Abstract
Homodimeric protein interactions are potent regulators of cellular functions, but are particularly challenging to study in vivo. We used a split synthetic renilla luciferase (hRLUC) complementation-based bioluminescence assay to study homodimerization of herpes simplex virus type 1 thymidine kinase (TK) in mammalian cells and in living mice. We quantified and imaged homodimerization of TK chimeras containing N-terminal (N-hRLUC) or C-terminal (C-hRLUC) fragments of hRLUC in the upstream and downstream positions, respectively (tail-to-head homodimer). This was monitored using luminometry (68-fold increase, and was significantly [P<0.01] above background light emission) and by CCD camera imaging of living mice implanted with ex vivo transfected 293T cells (2.7-fold increase, and is significantly [P<0.01] above background light emission). We also made a mutant-TK to generate N-hRLUC mutant TK and mutant TK-C-hRLUC by changing a single amino acid at position 318 from arginine to cysteine, a key site that has previously been reported to be essential for TK homo-dimerization, to support the specificity of the hRLUC complementation signal from TK homodimerization. Ex vivo substrate (8-3H Penciclovir) accumulation assays in 293T cells expressing the TK protein chimeras showed active TK enzyme. We also devised an experimental strategy by constructing variant TK chimeras (possessing extra N-hRLUC or C-hRLUC ‘spacers’) to monitor incremental lack of association of the tail-to-head TK homodimer. Application of this potentially generalizable assay to screen for molecules that promote or disrupt ubiquitous homodimeric protein–protein interactions could serve not only as an invaluable tool to understand biological networks but could also be applied to drug discovery and validation in living subjects.
Keywords: Bioluminescence, luciferase, Renilla, complementation, dimerization, thymidine kinase
Protein–protein interactions are of critical importance to most cellular functions, being integral to processes as diverse as enzymatic activity, signal transduction, immunological recognition, and DNA replication and repair (1, 2). Toward the common goal of understanding these ubiquitous interactions, several experimental techniques have been developed, including qualitative methods such as the yeast two-hybrid screen (3), immunoprecipitation (4), and gel-filtration chromatography (5), or quantitative biophysical methods such as analytical ultracentrifugation (6), calorimetry (7), and optical spectroscopy (8). Fluorescence resonance energy transfer (FRET) microscopy techniques (9, 10), including FRET anisotropy (11, 12), currently offer accurate cell culture methods to monitor dynamic real-time protein–protein interactions in living cells. However, several drawbacks of these techniques come to light when studying the oligomerization of identical protein subunits and none of these techniques can be optimally extended to imaging in living subjects. Similarly, β-galactosidase complementation has been used to monitor homodimerization of the epidermal growth factor receptor (EGFR) (13), and bioluminescence resonance energy transfer (BRET) has been used to monitor homodimerization of the β2-adrenergic (14), somatostatin (15), and δ-opioid (16) receptors, but only in cell culture.
The extreme abundance of localized temporary, or more stable protein homodimers (2895 entries in the Protein Data Bank were classified as homodimers by the year 2000 (17)) attests to their many functions in the cell and the important role they play in many biological processes. Here, we demonstrate a new generalizable application of an in vivo protein fragment-assisted complementation assay, based on a split bioluminescence reporter (synthetic humanized renilla luciferase [hRLUC]) complementation technology (18), for imaging of homodimeric protein–protein interactions in intact living subjects. The ability to detect, locate, and quantify protein homodimerization in the setting of a whole living animal model has important implications for a wide variety of biological research endeavors, drug discovery, and molecular medicine. In particular, the visual representation, characterization, and quantification of these biological processes in living subjects now creates unprecedented opportunities to complement available in vitro or cell culture methodologies, in order: 1) to characterize more fully known homodimeric protein–protein interactions (e.g., the reasons for, and the factors that drive their association) in the context of whole-body physiologically authentic environments, and 2) to accelerate the evaluation in living animal models of novel drugs that promote or inhibit active homodimeric protein assembly. The many noteworthy advantages of molecular imaging of biological processes in living subjects are reviewed by Massoud and Gambhir (19).
We previously identified suitable split sites in the molecule of hRLUC that generated inactive N-terminal 229-residue fragment (N-hRLUC) and C-terminal 82-residue fragment (C-hRLUC) of the reporter protein that, together, were able to produce significant recovered activity through assisted complementation (18). Here, we use the split hRLUC bioluminescence reporter system to record subunit homodimerization of a test protein, the mutant (sr39) herpes simplex virus type 1 thymidine kinase (HSV1-TK) (Fig. 1), in mammalian cells and to noninvasively image homodimerization in mouse tumor models. We also studied the inhibition of homodimerization using a TK mutant that has previously been reported not to dimerize (20).
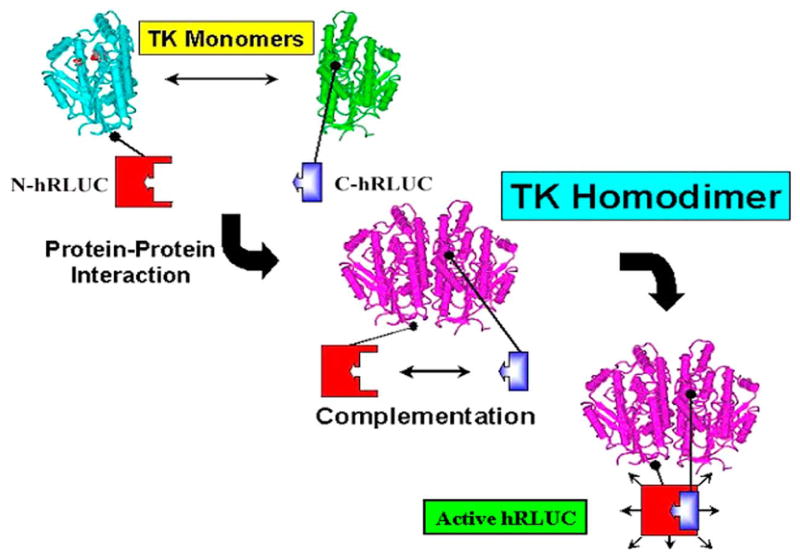
Figure 1. Schematic diagram showing the protein–fragment-assisted complementation strategy using split synthetic renilla luciferase (here abbreviated as hRLUC) to monitor the thymidine kinase (TK) homodimeric protein–protein interaction.
