Abstract
ALK is an established causative oncogenic driver in neuroblastoma, and is likely to emerge as a routine biomarker in neuroblastoma diagnostics. At present, the optimal strategy for clinical diagnostic evaluation of ALK protein, genomic and hotspot mutation status is not well-studied. We evaluated ALK immunohistochemical (IHC) protein expression using three different antibodies (ALK1, 5A4 and D5F3 clones), ALK genomic status using single-color chromogenic in situ hybridization (CISH), and ALK hotspot mutation status using conventional Sanger sequencing and a next-generation sequencing platform (Ion Torrent Personal Genome Machine (IT-PGM)), in archival formalin-fixed, paraffin-embedded neuroblastoma samples. We found a significant difference in IHC results using the three different antibodies, with the highest percentage of positive cases seen on D5F3 immunohistochemistry. Correlation with ALK genomic and hotspot mutational status revealed that the majority of D5F3 ALK-positive cases did not possess either ALK genomic amplification or hotspot mutations. Comparison of sequencing platforms showed a perfect correlation between conventional Sanger and IT-PGM sequencing. Our findings suggest that D5F3 immunohistochemistry, single-color CISH and IT-PGM sequencing are suitable assays for evaluation of ALK status in future neuroblastoma clinical trials.
Introduction
ALK is an established causative oncogenic driver in neuroblastomas. With a recent phase 1 trial documenting complete response to the ALK inhibitor crizotinib in two patients with neuroblastomas [1], it seems probable that ALK status (whether protein, genomic or both) will emerge as a routine biomarker in neuroblastoma diagnostics. Against this backdrop, we performed a study evaluating the various platforms for ALK protein and genomic status characterization.
ALK protein expression in neuroblastomas has been reported as an adverse prognostic factor by several groups [2]–[5]. At present, with regards to ascertainment of ALK immunohistochemical status, the major unresolved issue appears to be the choice of antibody, there being several available commercially (Table 1). In our earlier analysis, we observed ALK expression in only 1/54 (1.85%) of neuroblastomas [6]. This contrasts sharply with the aforementioned studies, in which the prevalence of strong ALK immunohistochemical expression is generally around 50% or higher [2]–[5]. We were thus interested in comparing the performance of different antibodies.
Table 1. Commercially available monoclonal ALK antibodies.
| Clone | Tumor type studied | Company | Reference |
| ALK1 | Neuroblastoma | DAKO Denmark A/S, Glostrup, Denmark | De Brouwer 2010 [2]; Yan 2013 [6] |
| D5F3 | Lung adenocarcinoma | Cell Signaling, Danvers, MA, USA | Ying 2013 [37] |
| RM-9108 | Neuroblastoma | Thermo Fisher Scientific, Fremont, CA, USA | Duijkers 2012 [3] |
| 5A4 | Neuroblastoma | Thermo Fisher Scientific, Fremont, CA, USA | Passoni 2009 [4] |
High-level ALK genomic amplification, although infrequent (prevalence 5% or less), has also been reported as an adverse prognostic factor in neuroblastomas in some studies [5], [7], [8]. We evaluated the performance of a chromogenic in situ hybridization (CISH) assay [9] for ascertaining ALK copy number status.
ALK sequence mutations have been reported in approximately 5–10% of neuroblastomas [7], [10]–[12], and these predominantly occur within the tyrosine kinase domain, the two hotspots being p.F1174 and p.R1275 [13]. The presence of ALK mutations appears to have clinical implications. For example, the presence of the p.F1174L mutation was associated with a worse prognosis in MYCN-amplified neuroblastomas in one study [2], and resistance to crizotinib in another preclinical in vitro study [14].
Although Sanger sequencing is considered the gold standard for mutational analysis [15], [16], there has been considerable interest in the use of next-generation sequencing (NGS) platforms for clinical diagnostics [17], [18]. To date, NGS analysis has been applied to neuroblastomas predominantly for discovery work [19]–[22]. From a clinical quality management perspective, we were interested to compare the performance of the Ion Torrent Personal Genome Machine (IT-PGM), a benchtop NGS platform, to Sanger sequencing in the detection of ALK mutations occurring at the p.F1174 and p.R1275 hotspots.
Materials and Methods
Study population and clinicopathological data
A total of 118 neuroblastoma samples (comprising 36 pre-treatment, 53 post-treatment, 26 relapsed or metastatic samples, and 3 with unknown treatment status) from 95 patients, were identified from the archives of the Department of Pathology and Laboratory Medicine, KK Women's and Children's Hospital, Singapore. Clinicopathological data including mean age at first biopsy, gender, stage and MYCN amplification status was extracted (Table 2).
Table 2. Clinicopathological characteristics of study cohort.
| Age at first biopsy (yrs, N = 95) | |
| Mean (Range) | 3.54 (0.02–10.94) |
| Gender (N = 95) | |
| Male | 60 |
| Female | 35 |
| Sample type (N = 118) | |
| Pre-treatment primary tumors | 36 |
| Post-treatment primary tumors | 53 |
| Relapsed/metastatic tumors | 26 |
| Treatment status unknown | 3 |
| Histology (N = 118) | |
| Neuroblastoma | 67 |
| Ganglioneuroblastoma | 16 |
| Ganglioneuroma | 3 |
| Neuroblastic tumor, unspecified | 32 |
| MYCN amplification status (N = 108) | |
| Amplified | 26 |
| Non-amplified | 81 |
| Equivocal | 1 |
Ethics Statement
Ethics approval was obtained from the Singhealth Centralised Institutional Review Board (CIRB Ref: 2012/450/B).
