Abstract
Although congenital heart disease (CHD) is the most common survivable birth defect, the etiology of most CHD remains unclear. Several lines of evidence from humans and vertebrate models have supported a genetic component for CHD, yet the extreme locus heterogeneity and lack of a distinct genotype-phenotype correlation has limited causative gene discovery. However, recent advances in genomic technologies are permitting detailed evaluation of the genetic abnormalities in large cohorts of CHD patients. This has lead to the identification of copy-number variation and de-novo mutations together accounting for up to 15% of CHD. Further, new strategies coupling human genetics with model organisms have provided mechanistic insights into the molecular and developmental pathways underlying CHD pathogenesis, notably chromatin remodeling and ciliary signaling.
1. Epidemiology
Congenital heart disease (CHD) is the most common survivable human birth defect, affecting approximately .5–.8% of live births. Several studies report an increasing incidence of CHD over time; for example a review of studies reported worldwide since 1930 revealed an increase in overall prevalence from 5.3/1,000 live births in 1960 to 9.1/1,000 live births in 1995. Most of the increase was reported in less severe lesions such as atrial septal defects (ASD), ventricular septal defects (VSD) and patent ductus arteriosus (PDA), and largely coincided with improved imaging leading to early diagnosis of CHD that was not life threatening in the immediate newborn period [1]. Notably however, one study did report a small increase in incidence of some forms of severe CHD including Tetralogy of Fallot (TOF) and atrioventricular canals (AVC) between 1995 and 1997 [2]. In contrast to the relatively stable incidence of CHD since 1995, the dramatic improvements in medical and surgical management of CHD has increased the prevalence of CHD in the entire population significantly, and it is estimated that the population of adults with CHD is growing ~5%/year [3]. Until recently, severe CHD including TOF and AVC almost always resulted in dramatically lower reproductive fitness, so it is interesting to speculate that the recent subtle increases in the incidence of some CHD are due to vertical transmission. Indeed, parents with CHD, especially mothers with CHD, have a higher rate of having a child with CHD [3].
2. Developmental Biology: genetic control of cardiac morphogenesis
CHD is a disorder resulting from abnormal heart development, so it is likely that defects in the genetic control of cardiac development underlie a majority of CHD. Compared to humans, the heart in many easily manipulated model organisms is accessible and the molecular and genetic mechanisms regulating cardiac morphogenesis are highly conserved. Studies have predominantly utilized four model systems: zebrafish, Xenopus, chick and mouse. Zebrafish and Xenopus are inexpensive, high throughput and accessible, but do not have a four-chambered heart. Avian hearts are accessible, easily manipulated and have four chambers, but avian genetics are still extremely limited. Finally, the most high-fidelity model for human heart disease is offered by the mouse, combining a four chambered heart and well-established genetics. From studies utilizing these model systems, a picture of heart development and its highly complex genetic control has emerged [4]. It is difficult to estimate the number of genes required for cardiac development, especially since some essential cardiac genes, such as those involved in Hedgehog signaling, are also essential in very early, essential developmental processes that can obscure the cardiac phenotype. Defects in these general developmental genes may result in very early lethality [5], or instead, manifest as complex syndromes that have CHD as an important component.
Cells destined to become the heart originate from the mesoderm and to a smaller extent the neural crest. The genetic network regulating cardiac and regional cardiac identity is complex, and any of the genes involved are candidates for CHD-causing genes [4]. Several of the genes that are specifically involved in cardiac cell specification, such as NKX2.5 [6], TBX1 [7]and TBX5 [8] have been linked to human CHD. The first major morphogenetic event in heart development is heart looping. The left-right (LR) asymmetry of the heart depends on global LR positional information generated by cilia during gastrulation. Defects in genes encoding ciliary function underlie many mouse models of heterotaxy [9], and ciliary defects have been identified in some humans with heterotaxy. The LR axis is initiated at a transient, evolutionary conserved ciliated structure called the Left-Right Organizer (LRO). LRO function depends on the extensive set of genes required for ciliogenesis, motility and signaling. The size of this gene set has been reported to range from 200 [10] to over 1,200 [11], and it is not yet known how many of the core set of cilia genes are required for LR development and cardiac morphogenesis. Subsequent to heart looping, the T-Box transcription factors TBX 2,3,18, and 20 play a prominent role in specifying ventricular identity. Patterning the AV canal specifies the valve-forming regions by identifying domains of endocardium that are fated to undergo epithelial-mesenchymal transformation (EMT) leading to formation of the atrioventricular valves. Genes involved in valve development include the extensive repertoire of genes that are globally required for EMT, such as the TGFp family, and others not required in EMT such as NFATC1 along with several other genes that lie within the “Down Syndrome critical region”. It is interesting to note that patients with Down Syndrome have a high frequency of atrioventricular canal defects that are phenotypically more uniform than AVCs seen in patients that do not have Down Syndrome, and that the Down Syndrome-associated AVCs usually respond remarkably well to surgical repair.