The N-terminal portion of hRLUC is attached to one TK monomer through a peptide linker, and the C-terminal portion of hRLUC is similarly attached to another TK monomer. Dimerization of the two TK monomers restores hRLUC activity through protein complementation and produces light in the presence of the hRLUC substrate coelenterazine.
METHODS
Chemicals, enzymes, and reagents
Restriction and modification enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). PCR amplification was performed with TripleMaster Taq DNA polymerase purchased from Brinkmann Eppendorf (Hamburg, Germany). The plasmid pCMV-hRL was purchased from Promega (Madison, WI), and pCMV-HSV1-sr39TK was a gift from Dr. Margaret Black (Washington State University, Pullman, WA). Superfect transfection reagents, plasmid extraction kits, and DNA gel extraction kits were purchased from Qiagen (Valencia, CA). Coelenterazine was purchased from Biotium (Hayward, CA). Bacterial culture media were purchased from BD Diagnostic Systems (Sparks, MD). All animal cell culture media, fetal bovine serum, the antibiotics streptomycin and penicillin, and plastic wares for growing cell cultures were purchased from Invitrogen (Carlsbad, CA). Rapamycin was purchased from Sigma (St. Louis, MO), and [8-3H]Penciclovir was obtained from Moravek Biochemicals (Brea, CA).
Construction of plasmids
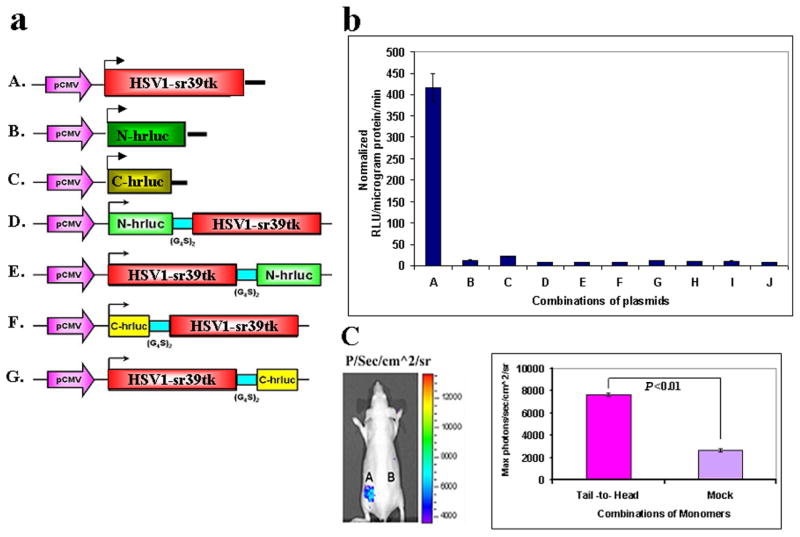
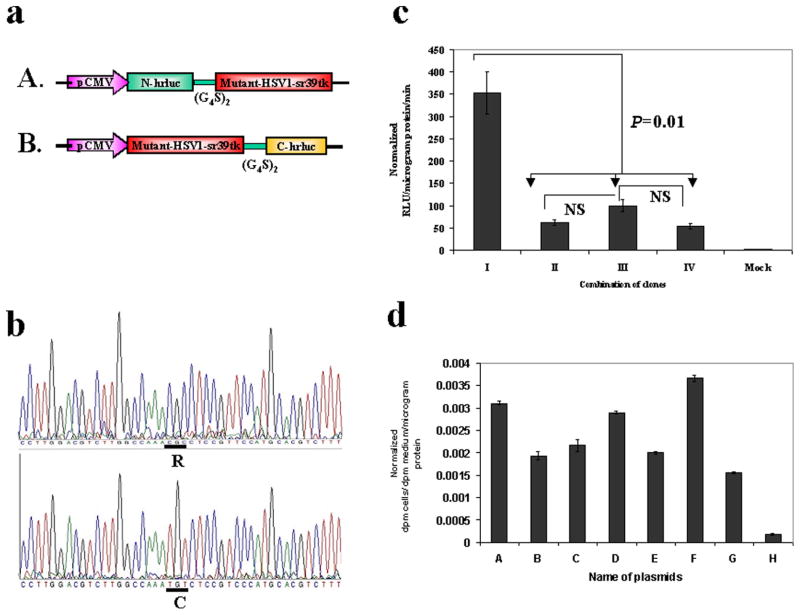
To construct the N-terminal fragment (1-687 bp) and the C-terminal fragment (688-933 bp) of the hrluc gene, either upstream or downstream of HSV1-sr39tk (tk), N-hrluc, or C-hrluc were amplified using the primers described previously (18), and pCMV-hRL as template, and tk was amplified using its 5′ end forward and its 3′ end reverse primers and pCMV-HSV1-sr39tk as template. For convenient cloning, the forward primer for the upstream gene was introduced with an NheI restriction enzyme site and a start codon, both satisfying partial Kozak consensus sequence requirements for expression enhancement. The reverse primer of the upstream gene and the forward primer of the downstream gene were introduced with a BamHI restriction enzyme site. The reverse primer of the downstream gene was introduced with an XhoI restriction enzyme site and a stop codon. The cDNA encoding the flexible linker (GGGGS)2 was placed between the upstream and downstream genes in each cassette. The digested fragments were cloned into a pcDNA3.1 (+) vector backbone at NheI/XhoI restriction enzyme sites. The N-hrluc and C-hrluc with start and stop codons cloned in the same vector backbone was used as controls (Fig. 2a). Similarly, an N-hrluc and C-hrluc fragment on both NH2 and COOH of the TK protein was also constructed (Fig. 3a). To generate N-hrluc and C-hrluc fragments with a mutant HSV1-sr39tk (tk), the tk gene was mutated at amino acid position 318 by using a site-directed mutagenesis kit purchased from Stratagene (La Jolla, CA) using the forward primer 5′cgtcttggccaaatgtctccgtcccatgc and the reverse primer 5′gcatgggacggagacatttggccaagacg synthesized at the Stanford Protein and Nucleic Acid Facility. Isolated plasmids were analyzed for the presence of inserts, and positive clones were confirmed for insert presence based on fragment size using restriction endonuclease analysis. The mutants were verified by sequencing and used for further studies (Fig. 5b, c).
