F-eGFP expressed under the control of bam-GAL4 (Fig. S2I). No nuclear ProtB-eGFP was detected in late spermatids in testes from bam-GAL4;UAS-ProtB-eGFP flies, perhaps because, as indicated by the eGFP transcript distribution, the protB-eGFP transcripts expressed under control of the β2t regulatory sequences are present in spermatids later than the protB-eGFP transcripts expressed under control of the bam-GAL4–UAS system (compare Fig. 6F,M, double arrows).
Together these data suggested that accumulation of ProtB-eGFP and Mst77F-eGFP is prevented in spermatocytes, either by rapid protein turnover or by translational repression directed by sequences within the protein-coding region of the protB and Mst77F loci.
Discussion
Expression of sperm chromosomal proteins is regulated at three levels
Protamines and Mst77F are the major chromatin components of mature sperm. The genes encoding ProtB and Mst77F are both transcribed in primary spermatocytes, and the mRNAs are translationally repressed until post-meiotic stages. Here we presented evidence that expression of one or both of the sperm head proteins ProtB and Mst77F is regulated at three levels: 1) transcription dependent on the tTAFs, 2) translational repression dependent on sequences in the 5′ UTR and/or ORF and 3) stage-specific protein accumulation regulated by sequences in the ORF. The results of transgenic fly assays are summarized in Table 1.
Transcription of protB and Mst77F is regulated by short upstream sequences and transcription of both depends on tTAF function
Reporter gene assays revealed that very short genomic regions proximal to the transcription start site were sufficient for proper tTAF-dependent transcription of protB (−105 bp) and Mst77F (−89 bp). We propose that as yet unknown proteins may cooperate with tTAFs to selectively activate transcription of chromatin condensation-relevant genes, complementary to our recently finding that selective cooperation of a bromodomain protein tBRD1 with tTAFs is important for nuclear shaping and nuclear positioning in spermatids bundles but not for chromatin condensation (Leser et al., 2012).
In addition, sequences in the 5′ UTR of both protB and Mst77F favored high expression levels of β-Galactosidase reporter constructs. However, we found no obvious sequence similarities between protB and Mst77F in their promoter proximal upstream regions or 5′ UTRs. We showed here that the tTAF Sa occupies the promoter of Mst77F and ProtB as was known only for three other genes fzo, dj and Mst87F. However, the canonical generally expressed TAFs are recruited by TBP, the DNA-binding part of the TBP complex. Sa has no proposed DNA-binding site (Hiller et al., 2004). The presumptive promoter region of Mst77F and ProtB is AT-rich, thus, we cannot exclude that TAF1-2, a splice variant of TAF1, binds to this region, as TAF1-2 binds to AT-rich sequences (Metcalf and Wassarman, 2006). TRF2, a variant of TBP expressed in spermatocytes, might be involved (Kopytova et al., 2006). TRF2 was proposed to act as core promoter-selective vector (Hochheimer et al., 2002). In addition, we did not find known promoter elements or other regulatory elements, such as the INR (initiator) or DRE (downstream regulatory element). Transcription of protB and Mst77F starts in early spermatocytes and depends directly or indirectly on the function of the testes-specific TAFs. In the case of protB, at least, the requirement for tTAF function appears to be direct, as binding of the tTAF Sa to the protB promoter. In the case of Mst77F, interpretation of results is made more complex by the 18 transcribed pseudogenes on the Y chromosome (Krsticevic et al., 2010). Krsticevic et al. (2010) showed that 28% of the Mst77 transcripts in testes derive from Mst77Y, for which the upstream sequences are not known (FlyBase), while 72% originate from Mst77F. This correlates well with our transcriptome data, in which 25% of the Mst77F transcripts were tTAF independent and 75% were tTAF dependent. Thus, we hypothesize that the autosomal Mst77F might directly depend on tTAFs, while expression of the Y chromosomal copies may be tTAF independent.
Efficient ProtB accumulation depends on the 5′ UTR and the ORF, while accumulation of Mst77F protein in spermatids is mainly regulated by sequences within the ORF
During spermiogenesis, numerous stored, translationally silent mRNAs are translationally activated in a precise temporal sequence. In protB mRNAs, TCEs that strongly restrict expression in spermatocytes were located in the 5′ UTR just downstream of the transcriptional start site. However, full repression of protein accumulation required sequences beyond the 5′ UTR, within the protB ORF or internal introns (see Table 1 for an overview of reporter constructs and their expression), raising the possibility that sequences within the ORF participate in translational repression and/or regulate protein turnover. In comparison to β-Galactosidase activity of 5′ UTR containing lacZ reporter genes in spermatids, however, the expression in spermatocytes was low, raising the possibility that the 5′ UTR of Mst77F confers higher efficiency of translation in spermatids than in spermatocytes.
