Abstract
Networks of protein interactions mediate cellular responses to environmental stimuli and direct the execution of many different cellular functional pathways. Small molecules synthesized within cells or recruited from the external environment mediate many protein interactions. The study of small molecule-mediated interactions of proteins is important to understand abnormal signal transduction pathways in cancer and in drug development and validation. In this study, we used split synthetic renilla luciferase (hRLUC) protein fragment-assisted complementation to evaluate heterodimerization of the human proteins FRB and FKBP12 mediated by the small molecule rapamycin. The concentration of rapamycin required for efficient dimerization and that of its competitive binder ascomycin required for dimerization inhibition were studied in cell lines. The system was dually modulated in cell culture at the transcription level, by controlling nuclear factor κB promoter/enhancer elements using tumor necrosis factor α, and at the interaction level, by controlling the concentration of the dimerizer rapamycin. The rapamycin-mediated dimerization of FRB and FKBP12 also was studied in living mice by locating, quantifying, and timing the hRLUC complementation-based bioluminescence imaging signal using a cooled charged coupled device camera. This split reporter system can be used to efficiently screen small molecule drugs that modulate protein-protein interactions and also to assess drugs in living animals. Both are essential steps in the preclinical evaluation of candidate pharmaceutical agents targeting protein-protein interactions, including signaling pathways in cancer cells.
INTRODUCTION
Biochemical pathways and networks constitute many different systems of dynamic assembly and disassembly of proteins with other proteins and nucleic acids (1). Most modern biological research is concerned with how, when, and where proteins interact with other proteins involved in biological processes. The demand for simple approaches to study protein-protein interactions, particularly on a large scale, has escalated recently with the completion of the genome project. To understand these ubiquitous protein interactions, several techniques have been developed, including the inducible yeast two hybrid system (2), immunoprecipitation (3), gel filtration chromatography (4), analytical ultracentrifugation (5), calorimetry (6), optical spectroscopy (7), split-ubiquitin system (8), Sos recruitment system (9–11), β-galactosidase complementation (12), G-protein fusion system (13, 14), and the fluorescence resonance energy transfer system (14, 15). The protein fragment-assisted complementation of dihydrofolate reductase and β-lactamase also has been used to study protein-protein interactions in bacteria and mammalian cells (15, 16). Each system has its own merits and demerits. The overall application of these systems to study protein-protein interactions in living small animal models of cancer is difficult because of insufficient sensitivity of detection systems and/or because of the low sensitivity of the assay systems.
In our previous studies, we reported an inducible yeast two hybrid system with firefly luciferase (17) and a split firefly luciferase complementation system (18) to study protein-protein interactions in cell lines and noninvasively in living animals using a cooled charged coupled device (CCD) camera. We also have reported recently protein fragment-assisted complementation of hRLUC to study real-time protein-protein interaction in cell lines (19). These optical imaging approaches and others that use different imaging modalities (e.g., positron emission tomography; Refs. 20, 21) can be used to study protein-protein interactions in cell culture and intact living small animals. We also have reported recently imaging intact firefly luciferase and intact renilla luciferase reporter gene expression in living mice using the substrates D-luciferin and coelenterazine, respectively, without any cross-reactivity (22).
There presently is a great deal of interest in developing pharmaceutical drugs that can specifically alter dysfunctional protein-protein interactions in diseased or transformed cancer cells (23–25). Small molecules that induce or stabilize the interactions of macromolecules have proven to be useful effectors of an extensive variety of biological processes (26). Many cellular functions are initiated by the induced interaction, or dimerization, of signaling proteins (26). The clustering of cell surface receptors by extracellular growth factors and the subsequent stepwise recruitment and activation of intracellular signaling proteins are prime examples of this process. The main impetus to develop artificial dimerizers that can alter signaling pathways is to achieve the important goal of managing abnormal cellular processes, particularly in cancer. Dimerizers currently are used mostly in laboratory research to study cell proliferation (27), transcription (28, 29), and apoptosis (28, 30). The use of these systems can be extended potentially into clinical practice, particularly in gene therapy. The preclinical evaluation of such dimerizers would benefit considerably from the availability of an efficient system to study the modulation of protein-protein interactions in cell lines and to do so noninvasively in living animals.
To evaluate such a model system, we used protein fragment-assisted complementation of split hRLUC in combination with rapamycin-mediated dimerization of the human protein FRB and its interacting partner, FKBP12 (Fig. 1) in cell culture and by noninvasive imaging in living animals. We also assessed the optimal dosage of rapamycin and the mode and frequency of its administration in living mice so as to yield an effective concentration for protein-protein heterodimerization at intended targets within the body.
Fig. 1.