Figure 2.
a) Schematic representation of the plasmid vector constructs made for transient expression of each of the four TK plus split hRLUC protein individually and as chimeras in a pcDNA3.1 (+) plasmid backbone, under control of a CMV promoter. The constructed four imaging cassettes contained N-hrluc (gene encoding the N-terminal of hRLUC protein) or C-hrluc (gene encoding the C-terminal of hRLUC protein) fused upstream or downstream of the TK gene, respectively. cDNA encoding intervening linker peptides of 10 residues each (GGGGS)2 were included to prevent conformational strain between each TK molecule and the fused split hRLUC fragment. A. HSV1-sr39tk (express full TK protein). B. N-hrluc with start and stop codons (express only the N-portion of the hRLUC protein). C. C-hrluc with start and stop codons (express only the C-portion of the hRLUC protein). D. N-hrluc is upstream of tk (expresses N-hRLUC in the tail position with respect to TK). E. N-hrluc is downstream of tk (expresses N-hRLUC in the head position). F. C-hrluc is upstream of tk (expresses C-hRLUC in the tail position). G. C-hrluc is downstream of tk (expresses C-hRLUC in the head position). b) Homodimerization-mediated, fragment-assisted complementation of the split hRLUC system with different negative controls in transiently transfected 293T cells. Graph shows luminometry results on cell lysates 24 h after cotransfection of constructs D plus G [(A) (Tail-to-Head)], E plus F [(B) (Head-to-Tail)], D plus F [(C) (Tail-to-Tail)], E plus G [(D) (Head-to-Head)], and controls include the vector combinations D plus C (E), B plus F (F), C plus E (G) and, B plus G (H) containing one vector with tk molecule with a portion of hrluc fragment and another portion of hrluc molecule without any protein to prove the recovered hRLUC signal was through the complementation of TK homodimerization. The combination of vectors, B plus C (I) contains hrluc fragments expressed without any protein. All combinations except D plus G (tail to head) showed the hRLUC signals that were not significantly different from mock transfected cells (J). The signals from the head-to-tail, tail-to-tail, and head-to-head combinations were only minimally above those of mock-transfected cells (not shown, <15 RLU/microgram protein/min). The error bar is the standard error of the mean for three samples. All results are normalized for transfection efficiency. c) Optical CCD imaging of living mice engrafted with transiently transfected 293T cells for in vivo assessment of TK homodimerization-mediated complementation of split hRLUC. The image of one representative mouse is shown as a visible light image superimposed on the CCD bioluminescence image with a scale in photons/s/cm2/steradian. Mice were imaged in the prone position after tail-vein injection of coelenterazine. Each mouse was imaged several minutes after subcutaneous implantation of 10×106 293T cells transiently cotransfected 24 h previously with the tail-to-head combination of monomers (site A) or with mock transfected cells (site B). There was significantly higher (P<0.01) recovered activity of hRLUC from site A as compared with site B, as shown in the accompanying graph. The error bar is the standard error of the mean for three samples.
Figure 3.
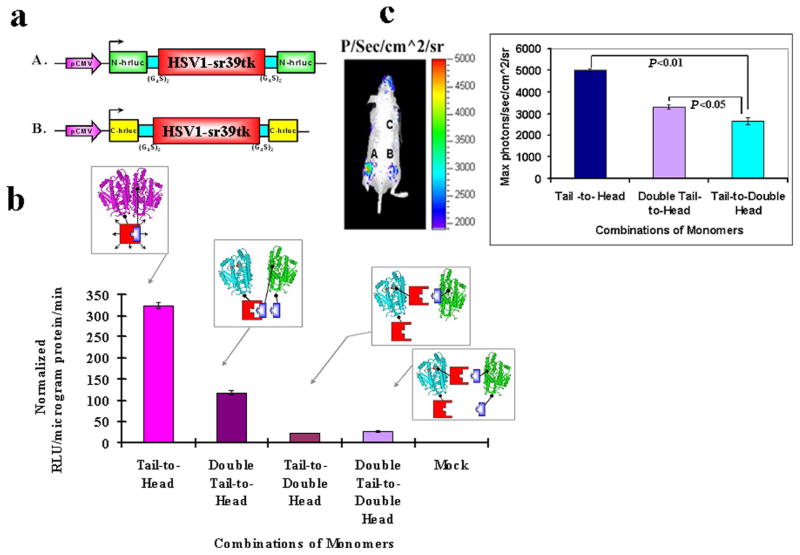
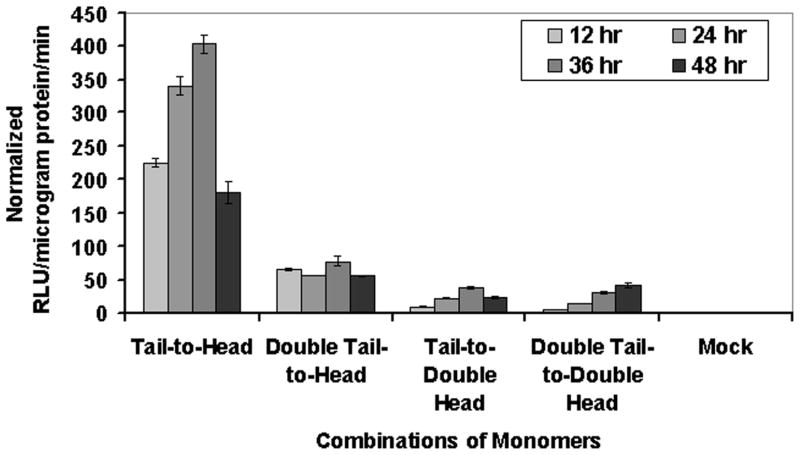
a) Schematic representation of the two additional vector constructs made for transient expression of TK plus split hRLUC protein chimeras in a pcDNA3.1 (+) plasmid backbone, under control of a CMV promoter. The constructed two imaging cassettes contained N-hrluc or C-hrluc fused upstream and downstream of the tk gene, respectively. cDNA encoding intervening linker peptides of 10 residues each (GGGGS)2 were included on either side of TK protein to prevent conformational strain between each TK molecule and each fused split hRLUC protein fragments. A. N-hrluc is upstream and downstream of tk (expresses N-hRLUC in the tail and head positions with respect to TK). B. C-hrluc is upstream and downstream of tk (expresses C-hRLUC in the tail and head positions with respect to TK). b) Inhibition of homodimerization-mediated, fragment-assisted complementation of the split hRLUC system in transiently transfected 293T cells. Graph shows luminometry results on cell lysates 24 h after cotransfection of constructs A plus D (both from Fig. 2a, Tail-to-Head combination), A from Fig. 2a plus B from Fig. 3a (double-tail-to-head combination), A