For Mst77F mRNA, the 5′ UTR appeared to be of minor importance for translational repression. In this case, protein accumulation was mainly controlled by the ORF, as has been reported earlier for Mst84Db (Gigliotti et al., 1997). Although most of our Mst77F fusion gene reporters contained the intron of Mst77F, Mst77FΔ141C-mCherry, which lacked the intron, also conferred proper translational repression and/or protein accumulation, making it unlikely that key sequences responsible for translational repression are located in the small intron. Likewise, the UAS-protB-eGFP tested under control of bam-GAL4 lacked the intron but was abruptly down regulated in early spermatocytes, indicating that the ORF is sufficient for repression of protB protein accumulation in spermatocytes.
The transcription of protamine genes in mice as controlled by short upstream sequences (−113 bp) (Zambrowicz et al., 1993) and protamine mRNA is also translationally regulated. In mice, the protamine genes are transcribed in round spermatids and translated several days later in elongating spermatids (for a review, see Kleene (2003). This translational repression depends on an 18-bp TCE in the 3′ UTR (Zhong et al., 2001), although in vivo RNA-binding proteins that regulate this repression have not yet been identified. In mammals, recent evidence points to microRNA levels as an important molecular control mechanism for translational repression (Dai et al., 2011). In Drosophila oogenesis, maternally provided mRNAs for early embryogenesis are stored and translationally repressed until the appropriate time during embryogenesis. Here, the 3′ UTR is responsible for translational repression by recruiting proteins, which, for example, prevent assembly of the cap-binding complex, or bind a cap-binding protein that cannot interact with eIF4G (Lasko, 2011, in press).
In Drosophila spermatogenesis, however, our data point toward translational repression elements in the 5′ UTR of protamine mRNA. This is a typical feature for translational control of certain transcripts during spermatogenesis in Drosophila (for a review, see Renkawitz-Pohl et al., 2005). The mRNAs that encode other post-meiotic chromatin components, such as Don Juan and Don Juan-like have also been shown to be translationally regulated by sequences in the 5′ UTR (Blumer et al., 2002; Hempel et al., 2006; Rathke et al., 2007). In addition, mRNAs encoding sperm-flagellar outer dense fiber proteins have been shown to be regulated by a conserved translational control element (TCE) of 12 nucleotides in the 5′ UTR in the Mst(3)CGP gene family (Kempe et al., 1993; Kuhn et al., 1988; Schäfer et al., 1990). In sharp contrast, chromatin-relevant mRNAs of Drosophila (dj, dj-like, protB, Mst77F, tpl94D) do not share this or another conserved sequence motif in the 5′ UTR. However, the 5′ UTR of the dj and dj-like mRNAs is characterized by predicted, extensive stem-and-loop structures (Hempel et al., 2006). Those structures might be binding sites for translational repression-mediating proteins.
Spermatocytes contain the machinery to repress translation and/or accumulation of ProtB and Mst77F
The bam-GAL4–UAS system drives transcription starting in late spermatogonia. With this system, we observed an initial burst of ProtB-eGFP and Mst77F-eGFP protein expression in early germ cells in a small distance from the apical tip of the testis. These proteins were not detected when transcription of protB-eGFP or Mst77F-eGFP was driven under the control of the β2-tubulin promoter and 5′ UTR, which initiates transcription and translation in spermatocytes. We speculate that the initial burst of ProtB-eGFP and Mst77F-eGFP expression in late spermatogonia in testes from bam-GAL4; UAS-ProtB-GFP or UAS-Mst77F-eGFP occurred because spermatogonia lack trans-acting factors required for translational repression or rapid protein turnover that normally restrict ProtB and Mst77F expression in spermatocytes and elongating spermatids. The abrupt decline in ProtB and Mst77F levels observed in young spermatocytes may reflect the onset of cell-type-specific expression of components of such trans-acting regulatory mechanisms.