Schematic diagram of rapamycin-mediated synthetic renilla luciferase (hRLUC) protein fragment-assisted complementation strategy. In this strategy, N-terminal and COOH-terminal portions of hRLUC fragments are attached to proteins X and Y, respectively, through a short peptide linker GGGGSGGGGS. The N and C portions of hRLUC fragments are closely approximated by the dimerization of proteins FRB and FKBP12 only in the presence of the small molecule rapamycin, and this, in turn, leads to recovered activity of the hRLUC protein.
MATERIALS AND METHODS
Chemicals, Enzymes and Reagents
Restriction and modification enzymes and ligase were purchased from New England Biolabs (Beverly, MA). PCR amplification using TripleMaster TaqDNA polymerase purchased from Brinkmann Eppendorf (Hamburg, Germany) was used to generate different fragments of the reporter gene hRluc and the genes for the rapamycin-dimerizing proteins FRB and FKBP12. The plasmids pCMV-hRL purchased from Promega (Madison, WI) and pFRB and pFKBP12 bought from Ariad Pharmaceuticals, Inc. (Cambridge, MA) were used as templates for the amplification of the aforementioned fragments. Rapamycin, ascomycin, tumor necrosis factor α (TNF-α), and antibiotics for bacterial culture were purchased from Sigma (St. Louis, MO). The nuclear factor κB (NFκB) promoter/enhancer elements were used from the vector pNFκB-Luc of Stratagene (La Jolla, CA). SuperFect transfection reagent, plasmid extraction kits, and DNA gel extraction kits were purchased from Qiagen (Valencia, CA). Coelenterazine was purchased from Biotium (Hayward, CA). Bacterial culture media were purchased from BD Diagnostic Systems (Sparks, MD). All of the animal cell culture media, fetal bovine serum, the antibiotics streptomycin and penicillin, and plastic wares for growing cell cultures were purchased from Invitrogen (Carlsbad, CA).
Construction of Plasmids
The N-terminal portion (amino acids 1–229) of the hRluc gene was amplified using the forward primer designed with NheI and the start codon, and the reverse primer designed with BamHI using phRL-CMV (Promega) as a template. Similarly, the COOH-terminal portion (amino acids 230–311) of the hRluc gene was amplified using the forward primer designed with BamHI and a linker sequence (GGGGSGGGGS), and the reverse primer was designed with XhoI and a stop codon. The restriction enzyme-digested fragments were cloned into a corresponding enzyme-digested vector backbone of pcDNA3.1(+) (Invitrogen). The human FRB fragment was amplified using the forward primer designed with BamHI and the linker sequence (GGGGSGGGGS), and the reverse primer designed with XhoI and a stop codon by using the template vector provided by Ariad Pharmaceuticals. The amplified fragment was cloned downstream of the N portion of hRluc fragment by using corresponding restriction enzymes. Similarly, the protein fragment FKBP12 was amplified using the forward primer designed with NheI and a start codon, and the reverse primer designed with BamHI by using the template provided by Ariad Pharmaceuticals. The digested fragment was cloned in the upstream of the C portion of the hRluc fragment by digesting with corresponding enzymes (Fig. 2). Additional details of the cloning methods and the primer sequences used are available on request.
Fig. 2.
Schematic representation of the plasmid constructs made and used in this study. Shown are the components of the genes (Nhrluc, N portion of synthetic renilla luciferase fragment; Chrluc, C portion of synthetic renilla luciferase fragment; FRB and FKBP12 are the rapamycin-binding human proteins) and promoters [CMV, cytomegalovirus early promoter/enhancer elements; nuclear factor κB (NFκB), tumor necrosis factor promoter sequences]. (G4S)2 is the amino acid sequence of the linkers used between the reporter fragments and the interacting proteins. A, pCMV-Nhrluc-FRB. B, pNFκB-Nhrluc-FRB. C, pCMV-FKBP12-Chrluc. D, pCMV-Nhrluc. E, pCMV-Chrluc.
Cell Culture
Human 293T embryonic kidney cancer cells and human HeLa cancer cells, purchased from American Type Culture Collection (Manassas, VA), were grown in MEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution. The N2a mouse neuroblastoma cells were obtained from V. P. Mauro (Scripps Research Institute, La Jolla, CA) and were grown in DMEM (high glucose) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Rat C6 glioma cells were maintained in glucose-deficient DMEM supplemented with 0.01% histidinol, 10% fetal bovine serum, and 1% penicillin/streptomycin/glutamine. Human U87 cancer cells purchased from American Type Culture Collection were grown in MEM supplemented with 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 0.15% sodium bicarbonate, 1% penicillin/streptomycin, and 10% fetal bovine serum.