from Fig. 3a plus D from Fig. 2a (Tail-to-Double Head combination), and A plus B (both from Fig. 3a, Double Tail-to-Double Head combination). The recovered activity of hRLUC after successful homodimerization of the tail-to-head TK monomer combination (shown in lilac) was mostly eliminated by the presence of extra N-hRLUC ‘spacers’ in the downstream position of the tail-to-double-head and double-tail-to-double-head monomer combinations. The double-tail-to-head combination resulted in an intermediate inhibition of TK homodimerization and a corresponding decrease in the light signal. Positions of the ‘spacers’ are shown in the accompanying schematic diagram (N-hRLUC in red, and C-hRLUC in blue). The signal from the Double tail-to-double-head was ≈50% of the signal obtained from Double tail-to-head and, Tail-to-double-head combinations was around 50% of Double tail-to-double head and that in turn only minimally above those of mock-transfected cells (not shown, <10 RLU/microgram protein/min). The error bar is the standard error of the mean for three samples. Results are normalized for transfection efficiency. c) Optical CCD imaging of living mice engrafted with transiently transfected 293T cells for in vivo assessment of inhibition of TK homodimerization mediated complementation of split hRLUC. The image of one representative mouse is shown as a visible light image superimposed on the CCD bioluminescence image with a scale in photons/s/cm2/steradian. Mice were imaged in the prone position after tail-vein injection of coelenterazine. Each mouse was imaged several minutes after subcutaneous implantation of 10×106 293T cells transiently cotransfected 24 h previously with the tail-to-head combination of monomers (site A), the double-tail-to-head combination of monomers (site B), and the tail-to-double-head combination of monomers (site C, shoulder region). There was significant decrease (P<0.05) in recovered activity of hRLUC from sites B and C as compared with site A, as shown in the accompanying graph. The error bar is the standard error of the mean for three samples.
Figure 5.
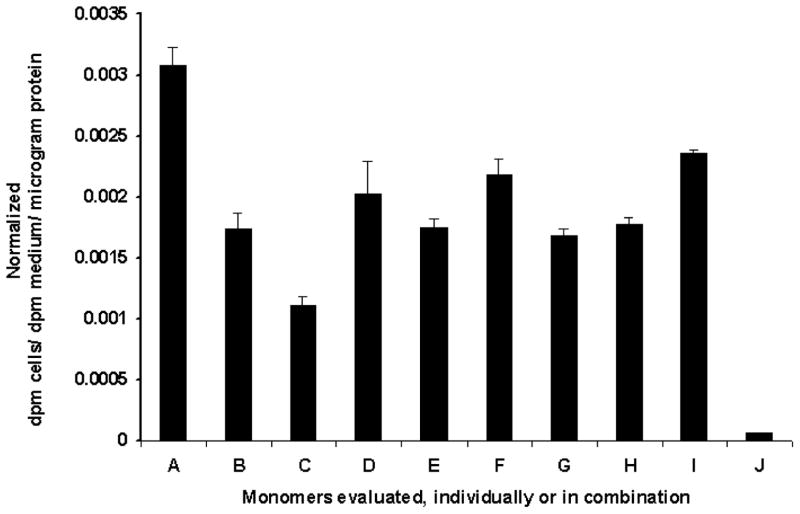
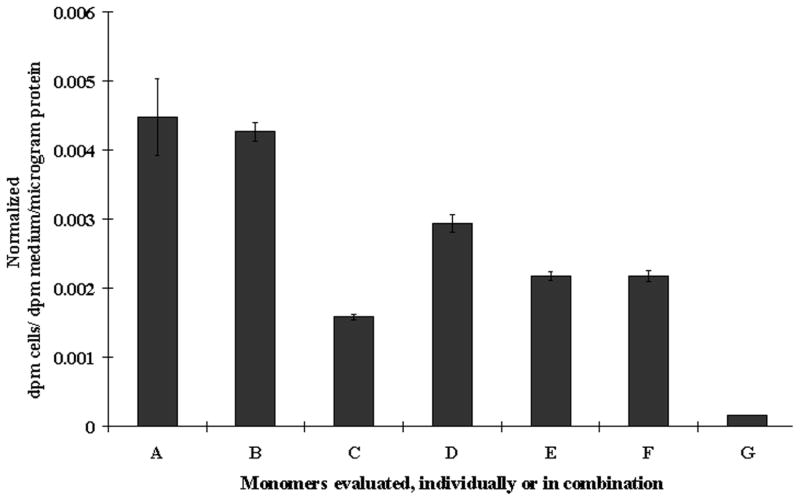
a) Schematic representation of the plasmid vector constructs made by mutant TK molecule with split hRLUC protein chimeras in a pcDNA3.1 (+) plasmid backbone, under control of a CMV promoter. The HSV1-sr39tk containing N-hrluc in the upstream and C-hrluc in the down stream position of vector constructs from Figure 2a (D and G) were mutated at amino acid position 318 by changing arginine to cysteine. b) Nucleic acid sequencing chromatogram indicating a portion with mutated nucleotide at amino acid position 318 (underlined/upper: normal tk; lower: mutated tk). c) Homodimerization-mediated fragment-assisted complementation of the split hRLUC system in transiently transfected 293T cells by using split hrluc with mutant HSV1-sr39tk. Graph shows luminometry results on cell lysates 24 h after cotransfection of different combination of vector constructs from Fig. 2a and Fig. 5a (I. constructs D plus G from Fig. 2a, II. constructs A of Fig. 5a plus G of Fig. 2a, III. constructs D of Fig. 2a plus B of Fig.5a, IV. constructs A plus B of Fig. 5a, and V. mock transfection). The result shows significant complemented hRLUC activity from the cells cotransfected with vector constructs containing tk molecule with upstream N-hrluc and another tk molecule with downstream C-hrluc. The cells cotransfected with other vector combinations containing upstream and downstream hrluc fragments with one or both mutated tk molecules (II, III and IV) shows signal that is 3- to 4-fold less than the tail-to-head combination (D plus G of Fig. 2a). The error bars represent the standard error for triplicate determinations. Results are normalized for transfection efficiency. d) [8-3H]Penciclovir accumulation in 293T cells 24 h after transient transfection with different chimeras individually (B: N-hrluc-tk, C: tk-C-hrluc, D: tk-N-hrluc, E: C-hrluc-tk, F: mutant-N-hrluc-tk, G: mutant-tk-C-hrluc). Net accumulation of probe in cells transfected with the intact nontruncated sr39TK (A) acted as control. Results are for net accumulation (in dpm cells/dpm medium/microgram of protein) of the probe over a 4-h incubation period. Accumulation was significantly above (P<0.01) mock levels (H) in all cases. The error bars represent the standard error for triplicate determinations. Results are normalized for transfection efficiency.