Tissue microarray construction
Tissue microarray construction (TMA) was performed for 112 samples. One tissue core (1.0 mm diameter) was punched from a representative tumor area from the donor tissue blocks and deposited into a recipient block using a manual tissue-arraying instrument (Beecher Instruments, Sun Prairie, Wisconsin, USA).
Immunohistochemistry
The following anti-ALK antibodies were used: ALK1 antibody (Dako Denmark A/S, Glostrup, Denmark), D5F3 antibody (Cell Signaling, Danvers, MA, USA) and 5A4 antibody (Leica Biosystems, Newcastle Upon Tyne, UK). Immunohistochemical staining was performed using the dilutions and platforms as stated in Table 3.
Table 3. Antibodies used in this study with corresponding platforms.
| Clone (dilution) | Company | Platforms | Antigen Retrieval |
| ALK1 (1∶25) | DAKO Denmark A/S, Glostrup, Denmark | Ventana Benchmark ULTRA (Ventana Medical Systems, AZ, USA) | CC1 solution (Ventana Medical Systems, AZ, USA) for 36 min |
| D5F3 (1∶100) | Cell Signaling, Danvers, MA, USA | Ventana Benchmark XT (Ventana Medical Systems, AZ, USA) | CC1 solution (Ventana Medical Systems, AZ, USA) for 64 min |
| 5A4 (1∶60) | Leica Biosystems, Newcastle Upon Tyne, UK | Ventana Benchmark XT (Ventana Medical Systems, AZ, USA) | CC1 solution (Ventana Medical Systems, AZ, USA) for 64 min |
Scoring (taking into account staining intensity and percentage positivity, modified from Passoni et al [4]) was performed as follows: 0, no staining; 1+, weak cytoplasmic staining, >1% of cells; 2+, moderate cytoplasmic staining, >50% of cells; 3+, strong cytoplasmic staining, >50% of cells. Similar to guidelines for HER2 IHC scoring [23], the following categories were defined: 0 and 1+ - negative; 2+ - equivocal; 3+ - positive.
Chromogenic in situ hybridization
ALK genomic copy number status was determined by using the ZytoDot 2C SPEC ALK break-apart probe (ZytoVision, Bremerhaven, Germany). The protocol is similar to that previously reported [9] with some minor modifications. After deparaffinization in xylene and ethanol, the slides were incubated in 3% hydrogen peroxide in methanol for 5 min to quench endogenous peroxidase activity. 15 min incubation in EDTA pretreatment buffer (ZytoVision) at 95°C in a water bath, followed by digestion with pepsin solution (ZytoVision) at RT for 6 min was performed. The sections were dehydrated in graded ethanol and air-dried. 10–20 ml of probe were applied to the tissue, covered with a coverslip, and sealed with rubber cement. The slides were denatured at 79°C for 5 min and hybridized overnight at 37°C in a TDH-500 slide denaturation/hybridization system (Hangzhou Allsheng Instruments Co., Zhejiang, China). After removal of the coverslips, the slides were washed for 5 min in Wash Buffer SSC (ZytoVision) at 75–80°C and rinsed in deionized water for 2 min at room temperature. Probe detection was performed with sequential incubation of rabbit-anti-DNP and anti-rabbit-AP antibody, each for 15 min at 37°C in a humidity chamber, followed by incubation with the color substrates AP-Red for 10 min at room temperature (ZytoVision). The tissue was counterstained with the Nuclear Blue Solution provided by the kit, washed in running tap water for 2 min and dehydrated in a short sequence of 100% ethanol (3×30 s) and xylene (2×30 s) before coverslipping.
Twenty non-overlapping cells were evaluated. Similar to guidelines for HER2 genomic status [23], the following categories were defined: average ALK copy number >6 signals/nucleus - positive; 4–6 signals/nucleus - equivocal; <4 signals/nucleus - negative.
Cell line-Derived Tumor Xenografts
All the animal experiments were carried out with approval from Institutional Animal Care and Use Committee (IACUC) at the Biological Resource Center, A-STAR, Singapore. 5×106 SKNF1, SH-SY5Y and SK-N-SH neuroblastoma cells (obtained from the American Type Culture Collection) in matrigel (1∶1) were implanted subcutaneously on the flank of four to six old female Balb/c nude mice (BRC, Singapore). Tumor growth was monitored over a period of time and tumor volume (mm3) calculated by the formula V = L*B2/2, where V = volume of tumor, L = length of tumor and B = breadth of tumor using electronic Vernier callipers (Mitutoyo, Japan). When the tumor volume was between 200–300 mm3, the animals were euthanized. The tumors were removed and fixed in 10% neutral buffered formalin solution.
DNA extraction
DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) samples using ReliaPrep gDNA FFPE kit, Promega. Percentage of tumor cells, as assessed by light microscopy, was >30% in all cases. Purified DNA was quantified using the Qubit (Invitrogen, Life Technologies).