3. Clinical CHD: the intersection of genes and heart development
a. “Syndromic” CHD
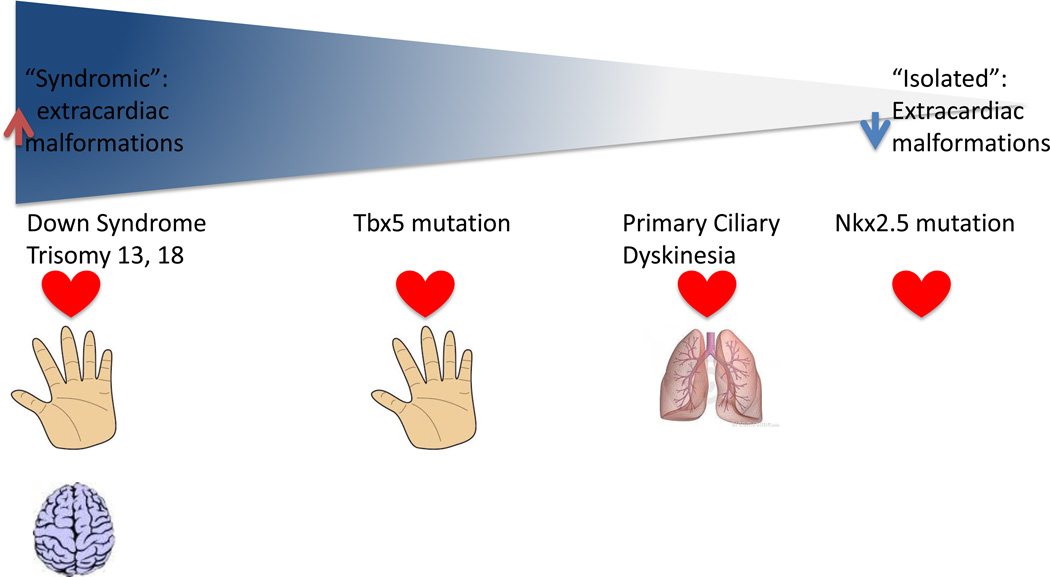
The genetic complexity underlying heart development also predicts extensive genetic heterogeneity in the cause of CHD. It is likely that the spectrum of “isolated” CHD and “syndromic” CHD is much more continuous than previously appreciated (Figure 1). At one extreme of the spectrum lies CHD due to aneuploidy and large chromosomal rearrangements altering gene dosage of many cardiac genes simultaneously. At the next level of the spectrum, isolated mutations in genes with a broad range of developmental roles, such as TBX5, can affect multiple organ systems and result in definable syndromes. Further along the spectrum lies CHD due to a genetic defect also affecting organs outside of the heart, but where the defects in other organ systems are obscured to the clinician because they are masked by the severity of the CHD and the invasive therapy required to manage it. One example of such a condition is complex CHD associated with primary ciliary dyskinesia (PCD): the primary manifestation of PCD is chronic respiratory disease, which is frequently not diagnosed in patients requiring extensive cardiac surgery [12]. Other CHD is associated with subtle extracardiac manifestations such as growth and developmental delays. Finally, some defects are truly cardiac-specific; the best examples to date are mutations in NKX2.5 resulting in ASDs and conduction delay.
Figure 1.
The clinical spectrum of congenital heart disease (CHD). CHD is associated with a continuous range of extracardiac phenotypes, extending from the chromosomal aneuploidies manifesting with multiple organ system involvement (left side of figure) through combinations of CHD and one other involved organ system such as Holt-Oram Syndrome and Primary Ciliary Dyskinesia to apparently isolated CHD (right side of figure).