The protB and Mst77F ORFs are required for translational activation and/or protein accumulation in late spermatids
Our results suggested that cell-type-specific mechanisms present in spermatocytes and early spermatids strongly inhibit accumulation of the ProtB and Mst77F proteins, acting at the level of translational repression, protein turnover, or both. These inhibitory mechanisms must be shut off or reversed to permit proper expression of the sperm chromatin proteins in late elongated spermatids. Translational activation of mRNAs at distinct time points is a general feature of post-meiotic protein expression during spermiogenesis. It remains to be elucidated whether accumulation of other sperm proteins synthesized from stored mRNAs also depends on sequences within the respective ORFs. We posit that specialized translational activation machinery mechanism(s) act at distinct time points of spermiogenesis (Fig. 7D), for example, to turn on protamine synthesis shortly before individualization. Indeed, we and others have previously characterized male sterile mutants of the eIF4G2 gene, also known as Off schedule (CG10192), which encodes an alternate version of the core translation initiation machinery component eIF4G that is expressed in spermatocytes and required for meiotic division and early spermatid differentiation in males. Among other effects, eIF4G2-mutant males have aberrant spermatids with large nuclei that arrest differentiation shortly after meiosis, long before ProtB and Mst77F synthesis starts (Baker and Fuller, 2007; Franklin-Dumont et al., 2007).
Fig. 7.
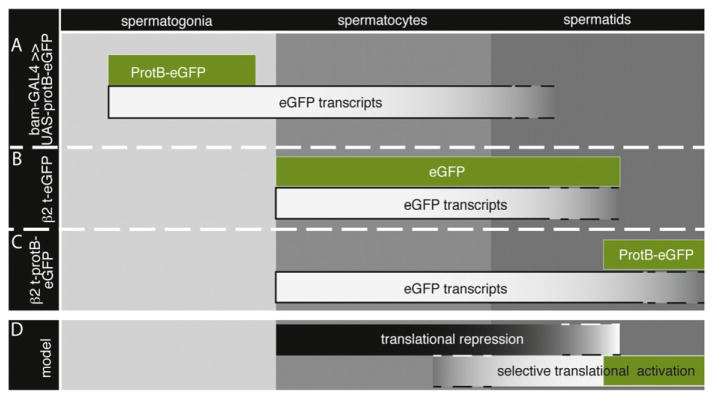
ProtB mRNA is regulated by translational repression and stage-specific translational activation. The autofluorescence of the eGFP reporter is shown in green; the eGFP transcript is shown in white boxes relative to the stages of spermatogenesis. (A) ProtB-eGFP is limited to spermatogonia, while the corresponding eGFP transcript persists during the spermatocyte stage. (B) β2t-eGFP leads to overlapping eGFP transcripts and protein distribution in spermatocytes and spermatids. (C) β2t-protB-eGFP leads to transcripts present in spermatocytes and spermatids, while ProtB-eGFP is synthesized in late spermatids. (D) This model proposes that many mRNAs are synthesized in spermatocytes and selectively translated during the spermatid phase; ProtB-eGFP (in green) is depicted as an example.
Translational control/activation is often regulated by cytoplasmic polyadenylation (Villalba et al., 2011). In Drosophila, both female and male germ lines contain cytoplasmic poly(A) polymerases (Benoit et al., 2008; Cui et al., 2008; Sartain et al., 2011). We showed that the 3′ UTR of the protamine mRNA is dispensable for correct expression of protamines. This agrees with the finding that the length of poly(A) tracks of protamine mRNAs is indistinguishable between wild-type and mutants of the poly(A) polymerase GLD-2. Nevertheless, Prot-eGFP was not detected in GLD-2 mutants that might arrest spermiogenesis too early to allow protamine translation/ accumulation. This agrees with our model that translation of protamine mRNAs takes place at a distinct developmental time point late in the spermatid elongation stage (Fig. 7A).
Thus, expression of ProtB and Mst77F in Drosophila male germ cells is controlled at least three levels. In spermatocytes, transcript expression is controlled by specialized cell-type-specific transcriptional machinery. In addition, protB and Mst77F mRNAs are translationally repressed until late spermatid stages, when ORF-dependent translational activation takes place. We expect this may also hold true for other stored mRNAs relevant for sperm morphogenesis. This regulation could involve cell-type-specific activation of translation, protein stabilization, or both.