Sanger sequencing
PCR amplification of ALK exons 23 (containing the p.F1174 hotspot) and 25 (containing the p.R1275 hotspot) was performed using the following primers (as reported by Chen et al) [10]: exon 23 forward primer – AAGATTTGCCCAGACTCAGC, exon 23 reverse primer – TGTCCTTGGCACAACAACTG; exon 25 forward primer – TAGTGATGGCCGTTGTACAC, exon 25 reverse primer - CCAGGAGATGATGTAAGGGA. The PCR product was analyzed by routine sequencing using the ABI 3730xl DNA sequencer (Applied Biosystems, Foster City, CA).
IT-PGM Sequencing
Sequencing was performing according to the manufacturer's protocol using 10 ng DNA, Ion AmpliSeq primer pool and Ion AmpliSeq Library Kit 2.0 Beta (Life Technologies) for sequencing of the ALK gene. Briefly, PCR enrichment of 5182 bp of the ALK coding region, corresponding to 66 amplicons and 100% of the gene, was performed. Template preparation was performed using the Ion OneTouch system (Life Technologies). The Ion Sphere Particles were recovered and enriched according to the manufacturer's protocol. Samples were loaded onto either a 314 or 316 chip.
Bioinformatics analysis
Bioinformatics analysis was performed using the vendor-provided data analysis pipeline (Torrent Suite Software version 4.0.2 and Ion Reporter version 4.0) and independently using a pipeline constructed with Bowtie2 [24] and GATK [25]. The Bowtie2-GATK pipeline maps the raw reads of each library to hg19 using the default settings of Bowtie2. Variants were called on this set of reads using GATK's haplotypecaller. Alignments were visualized using the Integrative Genomics Viewer [26].
Statistical analysis
Fisher's exact test for r×c tables was used to test for significance of associations between the parameters. All p reported are one-sided probabilities. All analysis was done using Stata version 11 (StataCorp LP).
Results
Immunohistochemistry and CISH
A total of 112 cases were analyzed by a TMA platform. In addition, 6 cases were analyzed by full section because these were more recent cases that had not been incorporated into the TMA.
Following ALK immunohistochemical staining, due to missing cores or the absence of tumor in the cores, complete results for all three antibodies were available for 105 cases. Following ALK CISH, in addition to missing cores or the absence of tumor in the cores, 12 cases were technically unsuccessful (no signal detected), while 2 cases had marked crush artefact resulting in interpretation difficulties. Therefore, complete results for all three antibodies and ALK CISH were available for 91 cases.
The IHC results for the three different antibodies are displayed in Table 4. The D5F3 antibody detected the most, while the ALK1 antibody detected the least, number of positive cases (percentage of positive cases for D5F3, 5A4 and ALK1 antibodies are 13.3% (14/105), 0.02% (2/105) and 0.01% (1/105) respectively.
Table 4. Sample type and ALK immunohistochemical expression (N = 105).
| ALK1 clone | 5A4 clone | D5F3 clone | |||||||||
| IHC Score | 0 | 1+ | 3+ | 0 | 1+ | 2+ | 3+ | 0 | 1+ | 2+ | 3+ |