b. Genotype-phenotype correlation (or the lack thereof)
The relationship between genetic defect and CHD is further complicated by tremendous genotype-phenotype variability observed in both humans and model organisms. A single point mutation in TBX5 can produce a range of cardiac defects associated with Holt-Oram syndrome ranging from a small ASD to a complete atrio-ventricular canal. Even more extreme phenotypic variability results from a point mutation in the Dnah11 gene in mouse: pups in a single litter are born with dextrotransposition of the great arteries (D-TGA), levo-transposition of the greater arteries (L-TGA), complete atrioventricular canal defects (CAVC), TOF and double outlet right ventricles (DORV) [13]. What is even more striking here is that the phenotypic variability occurs despite an absolutely inbred background and controlled environment, ruling out modifier genes and external environmental influences as the cause for the broad range of CHD. The mouse Df1/+ model of DiGeorge syndrome with heterozygous deletion of Tbx1 is another example: early in development, all Df1/+ embryos have a specific defect in the development of the 4th aortic arch. Remarkably, as development proceeds, a number of pups remodel their aorta to approximate normal, while others progress to severe aortic abnormalities [14,15]. Together, these observations suggest that the genetic defect(s) underlying any given case of CHD may be modified by a range of factors including genetic modifiers, external environmental factors, maternal uterine environment, epigenetic effects, and finally stochastic events.
4. DNA structural changes underlying CHD
CHD results from the full range of structural DNA abnormalities, ranging from duplication of entire chromosomes, copy-number variation of a wide range of sizes, small insertion/deletions and single-nucleotide mutations.
a. Copy number variation
It has long been established that large copy number variants (CNVs) contribute to CHD (reviewed in [16]). The first of these to be extensively studied is the deletion of 22q11.2 in DiGeorge/ velocardiofacial syndrome, which encompasses TBX1. Other examples of CNVs associated with syndromic CHD include 7q11.23, encompassing ELN (Williams-Beuren syndrome), 20p.12 (Alagille syndrome), 12q.25 (Holt-Oram syndrome). Large rare CNVs are also associated with non-syndromic isolated CHD, such as deletion of 8p23.1 [17], encompassing GATA4, and 1q21.1 [18]. The cardiac phenotypes associated with these CNVs are highly variable, underscoring the lack of strong genotype-phenotype correlation observed in other CHD mutations.
In addition to identification of rare recurrent CNVs, overall incidence of rare CNVs has demonstrated significant burden in CHD cases [19,20]. Multiple rare CNVs contribute to heterotaxy [21], congenital left-sided heart disease [22], TOF [23], and AVCs [24]. Studies have shown that rare genic CNVs account for 5–10% of different CHD phenotypes. Of note, these CNVs can either been inherited or de novo. De novo CNVs have been found in excess in sporadic CHD probands, including approximately 10% of sporadic TOF cases [23]. Global rates are estimated at 5% of de novo CNVs in the constellation of CHD phenotypes. This corresponds to a 2-fold increase when compared to published data for unaffected siblings of autism cases (approximately de novo CNV rate =2%) [25]. Exploration of genes disrupted in large CNVs may reveal further insights in the role of specific CNVs in the pathogenesis of CHD.
b. Point mutations and specific genotype-phenotype correlation in CHD
An extensive and ever growing list of point mutations in over 40 genes has been implicated in CHD [16]. The question arises whether the specific mutation correlates with the specific cardiac or extracardiac phenotype for some of these genes? Although over 40 mutations affecting DNA binding, transactivation activity, protein-protein interaction or nuclear localization in the cardiac-specific transcription factor NKX2.5 have been linked to CHD, most commonly ASDs with or without conduction abnormalities, no correlation between mutation or functional effect and CHD phenotype has been identified [26]. Another example of lack of genotype-phenotype correlation is provided by the observation that both gain-of-function and loss-of-function mutations in the T-box transcription factor TBX20 result in ASDs and valvar abnormalities [27]. Similar to CHD resulting from aneuploidy and CNV, the CHD resulting from different point mutations affecting genes in cardiac development point to exquisite dosage sensitivity in heart morphogenesis.
5. Genetic mechanisms causing CHD
a. Mendelian CHID
Evidence exists for both inherited and de-novo mutations leading to CHD. Large multigenerational pedigrees demonstrating Mendelian inheritance of CHD are rare. Autosomal recessive inheritance of CHD has been documented most extensively in populations with a high degree of consanguinity. Most of the reported multigenerational pedigrees of X-linked and dominantly inherited CHD include either non-lethal CHD such as ASDs, or CHD with variable phenotypes that include survivable CHD and more severe manifestations of the same spectrum. Examples include dominantly inherited left-sided obstructive disease ranging from bicuspid aortic valve to hypoplastic left heart due to defective Notch signaling [28], and X-linked heterotaxy due to mutations in the ZIC3 transcription factor [29]. Although Mendelian recessive inheritance appears to contribute only a small fraction of the overall population burden of CHD, it becomes a more prominent contribution to specific types of CHD, including heterotaxy and AVC [30].
b. De novo mutations
CHD often occurs sporadically and impairs reproductive fitness, suggesting a role for de novo mutations in its pathogenesis. Whole exome sequencing has allowed the identification of de novo point/insertion-deletion mutations previously unmappable by linkage analysis. Recently, the Pediatric Cardiac Genomics Consortium [31] performed whole-exome sequencing of 362 parent-offspring trios with an affected CHD proband. It was found that de novo point/insertion-deletion mutations in several hundred genes collectively contribute to at least 10% of severe CHD (Zaidi et al, Nature, in press 2013). Notably, similar odds ratios were seen across major classes of severe CHD: left ventricular obstructive lesions, conotruncal lesions and heterotaxy-associated CHD.