Cell Transfection and Renilla Luciferase Assay
Transfections were performed in 80% confluent 24-h-old cultures of 293T, HeLa, N2a, and U87 cells. For transfection, 250 ng/well of Nhrluc and Chrluc DNA were used in 12-well culture plates. Volumes of SuperFect used were as recommended by the manufacturer. For heterodimerization in cell culture, 40 nM rapamycin were added to each well immediately after transfection. For cell induction, 0.05 μg/ml TNF-α were added immediately after transfection along with the rapamycin. The cells were assayed after 24-h incubation at 37°C in 5% CO2. The luminometry assay for renilla luciferase activity was performed as per protocol published previously (22). In brief, the cells were lysed in 200 μl of 1 × passive lysis buffer supplied by Promega and were shook for 15 min at room temperature. The cell lysates were centrifuged for 5 min at 10,000 × g at 4°C. Twenty μl of supernatant were assayed by adding 1 μl of the substrate coelenterazine (1 mg/ml) and 100 μl of 0.05 M sodium phosphate buffer at pH 7.0, followed by photon counting in the luminometer (model T 20/20; Turner Designs, Sunnyvale, CA) for 10 s. By measuring the protein concentration in the cell lysates, the readings were normalized. Activity of hRLUC was represented as relative light units per microgram of protein per minute of counting.
Optical CCD Imaging in Living Mice
All of the animal handling was performed in accordance with University of California Animal Research Committee guidelines. For imaging living nude mice (nu/nu), we used 293T cells transiently cotransfected with Nhrluc-FRB and FKBP12-Chrluc. We studied three different modes of cell implantation—i.p., i.v., and s.c.—as outlined in the flow chart in Fig. 3. To study the consequences of i.p. implantation of cells, 5 × 106 cells were injected i.p. 3 h after ex vivo cotransfection. Four sets of three animals were imaged immediately after cell implantation by injecting 50 μg of coelenterazine via the tail vein. After obtaining these first baseline images, three of these sets of animals were reinjected via the tail vein with 10 μg, 25 μg, or 50 μg rapamycin. The fourth set was maintained as a control without rapamycin. Twenty-four h later, 50 μg coelenterazine was injected in the tail vein for follow-up imaging of the effects of the rapamycin administered the previous day. To study the consequences of the i.v. route of cell injection in two sets of three animals, 5 × 106 cells were injected via the tail vein 3 h after ex vivo cotransfection. The mice then were imaged immediately after tail vein injection of 50 μg coelenterazine. After this baseline imaging, three animals were injected in the tail vein with 50 μg rapamycin. The other three mice were maintained as controls without rapamycin. This protocol of injecting coelenterazine, imaging, and then injecting rapamycin was repeated every 24 h for 48 h. To study s.c. implantation of cells, two sets of three mice were used. For both sets, 5 × 106 cells were exposed to 20 nM of rapamycin in cell culture for 24 h before s.c. injection. All of these animals were imaged immediately after cell implantation on tail vein administration of 50 μg coelenterazine. The first set of mice were imaged every 24 h thereafter by injecting 50 μg coelenterazine, but no additional rapamycin was given. In the second set of mice, 50 μg rapamycin was injected in the tail vein after each imaging session. Twenty-four h later, 50 μg coelenterazine was injected in the tail vein for follow-up imaging of the effects of the rapamycin administered the previous day. This protocol of injecting coelenterazine, imaging, and then injecting rapamycin was repeated every 24 h for 96 h.
Fig. 3.
Flow chart showing the different experimental strategies used in this study.
Mice were anesthetized by i.p. injection of ~40 μl of a ketamine and xylazine (4:1) solution. All of the mice were imaged using a cooled CCD camera (Xenogen IVIS; Xenogen Corp., Alameda, CA). The animals were placed prone or supine in a light-tight chamber, and a gray scale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for 1 min. Images were obtained using Living Image Software (Xenogen Corp.) and Igor Image Analysis Software (Wavemetrics, Seattle, WA). To quantify the measured light, regions of interest were drawn over the area of the implanted cells, and the maximum photons/s/cm2/steradian (sr) were obtained as validated previously (22).
RESULTS
Rapamycin-Mediated Heterodimerization of FRB and FKBP12 Was Indirectly Revealed by Measuring Complemented Split hRLUC Activity
To study the small molecule rapamycin-mediated heterodimerization of the proteins FRB and FKBP12, 293T cells were cotransfected with vector constructs Nhrluc-FRB and FKBP12-Chrluc under transcriptional control of the constitutive cytomegalovirus (CMV) promoter. The heterodimerization of FRB and FKBP12 at 40-nM rapamycin concentration was measured indirectly by estimating the complemented split hRLUC activity. The results showed efficient complementation of split hRLUC fragments with a recovered activity significantly (P < 0.001) higher than in cells either transfected with the Nhrluc-FRB fragment alone or when this also was cotransfected with FKBP12-Chrluc (both without the addition of rapamycin), and also significantly (P < 0.001) higher than in cells transfected with Nhrluc-FRB that did receive rapamycin. All of the low signals measured did not differ significantly (P > 0.05) from those obtained from mock-transfected cells (Fig. 4, larger graph). The recovered hRLUC signal after the addition of 40 nM rapamycin was ~20% of that seen from cells transfected with the full hRluc gene (data not shown).