| Pre-treatment primary tumors | 28 | 0 | 0 | 20 | 5 | 3 | 0 | 11 | 8 | 6 | 3 |
| Post-treatment primary tumors | 49 | 0 | 1 | 41 | 7 | 1 | 1 | 17 | 19 | 9 | 5 |
| Relapsed/metastatic tumors | 23 | 1 | 0 | 15 | 5 | 3 | 1 | 4 | 10 | 4 | 6 |
| Treatment unknown | 3 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 2 | 0 |
| p (Fisher's exact test) | 0.519 | 0.165 | 0.322 | ||||||||
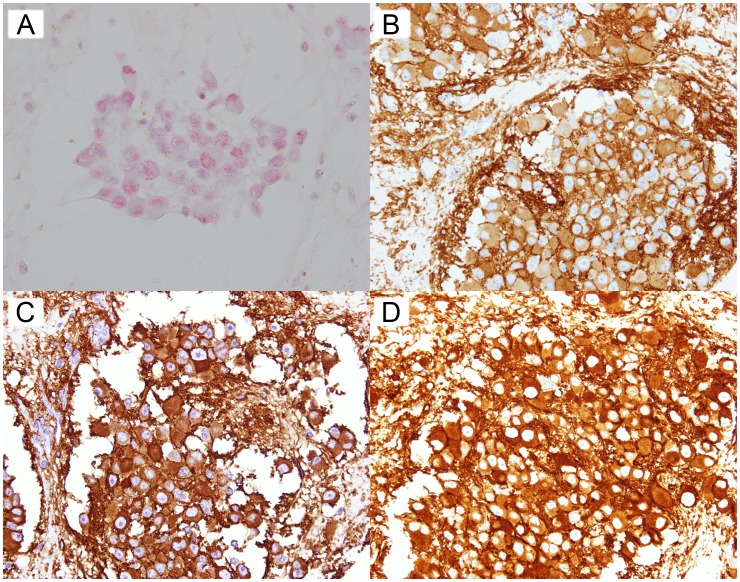
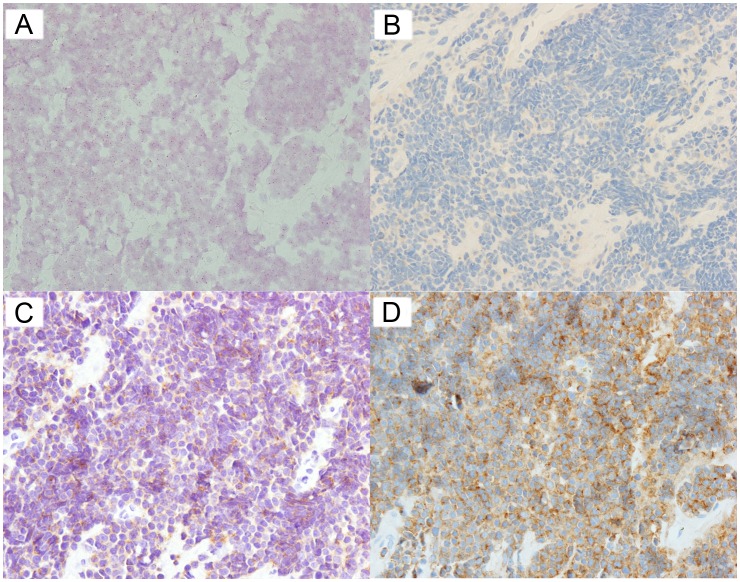
We detected one ALK amplified (positive) case, with all the tumor cells in the TMA core displaying more than 6 ALK copy numbers/nucleus. Of note, this was the only case in our cohort displaying 3+ ALK1 IHC expression (Figure 1). The remaining cases were ALK non-amplified (negative); there were no cases with equivocal ALK amplification status. Figure 2 illustrates the ALK IHC profiles of an ALK non-amplified case.
Figure 1. ALK CISH and IHC of an ALK amplified case.
(A) CISH (B) ALK1 IHC (C) 5A4 IHC (D) D5F3 IHC.
Figure 2. ALK CISH and IHC of an ALK non-amplified case.
(A) CISH (B) ALK1 IHC (C) 5A4 IHC (D) D5F3 IHC.
Tables 5, 6 and 7 show the correlation between ALK CISH and ALK IHC for the three antibodies. There were statistically significant positive correlations between ALK1 IHC expression and ALK amplification status; and 5A4 IHC expression and ALK amplification status. Of note, the majority of ALK D5F3 IHC-positive neuroblastomas did not possess ALK genomic amplification.
Table 5. Correlation between ALK1 IHC and ALK CISH (N = 91).
| ALK amplified | ALK non-amplified | |
| ALK1 IHC 3+ | 1 | 0 |
| ALK1 IHC 0–2+ | 0 | 90 |
p (Fisher's exact test) = 0.011.
Table 6. Correlation between 5A4 IHC and ALK CISH (N = 91).
| ALK amplified | ALK non-amplified | |
| 5A4 IHC 3+ | 1 | 1 |
| 5A4 IHC 0–2+ | 0 | 89 |
p (Fisher's exact test) = 0.022.
Table 7. Correlation between D5F3 IHC and ALK CISH (N = 91).
| ALK amplified | ALK non-amplified | |
| D5F3 IHC 3+ | 1 | 12 |
| D5F3 IHC 0–2+ | 0 | 78 |
p (Fisher's exact test) = 0.143.
Sanger and IT-PGM sequencing
53 patient samples were subjected to Sanger sequencing of ALK exons 23 and 25 for the detection of p.F1174 and p.R1275 mutations. We identified 3 cases with the p.F1174L mutation, and 2 cases with the p.R1275Q mutation (Table 8). The correlation between the mutation status and D5F3 IHC expression is shown in Table 9.
Table 8. ALK mutations detected by both Sanger sequencing and IT-PGM.
| Case Number | Coding DNA | Amino acid | Depth of Coverage (x) | Variant allele frequency (%) |
| 11011 | c.3522C>G | p.F1174L | 1992 | 50.45 |
| 145808 | c.3522C>A | p.F1174L | 782 | 15.09 |
| 70309 | c.3824G>A | p.R1275Q | 897 | 20.07 |
| 180713 | c.3824G>A | p.R1275Q | 1536 | 41.99 |
| 184213 | c.3522C>A | p.F1174L | 1996 | 56.21 |
Table 9. Correlation between ALK D5F3 IHC score and mutation status.
| p.F1174L | p.R1275Q | p.F1174 and p.R1275 wild-type | |
| D5F3 IHC 3+ | 1 | 2 | 5 |
| D5F3 IHC 2+ | 2 | 0 | 11 |
| D5F3 IHC 1+ | 0 | 0 | 21 |
| D5F3 IHC 0 | 0 | 0 | 12 |
p (Fisher's exact test) = 0.004.