6. Pathways implicated in CHD
Building a heart is an extremely complex developmental process, and utilizes a large repertoire of genes that must be precisely regulated with regard to dosage, timing and spatial expression. It is not surprising that even the incomplete genetic analysis of CHD to date points to extreme genetic heterogeneity. With this in mind, it may not be possible to make the essential connections between each very rare mutation and the resulting clinical phenotype. Instead, genetic discoveries are pointing to several biological pathways that have significant roles in CHD. In addition to the transcription factors such as NKX2.5 and the TBX family, growth factors including Nodal and TGFβ and Notch signaling that have long been thought to have a role in CHD, recent data has highlighted roles for cilia and chromatin remodeling in heart development and disease.
a. Cilia
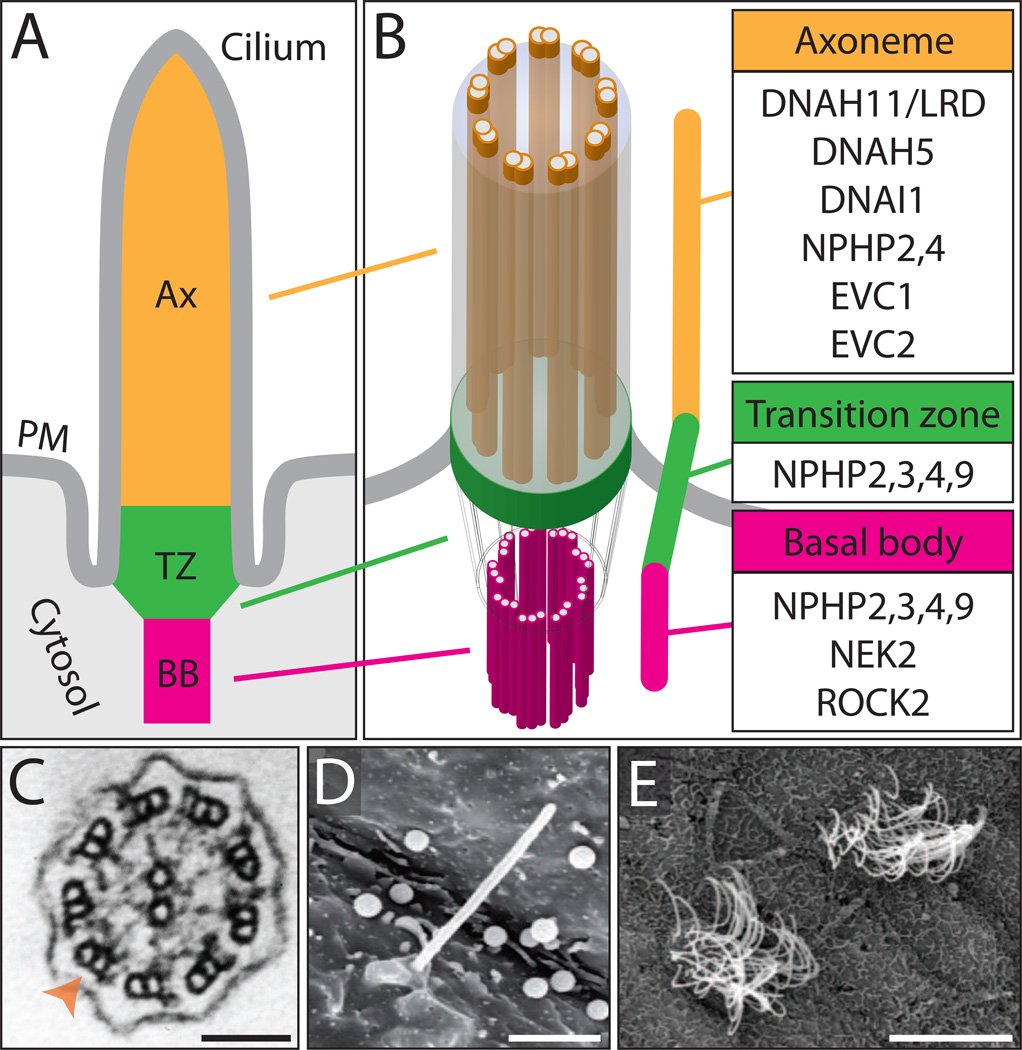
Cilia are found on the surface of most vertebrate cell types and serve a multitude of functions, including signaling and extracellular fluid propulsion. Defects affecting cilia structure and/or function have been intimately linked to a group of diverse human disorders coined ‘ciliopathies’. Recent studies have implicated cilia in the etiology of CHD and suggest that some CHD is part of the ciliopathy spectrum of disease (Figure 2)
Figure 2.