Fig. 4.
Larger graph, rapamycin-mediated complementation of synthetic renilla lucif-erase (hRLUC) activity in transiently transfected 293T cells. The 293T cells were transiently transfected with plasmid constructs Nhrluc-FRB (N) or Nhrluc-FRB plus FKBP12-Chrluc (N+C), either with rapamycin [N(+RAP) or N+C(+RAP)] or without rapamycin [N(−RAP) or N+C(−RAP)]. The cells cotransfected with Nhrluc-FRB plus FKBP12-Chrluc that received rapamycin showed significant recovered hRLUC signal. The error bars represent the SE for triplicate determinations. Inset graph, rapamycin was added to the lysates collected from the cells cotransfected with Nhrluc-FRB plus FKBP12-Chrluc to modulate extracellular dimerization of FRB and FKBP12. The cell lysates receiving rapamycin (Rap) showed significant dimerization-associated recovery of hRLUC complementation signal. The error bars represent the SE for triplicate determinations.
To study the potential for extracellular dimerization of FRB and FKBP12 by rapamycin, the lysates collected from cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc were incubated with 40 nM rapamycin at room temperature for 30 min and were assayed subsequently for hRLUC activity using luminometry. The cell lysates receiving rapamycin showed significant increase (P < 0.001) in hRLUC activity. Conversely, the cell lysates that did not receive rapamycin showed activity that was similar to that obtained from mock-transfected cells (Fig. 4, inset graph).
The Optimal Rapamycin Concentration for Heterodimerization of FRB and FKBP12 Was 10 nM and the Minimum Concentration of Ascomycin for Competitive Inhibition of Rapamycin Was 1 μM
To establish the optimal dose of rapamycin for efficient heterodimerization of FRB and FKBP12, 293T cells were cotransfected with Nhrluc-FRB and FKBP12-Chrluc, and escalating concentrations (from 0.0195 nM to 40 nM) of rapamycin were added to the cell medium. Activity of complemented hRLUC was found to increase with higher concentrations of rapamycin until 10 nM, at which point the activity plateaued (Fig. 5A). Ascomycin competes with rapamycin for binding to FKBP12. To study this competitive binding, 20 nM rapamycin were added to the cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc, and escalating concentrations (from 1 nM to 4 μM) of ascomycin were added subsequently to this mixture. The results showed that increasing the concentration of ascomycin led to a reduction in hRLUC activity, likely because it blocked the binding of rapamycin to FKBP12 and thus reduced the heterodimerization with FRB. Given the particular increments we used in this study, the smallest concentration of ascomycin required to initiate the competitive inhibition of rapamycin was found to be 1 μM (Fig. 5B).
Fig. 5.
A, graph showing the results obtained from the cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc on addition of different concentrations of rapamycin (0.0195–40 nM). The optimal concentration of rapamycin for efficient dimerization-associated recovery of complemented synthetic renilla luciferase (hRLUC) activity is >10 nM. Saturation of this activity is seen at 10 nM rapamycin. The error bars represent the SE for triplicate determinations. B, the cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc at a fixed concentration of rapamycin (20 nM) subsequently received different concentrations of ascomycin to demonstrate the competitive binding of these two compounds for FKBP12 and the associated reduction in the complemented hRLUC activity. The error bars represent the SE for triplicate determinations.
Rapamycin-Mediated Heterodimerization of FRB and FKBP12, and Split hRLUC Complementation Were Efficient in Different Cell Lines
To demonstrate a widespread applicability of FRB/FKBP12-assisted complementation of hRLUC fragments using the small molecule rapamycin, different cell lines (293T, U87, N2a, and HeLa) were cotransfected with Nhrluc-FRB and FKBP12-Chrluc and assayed after incubation with and without rapamycin for 24 h. The results showed significant (P < 0.01) increase in the hRLUC activity from cells receiving rapamycin in all of the cell lines studied. The degrees of restored hRLUC activity varied in the different cell lines studied, with the highest activity from 293T cells, followed by N2a, U87, and HeLa cells (Fig. 6). Cells cotransfected but not receiving rapamycin revealed only background activity similar to that from mock-transfected cells.
Fig. 6.

Graph showing the variable extents of synthetic renilla luciferase (hRLUC) complementation, modulated by rapamycin, obtained in the different cell lines (293T, U87, N2a, and HeLa) studied. Cells were cotransfected with Nhrluc-FRB and FKBP12- Chrluc exposed to 20-nM concentration of rapamycin and assayed after 24 h. The results show the highest signal from 293T, followed by N2a, U87, and HeLa cells. The error bars represent the SE for triplicate determinations.