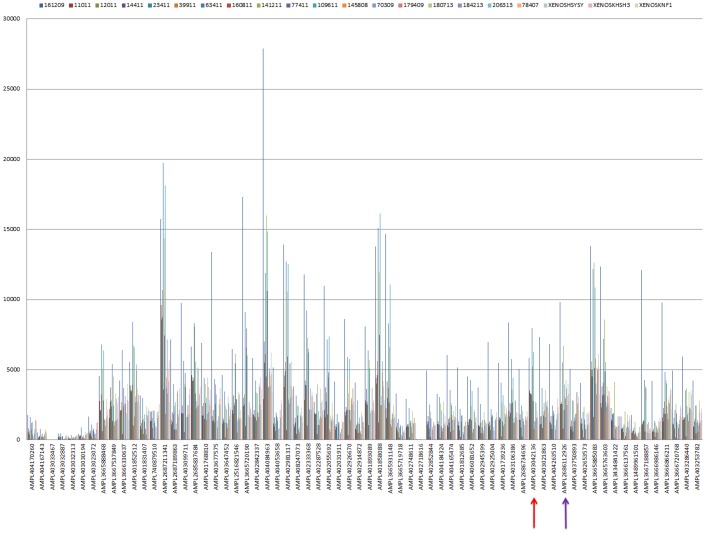
18 patient samples and 3 cell line-derived tumor xenografts were analyzed by IT-PGM. 19 of 21 samples showed adequate uniformity of coverage, with less than 2% of bases with zero coverage. The other 2 cases (145808 and 180713) had around 30% of bases with zero coverage. The depth of coverage across the ALK coding region for all 21 samples is illustrated in Figure 3.
Figure 3. Depth of coverage of amplicons from 5′ to 3′.
The p.F1174 and p.R1275 hotspots are covered by the amplicons marked with a red and purple arrow respectively.
Of particular interest to us, the depth of coverage for the amplicons encompassing the p.F1174 and p.R1275 hotspots ranged from 741 to 7968× (AMPL403042136), and 590 to 5059× (AMPL403750893) respectively.
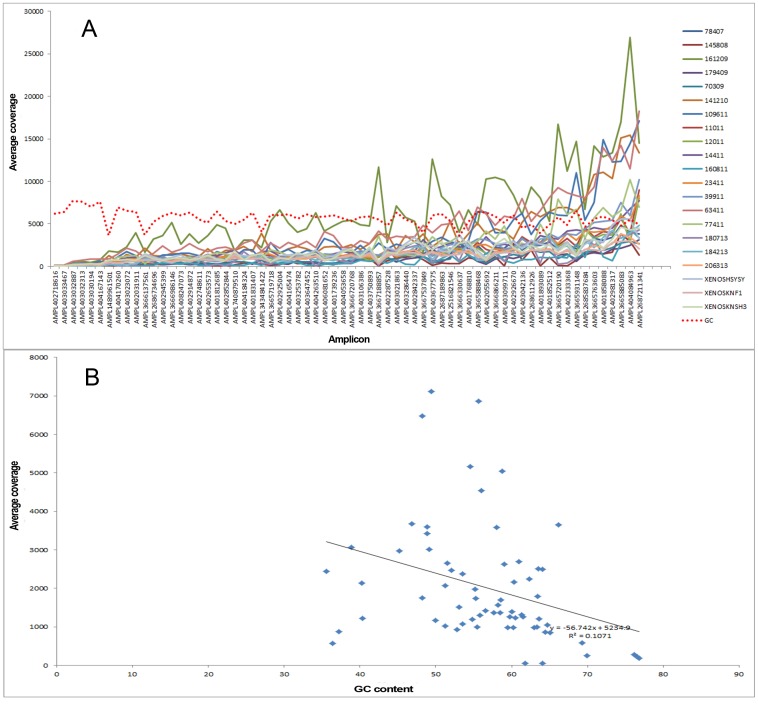
We were interested to study the correlation between depth of coverage and GC content, as undercoverage of GC-rich regions during the PCR amplification step of the library construction is a reported phenomenon resulting in coverage bias [27], [28]. We found a negative correlation between the depth of coverage and GC content, although this was not statistically significant (R2 = 0.1) (Figure 4).
Figure 4. Depth of coverage and GC content.
(A) The red dashed line indicates the GC content. The X-axis shows the amplicons ordered with increasing depth of coverage from left to right. (B) There is a negative correlation between depth of coverage and GC content; this is not statistically significant (R2 = 0.1).
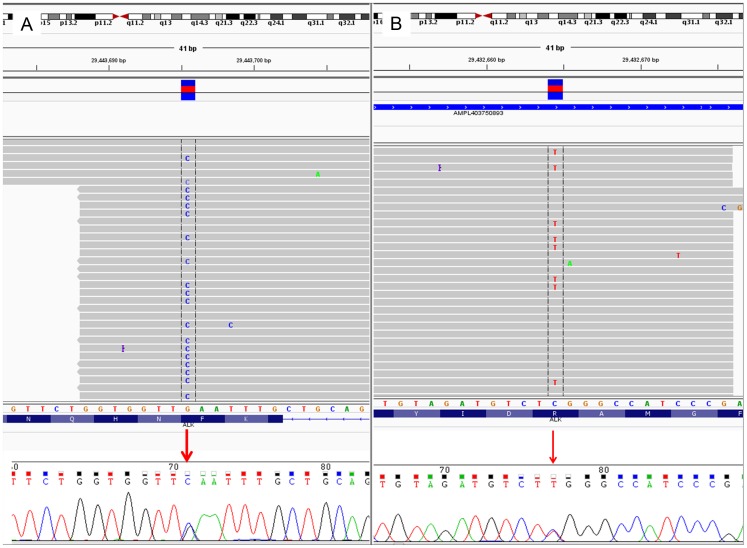
There was a perfect correlation between the results obtained by IT-PGM and Sanger sequencing for all 18 patient samples (3 cases with p.F1174L; 2 cases with p.R1275Q; 13 wild-type) and 3 cell line-derived tumor xenografts (2 with p.F1174L; 1 wild-type). A representative case is illustrated in Figure 5. Of note, our mutational analyses for the xenografts corroborate with that reported in the literature: SKNF1 – ALK wild-type (WT); SH-SY5Y and SK-N-SH – p.F1174L) [14], [29].