Cilia structure and ciliary components associated with CHD. (A, B) The ciliary compartment is defined by the axoneme (Ax), transition (TZ) and basal body (BB). The axoneme consists of 9 microtubule doublets arranged in a circle, which provide a cytoskeletal scaffold for the cilium, and surrounded by the ciliary membrane. The basal body serves to anchor, stabilize and regulate the formation of the cilium. The transition zone is positioned between the axoneme and basal body and serves as a docking site for ciliary trafficking. PM: plasma membrane. (B) Numerous ciliary components from the Ax, TZ and BB have been linked to CHD in both humans and model organisms. (C) Transmission electron microscopy (TEM) image of a transverse cross-section through the axoneme of a motile cilium from the skin epithelia of a Xenopus embryo. Orange arrowhead indicates the microtubule doublet ring structure. Scalebar, 100 nm. (D) Scanning electron microscopy (SEM) image of a motile cilium on the left-right organizer (LRO) of a Xenopus embryo. Scalebar, 2 µm (E) SEM image of multi-ciliated cells on the skin epithelia of a Xenopus embryo. Scalebar, 10 µm
Nephronophthisis-associated ciliopathies
Nephronophthisis (NPHP) and several associated ciliopathies, Meckel-Gruber syndrome (MGS), Joubert syndrome (JS), and Senior-Loken syndrome (SLS), are characterized by a spectrum of phenotypes attributed to ciliary defects, including renal cyst disease, retinal degeneration, and brain malformations. Mutations in 18 genes encoding proteins that localize to the cilium or basal body form a proteomic network, and have been linked to these four ciliopathies [32]. Among these genes, NPHP2/INVS, NPHP3, NPHP4 and NPHP9/NEK8 can also lead to heterotaxy, situs inversus and congenital heart malformations. A partial deletion of Nphp2/Invs in mouse results in situs inversus [33], while an interstitial deletion in humans has been linked to transposition of the great vessels [34]. A complete loss of Nphp3 in mouse results in situs inversus and CHD, while NPHP3 mutations in humans have been linked to numerous embryonic defects including situs inversus, structural heart disease and polydactyly [35]. Knockdown of nphp4 in zebrafish results in cardiac laterality defects, while NPHP4 mutations in humans are associated with heterotaxy [36]. Loss of Nphp9/Nek8 in mouse and zebrafish results in renal cyst formation and cardiac laterality defects [37], while a homozygous nonsense mutation in humans results in cystic kidneys and CHD [38].
Ellis-van Creveld syndrome
Ellis-van Creveld syndrome (EVCS) is a recessive skeletal dysplasia disorder that has been classified as a ciliopathy. EVCS is characterized by numerous defects, notably dwarfism, polydactyly and CHD. Among EVCS patients with CHD, 88% of affected individuals display an endocardial cushion defect and approximately 50% have an ASD producing a common atrium [39]. The causative genes, EVC1 and . [40], encode proteins that localize to the cilium and transduce cilia-dependent Hedgehog signaling by complexing with Smoothened in the cilium [41].
Heterotaxy
Various abnormalities in the lateral positioning and morphology of the heart and other major visceral organs result in heterotaxy syndrome, which is tightly correlated with CHD. Due to the role of motile cilia in determining lateral patterning, defects affecting ciliary motility machinery result in heterotaxy and CHD. In mice, mutations in components of the dynein motor complex, such as left-right dynein (Dnah11/Lrd) and dynein heavy chain 5 (Dnah5), result cardiac and visceral LR abnormalities. Not surprisingly, 6.5% of patients with PCD, a disorder defined by abnormal ciliary motility in the airway epithelia, also display heterotaxy, which emphasizes the role of motile cilia in both disorders [42]. Similarly, next-generation sequencing of 13 heterotaxy patients with ciliary dysfunction identified mutations in known dynein components: DNAI1, DNAH5 and DNAH11 [12].