Cell culture
Human embryonic kidney cancer 293T cells (ATCC, Manassas, VA) were grown in MEM medium supplemented with 10% FBS and 1% penicillin/streptomycin solution.
Cell transfection and in vitro renilla and firefly luciferase assay
Transfections were performed in 80% confluent 24 h old cultures of 293T cells. For transfection in 12-well culture plates, 250 ng/well of DNA were used for each monomeric construct used, giving a total of 500 ng/well when monomers were combined. For transfection with pCMV-hrluc alone, 500 ng/well were used. First, cotransfection was performed with constructs A plus D (tail-to-head combination), B plus C (head-to-tail combination), A plus C (tail-to-tail combination), and B plus D (head-to-head combination) (all as shown in Fig. 2a). Subsequently, for assessment of homodimerization inhibition, cotransfection was performed with constructs A plus D (both from Fig. 2a, tail-to-head combination), A from Fig. 2a plus B from Fig. 3a (double tail-to-head combination), A from Fig. 3a plus D from Fig. 2a (tail-to-double-head combination), and A plus B (both from Fig. 3a, double-tail-to-double-head combination). TK with a portion of the hrluc fragment cotransfected with another portion of the reporter without any protein was also studied (N-hrluc-HSV1-tk with C-hrluc and HSV1-tk-C-hrluc with N-hrluc). Similarly, mutant HSV1-tk with N and C portions of hrluc fragments with complement HSV1-tk with reporter portions were also studied. The HSV1-tk, mutant-HSV1-tk, with and without hrluc fragments were transfected individually and assayed for TK activity by [8-3H] Penciclovir uptake study as mentioned below. Volumes of Superfect used were as recommended by the manufacturer. The cells were assayed after 24 h incubation at 37°C and in 5% CO2. The luminometry assay for renilla luciferase activity was performed as per protocol published previously (21). In brief, the cells were lysed in 200 μl of 1X passive lysis buffer supplied by Promega and by shaking for 15 min at room temperature. The cell lysates were centrifuged for 5 min at 10,000 rpm at 4°C. Twenty μl of supernatant were assayed by adding 1 μl of the substrate coelenterazine (1mg/mL) and 100 μl of 0.05M sodium phosphate buffer at pH 7.0, followed by photon counting in the luminometer (Turner Designs, model T 20/20, Sunnyvale, CA) for 10 s. To normalize for transfection efficiency, 20 ng/well of firefly luciferase gene (fluc) in pcDNA3.1 (+) vector backbone was cotransfected along with the experimental DNA, and FLUC activity was estimated. For measuring the FLUC activity, 20 μl of cell lysates were added with 100 μl of LARII reagent of Promega (Madison, WI) by counting for 10 s in the luminometer. Measuring the total protein concentration in the cell lysates also normalized the readings. Activity of hRLUC was represented as relative light units (RLU) per microgram of protein per minute of counting.
Cell uptake studies
Uptake of [8-3H] Penciclovir (0.76 μCi/ml, 1.5 × 10−5 mg/ml) was assessed 24 h after transient cotransfection of 293T cells. The cells were incubated at 37°C for 4 h. At the end of this period, radioactivity in the medium was measured. The wells were washed with cold PBS, the cells were harvested, and the cell-associated radioactivity was determined with a Beckman LS-9000 Liquid Scintillation Counter with Biosafe II (Research Products International) scintillation fluid, as described previously (22). Samples of pCMV-fluc cDNA were cotransfected along with experimental samples, and FLUC activity was assayed to normalize for transfection efficiency. Triplicate samples were evaluated for all uptake studies. The same wells were also used for determining total protein content (22). Data are expressed as the net accumulation of probe in [dpm cells/dpm medium (at start of exposure)/μg protein] ± SE.
Bioluminescence imaging in living mice
All animal handling was performed in accordance with University of California Animal Research Committee guidelines. Nude mice (nu/nu) were used. In the first group of three mice, each mouse was imaged several minutes after subcutaneous implantation of 10 × 106 293T cells transiently cotransfected 24 h previously with the tail-to-head combination of monomers (site A) or with mock-transfected cells (site B). In the second group of three mice, the combinations of monomers were tail-to-head (site A), double-tail-to-head (site B), and tail-to-double-head (site C). Mice were anesthetized by intraperitoneal injection of ≈40 μl of a ketamine and xylazine (4:1) solutions. All mice were imaged using a cooled CCD camera (Xenogen IVIS, Xenogen Corp., Alameda, CA) immediately after tail vein injection of 10 μl of coelenterazine (stock solution, 2 μg/μl in methanol) diluted in 90 μl of PBS (pH 7.0). The animals were placed prone in a light-tight chamber, and a gray scale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for a period of 1 min. Images were obtained using Living Image Software (Xenogen Corporation, Alameda, CA) and Igor Image Analysis Software (Wavemetrics, Seattle, WA). To quantify the measured light, regions of interest were drawn over the area of the implanted cells, and the maximum photons/s/cm2/steradian (sr) were obtained as validated previously (21) and also corrected for transfection efficiency differences.