Figure 5. IGV (IT-PGM) and Sanger sequences of ALK hotspot mutations.
(A) c.3522C>G (p.F1174L) mutation. (B) c.3824G>A (p.R1275Q) mutation.
Correlation between ALK D5F3 IHC and hotspot mutational analysis results revealed that the majority of ALK D5F3 IHC-positive neuroblastomas did not possess the hotspot mutations.
Correlation of ALK IHC, copy number and mutation status with other clinicopathological parameters
We also correlated ALK IHC, copy number and mutation status with histology and MYCN copy number status (Tables S1, S2, S3, S4, S5, S6, S7, S8, S9 and S10 in File S1). We observed statistically significant positive correlations between ALK D5F3 IHC and MYCN copy number status; and ALK mutation status and MYCN copy number status (Tables S3 and S5 in File S1). No other statistically significant associations were observed.
Discussion
The evaluation of various diagnostic platforms for ascertainment of ALK status is an important topic in lung carcinoma [30], [31], and this subject will become increasingly prominent in neuroblastoma diagnostics. Our paper is an early study on this emerging theme.
In the lung cancer literature, much attention has been devoted to the comparison of different ALK antibodies for immunohistochemistry [30], [32]. The earliest antibody developed was the ALK1 clone [33], which has primarily been used diagnostically in the detection of ALK-rearranged anaplastic large-cell lymphomas. In 2010, a comparison study by Mino-Kenudson et al. [34] reported that the D5F3 clone appeared to be more sensitive than ALK1 in detecting ALK-rearranged lung adenocarcinomas. At present, the exact mechanism for this increased sensitivity is not known. Based on recent publications, D5F3 appears to be a fairly popular choice for ascertainment of ALK protein expression status in lung carcinomas [35]–[37].
Unlike in lung carcinomas where the sensitivity and specificity of ALK IHC can be compared against ALK rearrangement status (as evaluated by FISH or CISH), such a ‘gold standard’ reference does not exist in neuroblastomas, since the mechanism for ALK protein expression does not appear to involve a genetic event in the majority of cases (as noted previously, the prevalence of strong ALK protein expression is generally 50% or higher, whereas the prevalence of ALK genomic amplification and/or mutations is around 10%). We were thus not able to establish sensitivity and specificity of the various antibodies.
However, we observed a statistically significant positive correlation between ALK D5F3 IHC and MYCN copy number status, which is consistent with the literature reporting a positive correlation between ALK gene/protein expression and MYCN amplification [5], [38]. Since MYCN amplification is a well known adverse prognostic marker in neuroblastomas [39], this provides further support that the ALK D5F3 antibody is likely the most suitable among the three for ascertaining ALK protein expression status as a prognostic biomarker. It will be necessary to evaluate this antibody in prospective clinical trials to robustly determine its clinical utility as a prognostic and predictive therapeutic biomarker.
We have also shown that a single color ALK CISH assay enables detection of ALK genomic amplification in neuroblastomas. The advantages of employing CISH instead of FISH are well known, and include convenience resulting from use of a brightfield instead of a fluorescence microscope; a greater ability to correlate gene copy numbers with morphological features; and the dispensation of a requirement for micrograph storage, necessary in the case of FISH because of the photolability of the probes [40].
At present, compared to ALK immunohistochemical expression, ALK copy number status has not been as widely studied as a biomarker in neuroblastomas, probably because of the rarity of ALK genomic amplification (prevalence 5% or less) [5], [7], [8]. However, it is worth noting that ALK genomic amplification correlates with a poor outcome [5], [7], [8], and that an in vitro drug sensitivity study revealed that an ALK amplified cell line (NB1) displayed enhanced sensitivity to growth inhibition by crizotinib [14]. Our study shows that single color CISH can be employed in future clinical trials to ascertain the ALK copy number profile and its utility as a prognostic and predictive biomarker.
We were able to successfully utilize a NGS platform for the detection of the ALK hotspot mutations (p.F1174L and p.R1275Q), which are present in up to 10% of neuroblastomas [7], [10]–[12]. Although NGS might not be the most cost-effective method for identification of only two mutations in an uncommon tumor, it becomes a more viable alternative to conventional Sanger sequencing if the region of interest covers most, or all, of the entire ALK gene. Rare ALK mutations, with at present uncertain clinical significance, are known to occur outside of regions covered by amplicons for exons 23 and 25, where the p.F1174 and p.R1275 hotspots are located. These include truncations involving exons 2–3 [29] or exons 4–11 [41], and mutations in exon 20 (e.g. p.D1091N) and exon 22 (e.g. p.T1151M) [42]. We envisage that future diagnostic panels encompassing multiple genes will be the most cost-effective way forward for exploiting an NGS platform for neuroblastoma diagnostics.