Recent human genomics efforts have identified novel heterotaxy-causing genes, some of which have unanticipated roles in cilia structure and function. Rare genic CNVs in heterotaxy patients identified 5 genes (NEK2, GALNT11, ROCK2, NUP188, TGFBR2) that were verified by knockdown in Xenopus to produce laterality defects [43]. These genes were not previously described as “cilia” genes. Notably, however, further studies found that both NEK2 [44]and some nucleoporins [45] localize at the base of the cilium, while a study in zebrafish has demonstrated the function of Rock2 in ciliogenesis and LR development [46].
b. Chromatin remodeling
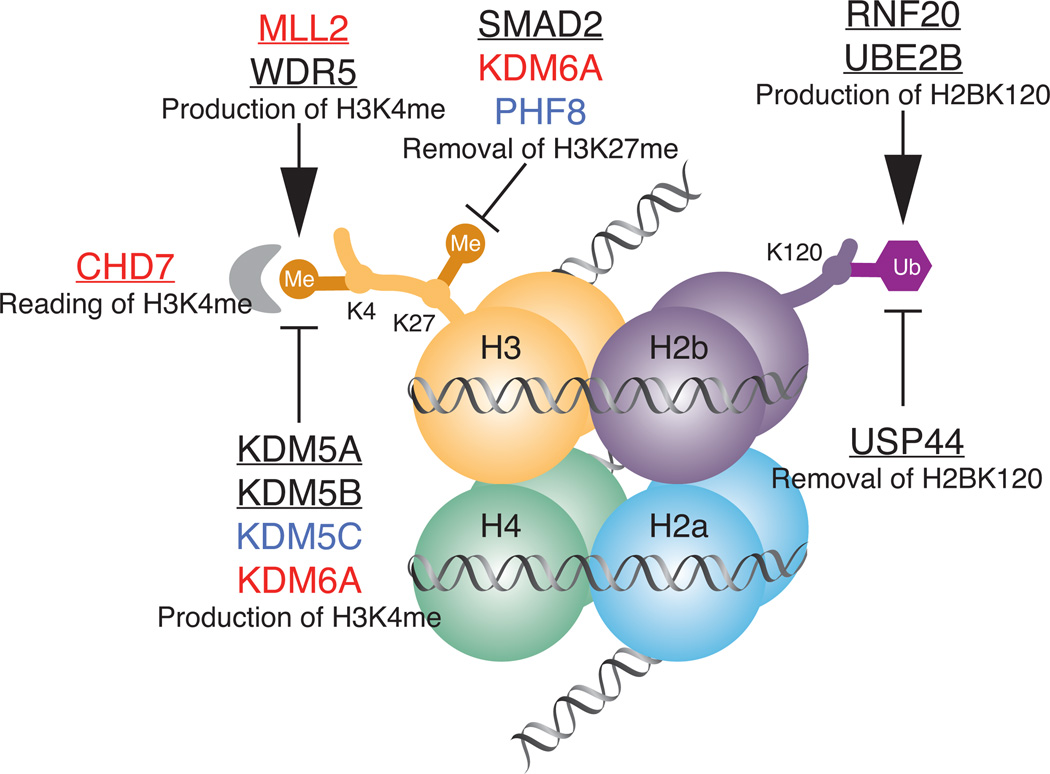
The analysis of de novo mutations in CHD demonstrated a significant increase in genes involved in production, removal or reading of H3K4 methylation (H3K4me), or ubiquitination of H2BK120, which is required for H3K4 methylation (Figure 3). In ES cells, H3K4me and H3K27me constitute ‘bivalent’ marks, which are found on the promoters and enhancers of key developmental genes poised for activation. These findings implicate marked dosage sensitivity of the histone methylation pathway in heart development [47,48].
Figure 3.
Chromatin remodeling genes in CHD. Nucleosome with histone octamer, H3K4 methylation and H2BK120 ubiquitination is shown. Genes associated with identified syndromes with CHD are shown in red, genes associated with identified syndromes without known CHD are shown in blue, genes with de-novo mutation in CHD patients are underlined.
Indeed, many chromatin modification genes have been previously found to be associated with CHD (Table 1). These include MLL2 and KDM6A (Kabuki syndrome) [49,50], CHD7 (CHARGE syndrome) [51], SETBP1 (Schinzel-Giedion) [52], and EHMT1 (Kleefstra syndrome) [53]. H3K4 methylation was initially identified through its role regulating HOX gene clusters, and it plays a central role in gene regulation in an extremely broad range of developmental processes extending greatly beyond HOX genes. Mutations in genes involved in H3K4 methylation have been implicated in human neurodevelopmental disorders including autism and Rett syndrome, and cognitive impairment is a prominent feature of the majority of human syndromes associated with mutations in histone modification genes. These observations suggest that some of the cognitive impairment, developmental delay and growth retardation that are frequently associated with CHD are caused by the broad developmental effects of underlying genetic mutation, instead of being preventable sequelae of caring for a child with CHD.