RESULTS
In vitro luminometry and in vivo bioluminescence imaging of TK homodimerization
We performed in vitro luminometry of cell lysates and in vivo whole-body bioluminescence imaging of intact cells for assaying hRLUC activity after transient cotransfection of two experimental plasmids with normalizing plasmid in different sets of 293T cells, each plasmid containing one of four vectors and fluc in a pcDNA3.1 (+) backbone (Fig. 2a). Cotransfection strategies were performed such that opposing split hRLUC domains would be expressed and made available to complement when spatially positioned in either a tail-to-head (i.e., N-hRLUC upstream and C-hRLUC downstream of the two TK monomers, respectively), head-to-tail (downstream and upstream, respectively), tail-to-tail, or head-to-head configuration with respect to the two TK monomers. Accordingly, we used four different combinations of vector constructs in these cotransfection experiments.
We obtained the greatest degree of in vitro recovered activity for hRLUC (415±25 RLU/μg protein/min [P<0.01] in contrast to the background signal of 4±1 RLU/μg protein/min) (Fig. 2b), and the highest in vivo imaging signal (7500±200 p/s/cm2/sr [P<0.01] that was significantly above background light emission of 2600±200 p/s/cm2/sr) (Fig. 2c), after interaction of the two TK chimeras in the tail-to-head configuration (tail-to-head homodimer).
Monitoring the lack of association of TK monomers
By contrast, we recorded very minimal levels of recovered hRLUC activity in vitro for the remaining three combinations of protein chimera configurations, varying from 1.5% to 3.2% of the activity seen in the tail-to-head homodimers (Fig. 2b). These combinations gave insufficient bioluminescence signal for in vivo imaging (data not shown), and unlike the tail-to-head homodimer, these combinations were unsuitable for detecting TK homodimers whether or not they were present (Fig. 2c). We observed a trend within these recorded levels of minimal activity for the relatively unsuccessful combinations of chimeras. The least favorable complementation was seen when N-hRLUC was in the downstream position of TK. This was made even worse when combined with a chimera carrying an opposing C-hRLUC, also in the downstream position.
In vitro luminometry and in vivo bioluminescence imaging of simulated inhibition of TK homodimerization
We designed a strategy for and assessed the effects of simulated homodimerization inhibition using in vitro luminometry and in vivo imaging. To this end, we constructed two further variant protein chimeras, one with an N-hRLUC domain attached at either end of the same TK molecule, and the other with C-hRLUC domains also sandwiching the TK molecule (Fig. 3a). The double N-hRLUC chimera was combined with the chimera containing C-hRLUC downstream of TK in cotransfection strategies as outlined above. Similarly, the double C-hRLUC chimera was combined with that containing N-hRLUC upstream of TK. The first combination represented our tail-to-head TK homodimer with the addition of another N-hRLUC in the head position. Given our previous finding that the presence of an N-hRLUC domain in the head position leads to ineffective homodimerization, it was reasoned that this new tail-to-double-head combination would inhibit the previously demonstrated homodimerization of the tail-to-head homodimer. The second combination represented our tail-to-head TK homodimer with the addition of another C-hRLUC in the tail position. It was reasoned here that the presence of an additional smaller C-hRLUC, albeit in the tail position, would perhaps lead to a lesser degree of homodimerization inhibition than for the first combination tested. Finally, a third combination used both new chimeras in a double-tail-to-double-head combination.
We compared these new combinations with the tail-to-head homodimer and demonstrated by in vitro luminometry that the extra N-hRLUC in the tail-to-double-head combination of monomers resulted in complete inhibition of hRLUC complementation (Fig. 3b). The double-tail-to-head combination resulted in an intermediate loss of complementation, observed on both luminometry and whole-body imaging, down to 56% and 64%, respectively, of that seen for the tail-to-head homodimer (Fig. 3b, c). We hypothesized that this smaller-sized C-hRLUC might have induced more modest deleterious conformational movements at the TK dimerization interface. As expected, also the double-tail-to-double-head combination inhibited completely the association of TK monomers. We also monitored the dynamic temporal changes of these simulated dimerization inhibitors over a 48 h period after transient transfection, the results of which are seen in (Fig. 4).
Figure 4. Graph shows temporal changes of luminometry results on cell lysates from 12 h to 48 h after cotransfection of constructs expressing the tail-to-head, double-tail-to-head (showing intermediate inhibition of TK homodimerization), tail-to-double-head, and double-tail-to-double-head combinations of monomers (both showing complete inhibition of TK homodimerization).
Results are normalized for transfection efficiency. The Tail-to-Head results are significantly higher (P<0.01) as compared with all other combination of monomers.
Controls to confirm TK homodimerization-associated recovery of hRLUC activity
We also designed strategies to prove the recovered hRLUC activity was generated through TK homodimerization. For that we made two independent experiments, one by cotransfection of 293T cells with TK molecule containing portions of hrluc gene with another complementing hrluc gene portions without any protein (Fig. 2a). The results showed significant recovery of hRLUC activity (P<0.001) only from the cells cotransfected with one TK molecule containing upstream N-hrluc and the other with downstream C-hrluc (415±25 RLU/μg protein/min). All other combinations showed signal that was not significantly different from mock-transfected cells (4±1 RLU/μg protein/min) (Fig. 2b).
In a second experiment, we generated a mutant HSV1-TK by changing an amino acid at position 318 from arginine to cysteine to prevent TK homodimerization as previously reported (20). By using this molecule, we generated one molecule containing mutant TK with upstream N-hrluc and another molecule of mutant TK containing downstream C-hrluc fragment (Fig. 5a, b). The constructs were cotransfected with another normal TK molecule containing complement portion of hrluc fragments. The results showed significant reduction (P<0.001) in the complementation associated recovery of hRLUC activity. The recovered activity from the cells cotransfected with one or both mutant TK containing complement hrluc fragments were 25 ± 5% of the cells transfected with both TK molecule with complement hrluc fragments (Fig. 5c). The mutant TK molecules were studied for TK enzyme activity by cell uptake study as mentioned in the methodology. The mutant TK molecule containing upstream N-hrluc fragments showed TK activity similar to HSV1-TK and the mutant TK molecule containing downstream C-hrluc fragment showed ~50% (Fig. 5d).