With regards to the IT-PGM sequencing, we observed several amplicons that showed low read depth (Figure 3). Amplicon read depth is an important, but to date not well-studied phenomenon, in NGS diagnostics. One of the well-known reasons that might account for low amplicon read depth, as mentioned above, is the presence of GC-extreme regions [27], [28]. However, this was not found to be the case in our study, as seen by the lack of significant statistical association between GC content and amplicon read depth. Based on our findings, optimization of the protocol, e.g. re-designing primers, will be necessary to achieve adequate amplicon read depth to the standard required for NGS diagnostics.
In conclusion, we have evaluated several platforms for the ascertainment of ALK status in neuroblastomas. Our study reveals the different staining properties of the commonly used ALK antibodies employed in routine diagnostics, in particular the increased sensitivity of the D5F3 antibody. It will be necessary for future clinical trials evaluating ALK as a prognostic and therapeutic predictive biomarker to determine the optimal antibody for routine diagnostics. This is particularly important for ALK IHC-positive but ALK copy number/mutation-negative neuroblastomas, in which the utility of ALK targeted therapeutics is at present unascertained. In addition, we find that single-color CISH and IT-PGM sequencing are suitable assays to ascertain ALK genomic copy and mutational status respectively, and these should also be employed and evaluated in future clinical trials.
Supporting Information
Tables S1–S10. Table S1 in File S1. Correlation between ALK1 IHC and MYCN status (N = 96). Table S2 in File S1. Correlation between 5A4 IHC and MYCN status (N = 98). Table S3 in File S1. Correlation between D5F3 IHC and MYCN status (N = 97). Table S4 in File S1. Correlation between ALK CISH and MYCN status (N = 83). Table S5 in File S1. Correlation between MYCN status and ALK mutation status (N = 48). Table S6 in File S1. Correlation between ALK1 IHC and histology (N = 104). Table S7 in File S1. Correlation between 5A4 IHC and histology (N = 106). Table S8 in File S1. Correlation between D5F3 IHC and histology (N = 105). Table S9 in File S1. Correlation between ALK CISH and histology (N = 91). Table S10 in File S1. Correlation between ALK mutational status and histology (N = 54).
(DOCX)
Acknowledgments
Funding support was provided by a grant from the National Medical Research Council (NMRC/CNIG/1095/2012).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Data can be requested from Dr Kenneth Chang (Kenneth.chang.te@kkh.com.sg). The authors are happy to provide the tissue microarray slides by mail, to researchers upon request.
Funding Statement
Funding support was provided by a grant from the National Medical Research Council (NMRC/CNIG/1095/2012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, et al. (2013) Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol 14: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, et al. (2010) Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res 16: 4353–4362. [DOI] [PubMed] [Google Scholar]
- 3. Duijkers FA, Gaal J, Meijerink JP, Admiraal P, Pieters R, et al. (2012) High anaplastic lymphoma kinase immunohistochemical staining in neuroblastoma and ganglioneuroblastoma is an independent predictor of poor outcome. Am J Pathol 180: 1223–1231. [DOI] [PubMed] [Google Scholar]
- 4. Passoni L, Longo L, Collini P, Coluccia AM, Bozzi F, et al. (2009) Mutation-independent anaplastic lymphoma kinase overexpression in poor prognosis neuroblastoma patients. Cancer Res 69: 7338–7346. [DOI] [PubMed] [Google Scholar]
- 5. Wang M, Zhou C, Sun Q, Cai R, Li Y, et al. (2013) ALK amplification and protein expression predict inferior prognosis in neuroblastomas. Exp Mol Pathol 95: 124–130. [DOI] [PubMed] [Google Scholar]
- 6. Yan B, Lim M, Zhou L, Kuick CH, Leong MY, et al. (2013) Identification of MET genomic amplification, protein expression and alternative splice isoforms in neuroblastomas. J Clin Pathol 66: 985–991. [DOI] [PubMed] [Google Scholar]
- 7. Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, et al. (2008) Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caren H, Abel F, Kogner P, Martinsson T (2008) High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem J 416: 153–159. [DOI] [PubMed] [Google Scholar]
- 9. Schildhaus HU, Deml KF, Schmitz K, Meiboom M, Binot E, et al. (2013) Chromogenic in situ hybridization is a reliable assay for detection of ALK rearrangements in adenocarcinomas of the lung. Mod Pathol 26: 1468–1477. [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, Takita J, Choi YL, Kato M, Ohira M, et al. (2008) Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455: 971–974. [DOI] [PubMed] [Google Scholar]
- 11. George RE, Sanda T, Hanna M, Frohling S, Luther W 2nd, et al. (2008) Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455: 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, et al. (2008) Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455: 967–970. [DOI] [PubMed] [Google Scholar]
- 13. Hallberg B, Palmer RH (2013) Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 13: 685–700. [DOI] [PubMed] [Google Scholar]