Table 1.
Defined syndromes caused by mutation in chromatin remodeling genes
| Syndrome | Gene | Face | Cardiac | Renal | Growth retardation (postnatal) |
Intellectual | Other | Hearing/ear |
|---|---|---|---|---|---|---|---|---|
| Kleefstra | EHMT1 | + | 50% | + | + | Autism (30%) |
||
|
Schinzel- Giedion |
SETBP1 | + | 43% | + | + | ++ | Increased neoplasia, highly lethal |
+ |
|
Claes- Jensen |
JARID1C/KDM5C | - | + | + | Seizures, behavioral |
|||
| Siderius | PHF8 | + | - | + | Cleft lip/palate |
+ | ||
| CHARGE | CHD7 | +(choanal atresia) |
50–85% | 20% | + | +(25% with no cognitive impairment) |
Genital hypoplasia |
Hypoplastic semicircular canals, deafness (100%) abnormal middle and external ear |
| Kabuki |
MLL2 KDM6A |
+ | 50% | 47% | 100% | + | Cleft palate/lip (40%) |
7. Conclusions & Future Directions
The explosion of genomic studies focusing on human CHD has given us a glimpse into the genetic complexity underlying a common birth defect. Significant hurdles to a complete understanding of the genetics of CHD remain, one of the most challenging being the extremely large number of genes being implicated in CHD coupled with extensive phenotypic variability. This will require extremely large patient cohorts in order to assign causality on the base of purely genetic statistical analysis. However, the coupling of gene discovery in humans with functional analysis in high-throughput model organisms such as mouse, zebrafish and Xenopus have begun to facilitate proving causality, as well as providing deep mechanistic insight into the function of CHD genes in cardiac development. Such hybrid studies represent a new paradigm for human birth defects research and provide hope for the development of improved diagnostics and therapeutics.
Acknowledgements
We thank M. Khokha for kindly contributing electron microscopy images. SY is supported by NIH training grant 2T32HD007094-35, MB is supported by NIH 1RO1 HL093280-01A1 and 1 UO1 1HL098162-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr. 2008;153:807–813. doi: 10.1016/j.jpeds.2008.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. **An excellent review of recent statistical trends that supports a genetic component for the etiology CHD
- 4.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, et al. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–86. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- 6.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 7.Guo T, McDonald-McGinn D, Blonska A, Shanske A, Bassett AS, Chow E, Bowser M, Sheridan M, Beemer F, Devriendt K, et al. Genotype and cardiovascular phenotype correlations with TBX1 in 1,022 velo-cardiofacial/DiGeorge/22q11.2 deletion syndrome patients. Hum Mutat. 2011;32:1278–1289. doi: 10.1002/humu.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 9. Nakamura T, Hamada H. Left-right patterning: conserved and divergent mechanisms. Development. 2012;139:3257–3262. doi: 10.1242/dev.061606. *A comprehensive review of the conserved mechanisms of LR development in mouse, zebrafish, chick and frog
- 10.Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 12. Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012;125:2232–2242. doi: 10.1161/CIRCULATIONAHA.111.079780. *Demonstrated an association between CHD and PCD in CHD patients, which has implications for the clinical management of such patients and highlights the role of motile cilia in both disorders
- 13.Icardo JM, Sanchez de Vega MJ. Spectrum of heart malformations in mice with situs solitus, situs inversus, and associated visceral heterotaxy. Circulation. 1991;84:2547–2558. doi: 10.1161/01.cir.84.6.2547. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 15. Lindsay EA, Baldini A. Recovery from arterial growth delay reduces penetrance of cardiovascular defects in mice deleted for the DiGeorge syndrome region. Hum Mol Genet. 2001;10:997–1002. doi: 10.1093/hmg/10.9.997. **An extensive review, which discusses many relevant mutations that have been associated with CHD
- 16.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. [PubMed] [Google Scholar]
- 18.Soemedi R, Wilson IJ, Bentham J, Darlay R, Topf A, Zelenika D, Cosgrove C, Setchfield K, Thornborough C, Granados-Riveron J, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91:489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soemedi R, Topf A, Wilson IJ, Darlay R, Rahman T, Glen E, Hall D, Huang N, Bentham J, Bhattacharya S, et al. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet. 2012;21:1513–1520. doi: 10.1093/hmg/ddr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, Gai X, Shaikh TH. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6:592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci U S A. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D’Amours G, Lemyre E, Cullum R, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8:e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. *Identified several de novo CNVs which are associated with atrioventricular septal defects (AVSDs)