Split hRLUC Protein Fragment-Assisted Complementation Was Modulated at the Transcriptional Level by Using NFκB Promoter/Enhancer Elements and at the Interaction Level by Using Rapamycin
To study the modulation of the split hRLUC complementation system at the expression level and protein-protein interaction level, 293T cells were cotransfected with Nhrluc-FRB driven by NFκB or CMV promoters and with FKBP12-Chrluc driven by the CMV promoter. Cell induction was achieved using TNF-α to activate the NFκB promoter (17), and rapamycin was added to initiate the dimerization of FRB and FKBP12 proteins. The cells cotransfected with pCMV-Nhrluc-FRB and pCMV-FKBP12-Chrluc showed significant (P < 0.01) restored hRLUC activity on receiving rapamycin alone and a 30–40% higher activity on receiving rapamycin and TNF-α. We reasonably speculate that this increased hRLUC activity with the addition of TNF-α was the result of an early up-regulated stimulatory activity of the CMV promoter, which has been reported previously to last for up to 48 h (31). The cells cotransfected with pNFκB-Nhrluc-FRB and pCMV-FKBP12-Chrluc showed a restored activity that was significantly (P < 0.001) higher when receiving TNF-α and rapamycin than the cells receiving only rapamycin or those receiving neither rapamycin nor TNF-α (Fig. 7).
Fig. 7.
Graph showing the results obtained from 293T cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc and dually modulated at the expression level [by controlling synthetic renilla luciferase (hRLUC) transcription with varying concentrations of tumor necrosis factor α (TNF-α)] and at the dimerization level (by controlling the rapamycin concentration). The cells cotransfected with constructs driven by the CMV promoter and receiving rapamycin [(Rap+/TNF-α−) and (Rap+/TNF-α+)] showed significant increase in the complemented hRLUC signal. The cells cotransfected with constructs driven by the nuclear factor κB promoter showed significant signal only when receiving TNF-α and rapamycin (Rap+/TNF-α+). The error bars represent the SE for triplicate determinations.
Split hRLUC Activity Recovered through Protein-Protein Interaction-Mediated Complementation Was Imaged in Living Mice
That rapamycin-mediated heterodimerization of FRB and FKBP12 was achieved successfully was indicated indirectly by measuring complemented hRLUC activity in mice implanted with the 293T cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc. Five million of these 293T cells were implanted i.p. or s.c. or injected i.v. 3 h after cotransfection in different groups of animals. The animals were imaged using a CCD camera immediately after implanting the cells by injecting 50 μg of coelenterazine via tail vein. The animals receiving i.p. implants also were imaged 24 h after injecting the tail vein with different concentrations of rapamycin (10 μg, 25 μg, and 50 μg) to show the optimal concentration required for protein dimerization. The group of animals receiving 10 μg rapamycin emitted a signal (2.38 × 103 p/s/cm2/sr) that was not significantly different from animals with mock-transfected cells. The group of animals receiving 25 μg and 50 μg rapamycin emitted a signal of 6.0 × 103 p/s/cm2/sr and 1.2 × 104 p/s/cm2/sr, respectively (Fig. 8).
Fig. 8.
Optical charged coupled device imaging of living mice implanted i.p. with transiently cotransfected 293T cells to study the concentrations of rapamycin required for efficient heterodimerization of FRB and FKBP12 by using the synthetic renilla luciferase protein fragment-assisted complementation strategy. At 24 h after cell implantation, the group of animals receiving 10 μg rapamycin emitted a signal (2.38 × 103 p/s/cm2/sr) that was similar to mock transfection levels. The group of animals that received 25 μg and 50 μg rapamycin emitted signals of 6.0 × 103 p/s/cm2/sr and 1.2 × 104 p/s/cm2/sr, respectively. See inset graphical display of these signals; the error bars are the SE for three mice.
The effect of rapamycin on s.c. implants was studied using 5 × 106 293T cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc. The animals were imaged immediately after grafting the cells and 24 h, 48 h, and 72 h after injecting the tail vein with repeated doses (50 μg) of rapamycin. The signals emitted from the animals receiving rapamycin at these different intervals were not significantly different from those seen in mock-transfected cells (data not shown).
The effect of rapamycin on i.v. injected 5 × 106 293T cells cotransfected with Nhrluc-FRB and FKBP12-Chrluc was assessed in mice by imaging immediately and after 24 h and 48 h. The animals receiving i.v. cells, but no rapamycin, showed only a background signal of 4 ± 1 × 103 p/s/cm2/sr at all of the time points studied. The animals receiving repeated injections of rapamycin emitted signals that were threefold (1.6 × 104 p/s/cm2/sr) and fivefold (3.0 × 104 p/s/cm2/sr) higher than background (P < 0.05) at 24 h and 48 h after injection of rapamycin, respectively (Fig. 9). The pattern observed for liver localization at and beyond 24 h of i.v. injection is not uncommon for gene-marked 293T cells.