Enzyme activity of TK
To address the concern that the reporter fragments might inactivate the interacting TK protein partners under investigation, we assessed the presence of properly folded functional intracellular TK molecules by performing in vitro substrate [8-3H] Penciclovir accumulation assays in 293T cells 24 h after either transient transfection with all four chimeras individually, or transient cotransfection with all four combinations of chimeras. Net accumulation of probe in cells transfected with the intact nontruncated sr39TK acted as control. No cells possessed net accumulation less than 59.6% [except cells transfected individually with tk molecule containing downstream N-hrluc fragment (35%)] of that observed in control cells expressing nonchimeric intact sr39TK, and accumulation was significantly above mock levels (Fig. 6). Our results indicate that there was no major loss of activity for the sr39TK domains in each of the chimeras we constructed, whether the TK dimerized or remained as a monomer.
Figure 6. [8-3H]Penciclovir accumulation in 293T cells 24 h after either transient transfection with the four chimeras individually.
(B: N-hrluc-tk, C: tk-C-hrluc, D: tk-N-hrluc, E: C-hrluc-tk), or transient cotransfection with the four combinations of chimeras (F: N-hrluc-tk plus tk-C-hrluc [tail-to-head], G: tk-N-hrluc plus C-hrluc-tk [head-to-tail], H: N-hrluc-tk plus C-hrluc-tk [tail-to-tail], I: tk-N-hrluc plus tk-C-hrluc [head-to-head]. Net accumulation of probe in cells transfected with the intact nontruncated sr39tk (A) acted as control. Results are for net accumulation (in dpm cells/dpm medium/microgram protein) of the probe over a 4 h incubation period. Accumulation was significantly (P<0.01) above mock levels (J). The error bars represent the standard error for triplicate determinations. Results are normalized for transfection efficiency.
We further performed in vitro substrate [8-3H] Penciclovir accumulation assays in 293T cells to ensure that the additional reporter domains in the new chimeras designed for testing homodimerization inhibition did not inactivate the TK enzyme. No cells possessed net accumulation less than 40% of that observed in control cells expressing nonchimeric intact TK (Fig. 7). This substrate accumulation was significantly above mock levels, for the two new chimeras individually or when combined with other chimeras, as outlined above.
Figure 7. [8-3H]Penciclovir accumulation in 293T cells 24 h after either transient transfection with the two additional chimeras individually.
(B: N-hrluc-tk-N-hrluc, C: C-hrluc-tk-C-hrluc), or transient cotransfection with the three combinations of chimeras (D: N-hrluc-tk-N-hrluc plus tk-C-hrluc [tail-to-double-head], E: C-hrluc-tk-C-hrluc plus N-hrluc-tk [double-tail-to-head], F: N-hrluc-tk-N-hrluc plus C-hrluc-tk-C-hrluc [double-tail-to-double-head]). Net accumulation of probe in cells transfected with the intact nontruncated sr39tk (A) acted as control. Results are for net accumulation (in dpm cells/dpm medium/microgram protein) of the probe over a 4 h incubation period. Accumulation was significantly above (P<0.01) mock levels (G). The error bars represent the standard error for triplicate determinations. Results are normalized for transfection efficiency.
DISCUSSION
Advances in bioluminescence imaging using cooled charged coupled device (CCD) cameras are providing highly sensitive means for imaging and quantifying very low levels of visible light from intact living subjects (19, 21, 23). The specific advantages of bioluminescence imaging are summarized by Massoud and Gambhir (19). We previously reported an inducible yeast two-hybrid system with firefly luciferase (24) and a split firefly luciferase complementation system (25) to study protein–protein interactions in cell lines, and to noninvasively image interactions in living mice. More recently, we developed an inducible hRLUC protein fragment-assisted complementation-based bioluminescence assay to quantitatively measure real-time protein–protein interactions in mammalian cells in the presence of the substrate coelenterazine (18). The designing of a protein fragment-assisted complementation assay entails the interaction of two proteins chimeras, each composed of one protein partner fused in frame with a split segment of a reporter protein. The interaction is driven by and brings together the two protein partners, leading to recovery of reporter activity through protein complementation of the two trailing split reporter segments (Fig. 1). We have previously shown that the split hRLUC system has higher sensitivity than a similar split firefly luciferase system for imaging stable heterodimeric protein–protein interactions of high affinity (18).
Wild-type HSV1-TK phosphorylates a wide range of nucleoside analogs, allowing selective anti-herpetic and viral vector-based gene therapies. The mutant HSV1-sr39TK (TK) was derived by site-directed mutagenesis to result in an enzyme more and less effective at phosphorylating acyclovir/ganciclovir and thymidine, respectively, with consequent gain in imaging signal when used as a reporter protein for positron emission tomography (22). We chose this enzyme as our test protein for three reasons: First, when crystallized in the presence of thymidine or other nucleoside analog substrates, HSV1-TK (376 residues, 43 kDa) forms asymmetrical homodimeric units related by C2 symmetry (26) (Fig. 1). Second, homo-FRET anisotropy had been used previously to follow homodimerization of green fluorescent protein (GFP)-tagged molecules of HSV1-TK (27), and although a complex assay to perform in single cells, this has provided a framework for our new strategy using bioluminescence imaging in intact cells and in living subjects. Finally, the activity of TK itself is easily determined in vitro and in vivo (22), which allows assessment of any adverse effects of fusing split hRLUC fragments to the interacting TK monomers, such as inactivation due to improper folding or inappropriate intracellular trafficking. An important clarification in our choice of TK as test protein has to be made. Compelling arguments were recently put forward to suggest that although HSV1-TK always crystallizes as a homodimer, it might not be an obligate homodimer in vivo (20). Instead, it likely exists in a monomer/dimer equilibrium (thus, defining it as a transient homodimer), being mainly monomeric at low protein concentrations. Furthermore, HSV1-TK does not have to dimerize to become active; the dimer is enzymatically even less efficient than the monomer. Indeed, the precise advantages conferred by the in vivo homodimeric form of this enzyme are unknown (20). Although a test protein that met the above criterion and also was an obligate dimer may have been more useful, we felt the use of TK was a good starting point for this work. Future studies will need to explore the generalizability of this assay for other protein homodimers including obligate dimers.