Supplementary Material
Acknowledgments
We thank Nadine Müller and Ruth Hyland for excellent technical assistance and Katja Gessner for competent secretarial assistance and Karen Brune for excellent editing. We thank our colleagues Mireille Schäfer and Klaus Steger for critical reading of the manuscript as well as Mireille Schäfer for the LacZ clone to synthesize antisense RNA. This work was supported by the Deutsche Forschungsgemeinschaft within the International Graduate School GK 767 (“Transcriptional control during developmental processes”), the FOG 531 (“Chromatin mediated biological decisions” Re 628/12-3), LOEWE-MIBIE (“Männliche Infertilität bei Infektion & Entzündung”), the TRR 81 “Chromatin changes in differentiation and malignancies” (to R.R.-P.), a short-term fellowship of the Boehringer Ingelheim Fund (to B.B.), NIH Grant 3 RO1 GM061986 (to M.T.F.), and a Leukemia and Lymphoma Society Special Fellowship Grant LLS-3598-06 and the NIH career award K99/R00HD055052 (to X.C.).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.02.018.
References
- Awe S, Renkawitz-Pohl R. Histone H4 acetylation is essential to proceed from a histone- to a protamine-based chromatin structure in spermatid nuclei of Drosophila melanogaster. Syst Biol Reprod Med. 2010;56:44–61. doi: 10.3109/19396360903490790. [DOI] [PubMed] [Google Scholar]
- Baker CC, Fuller MT. Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development. 2007;134:2863–2869. doi: 10.1242/dev.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Benson E, Gudmannsdottir E, Newton F, White-Cooper H. Post-meiotic transcription in Drosophila testes. Development. 2008;135:1897–1902. doi: 10.1242/dev.021949. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer N, Schreiter K, Hempel L, Santel A, Hollmann M, Schafer MA, Renkawitz-Pohl R. A new translational repression element and unusual transcriptional control regulate expression of don juan during Drosophila spermatogenesis. Mech Dev. 2002;110:97–112. doi: 10.1016/s0925-4773(01)00577-9. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;CB13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller M. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu C, Prado JR, Eun SH, Fuller MT. Sequential changes at differentiation gene promoters as they become active in a stem cell lineage. Development. 2011;138:2441–2450. doi: 10.1242/dev.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008;178:2017–2029. doi: 10.1534/genetics.107.084558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Tsai-Morris CH, Sato H, Villar J, Kang JH, Zhang J, Dufau ML. Testis-specific miRNA-469 up-regulated in gonadotropin-regulated testicular RNA helicase (GRTH/DDX25)-null mice silences transition protein 2 and protamine 2 messages at sites within coding region: implications of its role in germ cell development. J Biol Chem. 2011;286:44306–44318. doi: 10.1074/jbc.M111.282756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Dumont TM, Chatterjee C, Wasserman SA, Dinardo S. A novel eIF4G homolog, off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development. 2007;134:2851–2861. doi: 10.1242/dev.003517. [DOI] [PubMed] [Google Scholar]
- Fuller M. Spermiogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 71–147. [Google Scholar]
- Fuller MT, Regan CL, Green LL, Robertson B, Deuring R, Hays TS. Interacting genes identify interacting proteins involved in microtubule function in Drosophila. Cell Motil Cytoskeleton. 1989;14:128–135. doi: 10.1002/cm.970140122. [DOI] [PubMed] [Google Scholar]
- Gigliotti S, Balz V, Malva C, Schafer MA. Organisation of regulatory elements in two closely spaced Drosophila genes with common expression characteristics. Mech Dev. 1997;68:101–113. doi: 10.1016/s0925-4773(97)00137-8. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hempel L, Rathke C, Raja S, Renkawitz-Pohl R. In Drosophila, don juan and don juan like encode proteins of the spermatid nucleus and the flagellum and both are regulated at the transcriptional level by the TAF II80 cannonball while translational repression is achieved by distinct elements. Dev Dyn. 2006;235:1053–1064. doi: 10.1002/dvdy.20698. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle M, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin T, Marino S, Fuller M. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hiller M, Lin T, Wood C, Fuller M. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime G, Brill J, Fuller M. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109(Pt 12):2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Jayaramaiah Raja S, Renkawitz-Pohl R. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol. 2005;25:6165–6177. doi: 10.1128/MCB.25.14.6165-6177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe E, Muhs B, Schäfer M. Gene regulation in Drosophila spermatogenesis: analysis of protein binding at the translational control element TCE. Dev Genet. 1993;14:449–459. doi: 10.1002/dvg.1020140606. [DOI] [PubMed] [Google Scholar]
- Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103:217–224. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- Klemenz R, Weber U, Gehring W. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, Korochkin LI, Tora L, Georgiev PG, Georgieva SG. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295–307. doi: 10.1534/genetics.109.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schäfer U, Schäfer M. Cis-acting regions sufficient for spermatocyte-specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9. EMBO J. 1988;7:447–454. doi: 10.1002/j.1460-2075.1988.tb02832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip Rev RNA. 2011;2:408–416. doi: 10.1002/wrna.70. [DOI] [PubMed] [Google Scholar]
- Lasko P. mRNA Localization and translational control in Drosophila oogenesis. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a012294. http://dx.doi.org/10.1101/cshperspect.a012294. in press. [DOI] [PMC free article] [PubMed]
- Leser K, Awe S, Barckmann B, Renkawitz-Pohl R, Rathke C. The bromodomain-containing protein tBRD1 is specifically expressed in spermatocytes and is essential for male fertility. Biol Open. 2012;1:597–606. doi: 10.1242/bio.20121255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf CE, Wassarman DA. DNA binding properties of TAF1 isoforms with two AT-hooks. J Biol Chem. 2006;281:30015–30023. doi: 10.1074/jbc.M606289200. [DOI] [PubMed] [Google Scholar]
- Michiels F, Buttgereit D, Renkawitz-Pohl R. An 18-bp element in the 5′ untranslated region of the Drosophila beta 2 tubulin mRNA regulates the mRNA level during postmeiotic stages of spermatogenesis. Eur J Cell Biol. 1993;62:66–74. [PubMed] [Google Scholar]
- Morris CA, Benson E, White-Cooper H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protocols. 2009;4:1807–1819. doi: 10.1038/nprot.2009.192. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–1700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- Rathke C, Barckmann B, Burkhard S, Jayaramaiah-Raja S, Roote J, Renkawitz-Pohl R. Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur J Cell Biol. 2010;89:326–338. doi: 10.1016/j.ejcb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Renkawitz-Pohl R, Hollmann M, Hempel L, Schäfer MA. Spermatogenesis. In: Lawrence I, Gilbert KI, Sarjeet S, Gill, editors. Comprehensive Molecular Insect Science, Reproduction and Development. Vol. 1. Elsevier; 2005. pp. 157–177. [Google Scholar]
- Russell SR, Kaiser K. Drosophila melanogaster male germ line-specific transcripts with autosomal and Y-linked genes. Genetics. 1993;134:293–308. doi: 10.1093/genetics/134.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Blumer N, Kampfer M, Renkawitz-Pohl R. Flagellar mitochondrial association of the male-specific Don Juan protein in Drosophila spermatozoa. J Cell Sci. 1998;111(Pt 22):3299–3309. doi: 10.1242/jcs.111.22.3299. [DOI] [PubMed] [Google Scholar]
- Santel A, Kaufmann J, Hyland R, Renkawitz-Pohl R. The initiator element of the Drosophila beta2 tubulin gene core promoter contributes to gene expression in vivo but is not required for male germ-cell specific expression. Nucleic Acids Res. 2000;28:1439–1446. doi: 10.1093/nar/28.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain CV, Cui J, Meisel RP, Wolfner MF. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development. 2011;138:1619–1629. doi: 10.1242/dev.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Schäfer M, Kuhn R, Bosse F, Schäfer U. A conserved element in the leader mediates post-meiotic translation as well as cytoplasmic polyadenylation of a Drosophila spermatocyte mRNA. EMBO J. 1990;9:4519–4525. doi: 10.1002/j.1460-2075.1990.tb07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C, Boulet A, Lipshitz H. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H, Consortium F. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Chalopin DS, Lopes HF, Long M, Karr TL. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics. 2010;186:431–433. doi: 10.1534/genetics.110.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opinion Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Schäfer M, Alphey L, Fuller M. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Harendza CJ, Zimmermann JW, Brinster RL, Palmiter RD. Analysis of the mouse protamine 1 promoter in transgenic mice. Proc Natl Acad Sci USA. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Klyne G, Benson E, Gudmannsdottir E, White-Cooper H, Shotton D. FlyTED: the Drosophila testis gene expression database. Nucleic Acids Res. 2010;38:D710–715. doi: 10.1093/nar/gkp1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Kafer K, Braun RE. A highly conserved sequence essential for translational repression of the protamine 1 messenger rna in murine spermatids. Biol Reprod. 2001;64:1784–1789. doi: 10.1095/biolreprod64.6.1784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.