Fig. 9.
Optical charged coupled device imaging of living mice carrying i.v. injected 293T cells transiently cotransfected with Nhrluc-FRB and FKBP12-Chrluc. The animals not receiving rapamycin showed only a mean background signal of 4 ± 1 × 103 p/s/cm2/sr at all of the time points studied. The animals receiving repeated injections of rapamycin emitted signals, originating from the region of the liver, that were threefold (mean, 1.6 × 104 p/s/cm2/sr) and fivefold (mean, 3.0 × 104 p/s/cm2/sr) higher than background (P < 0.05) at 24 h and 48 h after the injection of rapamycin, respectively. (R−, animals not receiving rapamycin; R+, animals receiving rapamycin).
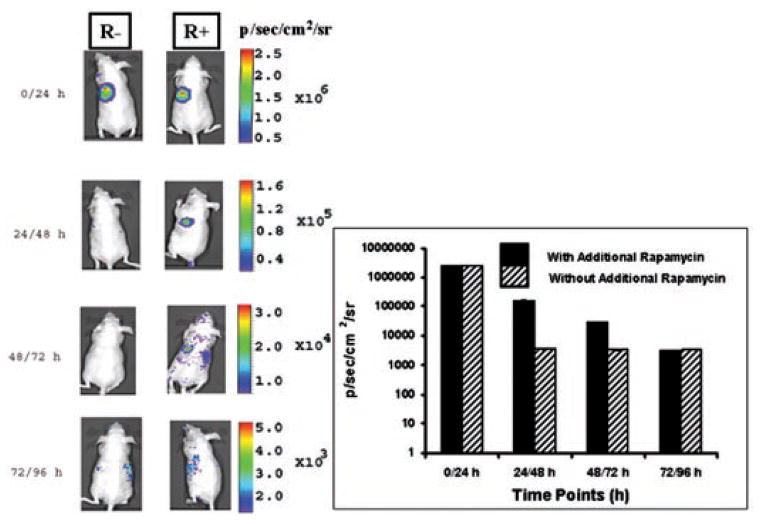
We performed an additional study by pre-exposing the 293T cells to a 20-nM concentration of rapamycin in cell culture after cotransfection with Nhrluc-FRB and FKBP12-Chrluc. The animals were s.c. implanted with 5 × 106 of these cells and imaged immediately by injecting 50 μg coelenterazine via the tail vein. One group was maintained as a control by not injecting additional doses of rapamycin, and another group was injected with 50 μg rapamycin every 24 h and imaged immediately on injection. At time point zero, control and experimental groups emitted equal signals (2.5 ± 0.5 × 106 p/s/cm2/sr). After 24 h, the control group showed a signal that was equal to background, and the group of animals receiving an additional dose of rapamycin (50 μg) emitted a signal (1.6 ± 0.3 × 105 p/s/cm2/sr) that was significantly (P < 0.001) higher than that obtained from the control group. The experimental group continued to emit significantly higher signals even 72 h after cell implantation (Fig. 10). The animals that received rapamycin every 24 h showed a signal reduction during the subsequent course of the experiment possibly because of the following factors: (a) a reduction in transcription by transiently transfected cells; (b) the degree of clearance of rapamycin from the cells (owing to the ex vivo pre-exposure to this compound) outweighing the amount of rapamycin reaching the cells on reinjection in vivo; (c) the injected dose of rapamycin may have been too low; and (d) poor neovascularization at the sites of cell implants over the short time course of these experiments.
Fig. 10.
Optical charged coupled device imaging of living mice carrying s.c. injected 293T cells transiently cotransfected with Nhrluc-FRB and FKBP12-Chrluc. The cells were pre-exposed in cell culture to 20 nM rapamycin for 24 h after cotransfection, and 5 × 106 of these cells then were implanted s.c. The groups of animals were imaged immediately (time 0) and 24 h, 48 h, and 96 h after injecting repeated doses of 50 μg rapamycin. The results showed significant increase in complemented synthetic renilla luciferase signal only from the group receiving repeated doses of rapamycin (R+, animals receiving rapamycin; R−, animals not receiving rapamycin). See inset graphical display of these signals; the error bars are the SE for three mice.