- 14. Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, et al. (2011) Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med 3: 108ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, et al. (2012) Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 30: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, et al. (2013) ACMG clinical laboratory standards for next-generation sequencing. Genet Med 15: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCourt CM, McArt DG, Mills K, Catherwood MA, Maxwell P, et al. (2013) Validation of next generation sequencing technologies in comparison to current diagnostic gold standards for BRAF, EGFR and KRAS mutational analysis. PLoS One 8: e69604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh RR, Patel KP, Routbort MJ, Reddy NG, Barkoh BA, et al. (2013) Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 15: 607–622. [DOI] [PubMed] [Google Scholar]
- 19. Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, et al. (2012) Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, et al. (2012) Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483: 589–593. [DOI] [PubMed] [Google Scholar]
- 21. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, et al. (2013) The genetic landscape of high-risk neuroblastoma. Nat Genet 45: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, et al. (2013) Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 45: 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, et al. (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31: 3997–4013. [DOI] [PubMed] [Google Scholar]
- 24. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, et al. (2013) Characterizing and measuring bias in sequence data. Genome Biol 14: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benjamini Y, Speed TP (2012) Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 40: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okubo J, Takita J, Chen Y, Oki K, Nishimura R, et al. (2012) Aberrant activation of ALK kinase by a novel truncated form ALK protein in neuroblastoma. Oncogene 31: 4667–4676. [DOI] [PubMed] [Google Scholar]
- 30. Weickhardt AJ, Aisner DL, Franklin WA, Varella-Garcia M, Doebele RC, et al. (2013) Diagnostic assays for identification of anaplastic lymphoma kinase-positive non-small cell lung cancer. Cancer 119: 1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim H, Yoo SB, Choe JY, Paik JH, Xu X, et al. (2011) Detection of ALK gene rearrangement in non-small cell lung cancer: a comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of ALK protein expression. J Thorac Oncol 6: 1359–1366. [DOI] [PubMed] [Google Scholar]
- 32. Selinger CI, Rogers TM, Russell PA, O'Toole S, Yip P, et al. (2013) Testing for ALK rearrangement in lung adenocarcinoma: a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol 26: 1545–1553. [DOI] [PubMed] [Google Scholar]
- 33. Pulford K, Lamant L, Morris SW, Butler LH, Wood KM, et al. (1997) Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood 89: 1394–1404. [PubMed] [Google Scholar]
- 34. Mino-Kenudson M, Chirieac LR, Law K, Hornick JL, Lindeman N, et al. (2010) A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 16: 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conklin CM, Craddock KJ, Have C, Laskin J, Couture C, et al. (2013) Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 8: 45–51. [DOI] [PubMed] [Google Scholar]
- 36. Li Y, Pan Y, Wang R, Sun Y, Hu H, et al. (2013) ALK-rearranged lung cancer in Chinese: a comprehensive assessment of clinicopathology, IHC, FISH and RT-PCR. PLoS One 8: e69016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ying J, Guo L, Qiu T, Shan L, Ling Y, et al. (2013) Diagnostic value of a novel fully automated immunochemistry assay for detection of ALK rearrangement in primary lung adenocarcinoma. Ann Oncol 24: 2589–2593. [DOI] [PubMed] [Google Scholar]
- 38. Hasan MK, Nafady A, Takatori A, Kishida S, Ohira M, et al. (2013) ALK is a MYCN target gene and regulates cell migration and invasion in neuroblastoma. Sci Rep 3: 3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang M, Weiss WA (2013) Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 3: a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan B, Yau EX, Bte Omar SS, Ong CW, Pang B, et al. (2010) A study of HER2 gene amplification and protein expression in gastric cancer. J Clin Pathol 63: 839–842. [DOI] [PubMed] [Google Scholar]
- 41. Cazes A, Louis-Brennetot C, Mazot P, Dingli F, Lombard B, et al. (2013) Characterization of rearrangements involving the ALK gene reveals a novel truncated form associated with tumor aggressiveness in neuroblastoma. Cancer Res 73: 195–204. [DOI] [PubMed] [Google Scholar]
- 42. Azarova AM, Gautam G, George RE (2011) Emerging importance of ALK in neuroblastoma. Semin Cancer Biol 21: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S10. Table S1 in File S1. Correlation between ALK1 IHC and MYCN status (N = 96). Table S2 in File S1. Correlation between 5A4 IHC and MYCN status (N = 98). Table S3 in File S1. Correlation between D5F3 IHC and MYCN status (N = 97). Table S4 in File S1. Correlation between ALK CISH and MYCN status (N = 83). Table S5 in File S1. Correlation between MYCN status and ALK mutation status (N = 48). Table S6 in File S1. Correlation between ALK1 IHC and histology (N = 104). Table S7 in File S1. Correlation between 5A4 IHC and histology (N = 106). Table S8 in File S1. Correlation between D5F3 IHC and histology (N = 105). Table S9 in File S1. Correlation between ALK CISH and histology (N = 91). Table S10 in File S1. Correlation between ALK mutational status and histology (N = 54).
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Data can be requested from Dr Kenneth Chang (Kenneth.chang.te@kkh.com.sg). The authors are happy to provide the tissue microarray slides by mail, to researchers upon request.