- 24.Priest JR, Girirajan S, Vu TH, Olson A, Eichler EE, Portman MA. Rare copy number variants in isolated sporadic and syndromic atrioventricular septal defects. Am J Med Genet A. 2012;158A:1279–1284. doi: 10.1002/ajmg.a.35315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reamon-Buettner SM, Borlak J. NKX2-5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD) Hum Mutat. 2010;31:1185–1194. doi: 10.1002/humu.21345. [DOI] [PubMed] [Google Scholar]
- 27.Posch MG, Gramlich M, Sunde M, Schmitt KR, Lee SH, Richter S, Kersten A, Perrot A, Panek AN, Al Khatib IH, et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47:230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 29.Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 30. Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. *A large multi-institutional effort to identify and correlate genetic lesions with clinical outcome in CHD patients. The results of this effort are a publicly available resource
- 31. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. *Large-scale proteomic analysis that identified many ciliopathy-associated proteins as a biochemical network within the ciliary transition zone
- 32.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama T, Copeland NG, Jenkins NA, Montgomery CA, Elder FF, Overbeek PA. Reversal of left-right asymmetry: a situs inversus mutation [see comments] Science. 1993;260:679–682. doi: 10.1126/science.8480178. [DOI] [PubMed] [Google Scholar]
- 34.van Bon BW, Koolen DA, Pfundt R, van der Burgt I, de Leeuw N, de Vries BB. Transposition of the great vessels in a patient with a 2.9 Mb interstitial deletion of 9q31.1 encompassing the inversin gene: clinical report and review. Am J Med Genet A. 2008;146A:1225–1229. doi: 10.1002/ajmg.a.32289. [DOI] [PubMed] [Google Scholar]
- 35.Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.French VM, van de Laar IM, Wessels MW, Rohe C, Roos-Hesselink JW, Wang G, Frohn-Mulder IM, Severijnen LA, de Graaf BM, Schot R, et al. NPHP4 variants are associated with pleiotropic heart malformations. Circ Res. 2012;110:1564–1574. doi: 10.1161/CIRCRESAHA.112.269795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning DK, Sergeev M, van Heesbeen RG, Wong MD, Oh JH, Liu Y, Henkelman RM, Drummond I, Shah JV, Beier DR. Loss of the ciliary kinase Nek8 causes left-right asymmetry defects. J Am Soc Nephrol. 2013;24:100–112. doi: 10.1681/ASN.2012050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank V, Habbig S, Bartram MP, Eisenberger T, Veenstra-Knol HE, Decker C, Boorsma RA, Gobel H, Nurnberg G, Griessmann A, et al. Mutations in NEK8 link multiple organ dysplasia with altered Hippo signalling and increased c-MYC expression. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt070. [DOI] [PubMed] [Google Scholar]
- 39.Hills CB, Kochilas L, Schimmenti LA, Moller JH. Ellis-van Creveld syndrome and congenital heart defects: presentation of an additional 32 cases. Pediatr Cardiol. 2011;32:977–982. doi: 10.1007/s00246-011-0006-9. [DOI] [PubMed] [Google Scholar]
- 40.Tompson SW, Ruiz-Perez VL, Blair HJ, Barton S, Navarro V, Robson JL, Wright MJ, Goodship JA. Sequencing EVC and EVC2 identifies mutations in two-thirds of Ellis-van Creveld syndrome patients. Hum Genet. 2007;120:663–670. doi: 10.1007/s00439-006-0237-7. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Chen W, Chen Y, Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res. 2012;22:1593–1604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038. **CNV analysis of heterotaxy patients, identified several novel candidates that link cilia to the etiology of heterotaxy. One of the first studies to identify genetic lesions causing heterotaxy in humans by complimenting human genetics with high-throughput validation in model organisms
- 43.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spalluto C, Wilson DI, Hearn T. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur J Cell Biol. 2012;91:675–686. doi: 10.1016/j.ejcb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang G, Cadwallader AB, Jang DS, Tsang M, Yost HJ, Amack JD. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer’s vesicle in zebrafish. Development. 2011;138:45–54. doi: 10.1242/dev.052985. **Identified and characterized the role of novel chromatin modifications during cardiac differentiation and development **(Zaidi et al, Nature In press): Whole exome study of CHD patients that identified novel de novo mutations implicating chromatin modifications in the etiology of CHD
- 47.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 49.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, Tsurusaki Y, Nakashima M, Saitsu H, Niikawa N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 51.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 52.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 53.Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, de Brouwer AP, Nillesen W, Sistermans EA, Hamel BC, de Bruijn D, et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]