DISCUSSION
In this study we used a regulated heterodimerization kit containing the proteins FKBP12- rapamycin-associated protein (also known as mTOR or RAFT) and FRB (both obtained from Ariad Pharmaceuticals), along with split hRLUC fragments developed and validated previously in our laboratory, to investigate the regulation of protein-protein interactions by a small molecule in cells and to noninvasively image this modulation in living mice. Although many techniques are available to study protein-protein interactions in cell lines, few (e.g., the inducible yeast two hybrid system and split firefly luciferase complementation system; Refs. 17, 18) can be extrapolated successfully for use in living animals. The inducible yeast two hybrid system has limited applications, being used specifically with nuclear proteins or proteins made to mimic nuclear localization signals. Moreover, in our hands, the split firefly luciferase complementation system has to date shown a low efficiency with rapamycin-mediated heterodimerization of the proteins FRB and FKBP12.6 In this study, we succeeded in applying a previously developed, more sensitive split hRLUC protein fragment-assisted complementation system to study small molecule rapamycin-mediated heterodimerization of the proteins FRB and FKBP12 in different cell lines and to noninvasively image these in living mice (19). We also showed that this system could be titrated by changing the concentration of interacting molecules in cells and in living mice, which is an essential principle in the preclinical drug screening and validation process. Small molecule rapamycin-mediated protein-protein interactions have been studied previously in cell culture by using β-galactosidase complementation (12). In this study, for the first time, we detected and imaged rapamycin-mediated protein-protein interactions in cell culture and in living animals. The development of this technique opens up the possibility of screening of agonists or antagonists of protein-protein interactions in cell culture and in living small animal models of cancer. This will be particularly useful because the efficacy of a given small molecule can be assessed directly in the living subject with all of the pharmacokinetic issues that apply to the compound considered fully.
The evaluation of small molecule-mediated protein-protein interactions can be relatively efficient and unproblematic in cell culture, even when exposing the cells to low concentrations of the small molecule in question. However, similar investigations in a living animal system would critically depend on many factors, including the availability of an efficient and sensitive reporter system and a highly sensitive imaging modality to help achieve a detectable signal (32). Moreover, the compound under investigation would need to circulate in the vascular compartment with enough concentration and for a suitable period to permit sufficient quantity to reach the target site, and this, in turn, would allow sufficient chemical interaction to occur at the target. In this study, we used transiently transfected 293T cells for in vivo studies. We achieved significant enhancement of the imaging signal from animals receiving rapamycin when cells were implanted i.p. or injected i.v. No significant signal gain was detectable in animals with s.c. cell implants, likely because of the relatively poor vascularization in this body compartment. We reasonably speculate that the establishment of longer-term s.c. xenografts using stably transfected engrafted cells would likely result in a greater blood supply to these engrafted cells and a consequent improvement in imaging signal. The potential effects, deleterious or otherwise, of higher doses of rapamycin on experimental animals currently are unknown.
To date, each of the several techniques developed to study protein-protein interactions has advantages and limitations (32). The formation of homotetramers by intracistronic complementation of mutants has been used previously to identify some reporter proteins for studying protein interactions (15). Larger reporter proteins may be hindered sterically during the complementation process (33). The selection of irreversible mutants with no self-complementation properties is important to develop an intracistronic complementation system. Conversely, a protein fragment-assisted complementation assay uses rationally split fragments of a reporter protein that do not exhibit self-complementation properties. The enzyme hRLUC, an Mr 36,000 monomeric bioluminescence imaging reporter protein, is the smallest protein identified to date for studying protein-protein interactions through a protein fragment-assisted complementation strategy. This reporter protein, when rationally split at particular sites, functions efficiently in cell culture and in living animals, as demonstrated with several different protein partners studied to date (19). One limitation associated with the use of hRLUC is its relatively rapid reaction kinetics, requiring early time point measurements (22). Nevertheless, this split reporter system appears highly suitable to study protein-protein interactions in cells and in living animals because of its optical bioluminescence nature and its signal, which is amplifiable through an enzymatic process.
In this study, we assessed a known rapamycin-mediated protein-protein interaction system in cancer cells and in living animals. The split imaging reporter complementation-based system used in conjunction can be extended to study other protein-protein interactions with known or unknown protein partners. Therefore, the screening of new dimerizer drugs for many protein interactions now should be possible. The development of advanced systems to quantify optical reporter signals from cell culture and living subjects may lead to many new applications in the study of fundamental biological processes in cancer cells and the development of drugs to modulate them.
Acknowledgments
Grant support: NIH grants P50 CA86306 (S. S. Gambhir), R01 CA82214 (S. S. Gambhir), SAIRP R24 CA92865 (S. S. Gambhir), Department of Energy Contract DE-FC03-87ER60615 (S. S. Gambhir), CaP Cure (S. S. Gambhir), and a Mid-Career Award for Established Practitioners from The Health Foundation, United Kingdom (T. F. Massoud).
We thank Frank Berger for his valuable assistance in handling the animals, and Berj Demirjian for helping to start this research when a rotation student in the laboratory.
Footnotes
Unpublished observations.
References
- 1.Michnick SW. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr Opin Struct Biol. 2001;11:472–7. doi: 10.1016/s0959-440x(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 2.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 3.Williams NE. Immunoprecipitation procedures. Methods Cell Biol. 2000;62:449–53. [PubMed] [Google Scholar]
- 4.Bollag DM. Gel-filtration chromatography. Methods Mol Biol. 1994;36:1–9. doi: 10.1385/0-89603-274-4:1. [DOI] [PubMed] [Google Scholar]
- 5.Hansen JC, Lebowitz J, Demeler B. Analytical ultracentrifugation of complex macromolecular systems. Biochemistry. 1994;33:13155– 63. doi: 10.1021/bi00249a001. [DOI] [PubMed] [Google Scholar]
- 6.Doyle ML. Characterization of binding interactions by isothermal titration calorimetry. Curr Opin Biotechnol. 1997;8:31–5. doi: 10.1016/s0958-1669(97)80154-1. [DOI] [PubMed] [Google Scholar]
- 7.Lakey JH, Raggett EM. Measuring protein-protein interactions. Curr Opin Struct Biol. 1998;8:119–23. doi: 10.1016/s0959-440x(98)80019-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–4. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronheim A. Improved efficiency sos recruitment system: expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res. 1997;25:3373–4. doi: 10.1093/nar/25.16.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broder YC, Katz S, Aronheim A. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr Biol. 1998;8:1121– 4. doi: 10.1016/s0960-9822(98)70467-1. [DOI] [PubMed] [Google Scholar]
- 12.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by β-galactosidase complementation. Proc Natl Acad Sci USA. 1997;94:8405–10. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhard KN, Jacoby JJ, Fu XY, Jahn R, Dohlman HG. Use of G-protein fusions to monitor integral membrane protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1075–9. doi: 10.1038/80274. [DOI] [PubMed] [Google Scholar]
- 14.Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem. 2001;73:2516–21. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- 15.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of β-lactamase enzyme fragments. Proc Natl Acad Sci USA. 2002;99:3469–74. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelletier JN, Campbell-Valois FX, Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci USA. 1998;95:12141–6. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray P, Pimenta H, Paulmurugan R, et al. Noninvasive quantitative imaging of protein-protein interactions in living subjects. Proc Natl Acad Sci USA. 2002;99:3105–10. doi: 10.1073/pnas.052710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99:15608–13. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–9. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luker GD, Sharma V, Pica CM, et al. Noninvasive imaging of protein-protein interactions in living animals. Proc Natl Acad Sci USA. 2002;99:6961–6. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luker GD, Sharma V, Pica CM, Prior JL, Li W, Piwnica-Worms D. Molecular imaging of protein-protein interactions: controlled expression of p53 and large T-antigen fusion proteins in vivo. Cancer Res. 2003;63:1780–8. [PubMed] [Google Scholar]
- 22.Bhaumik S, Gambhir SS. Optical imaging of renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veselovsky AV, Ivanov YD, Ivanov AS, Archakov AI, Lewi P, Janssen P. Protein-protein interactions: mechanisms and modification by drugs. J Mol Recognit. 2002;15:405–22. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]
- 24.Grunwald V, Hidalgo M. Developing inhibitors of the epidermal growth factor receptor for cancer treatment. J Natl Cancer Inst. 2003;95:851– 67. doi: 10.1093/jnci/95.12.851. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro P. Ras-MAP kinase signaling pathways and control of cell proliferation: relevance to cancer therapy. Crit Rev Clin Lab Sci. 2002;39:285–330. doi: 10.1080/10408360290795538. [DOI] [PubMed] [Google Scholar]
- 26.Crabtree GR, Schreiber SL. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem Sci. 1996;21:418–22. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 27.Blau CA, Peterson KR, Drachman JG, Spencer DM. A proliferation switch for genetically modified cells. Proc Natl Acad Sci USA. 1997;94:3076–81. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amara JF, Clackson T, Rivera VM, et al. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci USA. 1997;94:10618–23. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci USA. 1996;93:4604–7. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacCorkle RA, Freeman KW, Spencer DM. Synthetic activation of caspases: artificial death switches. Proc Natl Acad Sci USA. 1998;95:3655– 60. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritter T, Brandt C, Prosch S, et al. Stimulatory and inhibitory action of cytokines on the regulation of hCMV-IE promoter activity in human endothelial cells. Cytokine. 2000;12:1163–70. doi: 10.1006/cyto.2000.0689. [DOI] [PubMed] [Google Scholar]
- 32.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545– 80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 33.Rossi FM, Blakely BT, Blau HM. Interaction blues: protein interactions monitored in live mammalian cells by β-galactosidase complementation. Trends Cell Biol. 2000;10:119–22. doi: 10.1016/s0962-8924(99)01707-9. [DOI] [PubMed] [Google Scholar]