We studied the complementation-based recovery of split hRLUC activity after transient cotransfection of various combinations of plasmid vectors in 293T cells. We first designed four different vector constructs to evaluate the influence of the positions and orientations of the two split hRLUC protein fragments relative to the TK monomers. The assumption that such spatial and topological features concerning the position of split hRLUC fragments might be pertinent to both complementation of the reporter and the actual TK homodimerization itself stems from the presence of an underlying asymmetrical shape to the TK homodimer and the isologous dimerization interface, which was skewed more toward the C-terminal of each monomer (26). The constructed four recombinant protein chimeras contained N-hRLUC or C-hRLUC fused upstream or downstream of TK respectively (Fig. 2a). Our molecular cloning strategy included cDNA encoding intervening linker peptides of an appropriate length so as to prevent conformational strain between the TK molecules and the fused split hRLUC fragments (18). Importantly, we chose not to truncate the TK molecule at its C-terminal when constructing the chimeras because deletion of the last 6 amino acids results in enzyme inactivation (28). This may also hinder homodimerization owing to the loss of the Glu-371 interface residue that makes contact with Asn-306 and Val-307 of the opposite monomer (28). We theorized that for each of the novel protein chimeras that we developed, there would likely be a low chance of misfolding for either component protein. This was based on prior findings that fusion of GFP downstream of HSV1-TK did not induce substantial alteration in the tertiary structure of GFP itself (27) and independent knowledge that the folding of a GFP domain fused downstream of a partner protein, via a linker of similar length and neutral amino acid composition to the one used in our study, is directly related to the proper folding of the upstream protein domain (29). Limited or absent conformational strain on account of the flexible linkers would also likely help in this regard. Nevertheless, we were able to directly assay the in vitro activity of TK in conjunction with monitoring its homodimerization indirectly via hRLUC complementation.
Complementation-based restoration of reporter enzyme activity by two interacting proteins requires correctly folded portions of the proteins in close proximity (18). This applies to both the test TK monomers driving the interaction and the trailing split reporter fragments. We reasonably speculated that for the tail-to-head configuration, the positive results obtained from luminometry of cell lysates and the in vivo bioluminescence images reflected these molecular biophysical events that characterize successful homodimerization, especially when bearing in mind the maintained enzymatic activity of the TK domains within these chimeras. Further support of this is based on the observations we made that a mutation that has previously been reported to prevent homodimerization led to a marked drop in the complementation signal.
By contrast, we recorded very minimal levels of recovered hRLUC activity in vitro for the remaining three combinations of protein chimera configurations, varying from 1.5% to 3.2% of the activity seen in the tail-to-head homodimers. This may have been due to 1) steric interference by large-sized split reporter domains insinuating themselves between the TK monomer interfaces, 2) ineffective orientations of adequately approximated two split hRLUC proteins, or 3) an inability of the split hRLUC proteins to approximate because of poor positioning on the TK monomers, rendering them too far apart despite adequate TK homodimerization. This is reminiscent of similar stringent steric conditions governing homo-FRET reactions that have also previously undermined the imaging of EGFR homodimerization in single cells (30). Reassuringly, the bioluminescence complementation-based assay that we describe for detecting protein homodimerization is extremely sensitive to low levels of light and therefore is able to indicate even very minimal levels of protein complementation on luminometry despite seemingly unfavorable steric constraints, because the background levels of light emission are almost at zero.
Other than the concern over potential loss of TK enzyme activity addressed in this study with the [8-3H]Penciclovir accumulation studies, another concern might be that the presence of additional reporter fragments in each chimera may hinder the correct physiological intracellular translocation of the interacting proteins. Reassuringly, GFP, which is of larger molecular weight than N-hRLUC or C-hRLUC, rarely hinders the trafficking of a test protein to which it is tagged (31) as shown, for example, when fused to EGFR monomers translocating to the cell membrane (30). We reasonably speculate that the lower molecular weight split hRLUC fragments would therefore likely be useful for imaging protein–protein interactions originating in diverse cellular compartments, although this remains to be investigated more fully.
We have demonstrated the use of an in vivo protein fragment-assisted complementation assay, based on a split hRLUC reporter technology, for bioluminescence imaging of homodimeric protein–protein interactions in cells and intact living subjects. We imaged in living mice the homodimerization of TK monomers tagged with split hRLUC reporter proteins. This technique critically depends on various factors, such as the size, orientation, and position of the split hRLUC reporter domains in relation to the test proteins. Our detailed characterization of this system should help broaden its application and make it a generalizable methodology for the study of homodimeric protein–protein interactions. The high sensitivity of this assay for detecting, locating, and quantifying transient homodimerization of proteins, combined with the advantages of doing so in a living subject environment, should make it of potential value in four areas.
Investigating the reasons for protein homodimerization
For many transient homodimers, especially when dimerization is not obviously linked to function, the biological rationale for the physiological monomer/dimer equilibrium is unknown, unclear, or speculative (32). New insights may potentially be gleaned as well from the workings of other homodimers that are better characterized by experiment (17). These homodimers include enzymes where each monomer provides the active site, or receptors where enhanced activity is conferred by tightening a signaling complex or where specificity is provided by a contribution to defining a ligand binding pocket (33).
Studying the factors that drive the association or dissociation of homodimer subunits
Linking the unique information gained from in vivo imaging with knowledge of biophysical phenomena—governed by the shape, chemical complementarities, and flexibility of the molecules involved (2)—potentially offers greater understanding of the conditions under which homodimeric subunits interact.
The assessment of known interactions within their intracellular physiological context
Likely, this is where the greatest value of this technique lies. Rather than screening proteomes for unknown interactions, the protein-fragment-assisted complementation assay that we present is can complement and work in concert with other techniques (e.g., FRET, BRET, etc.) in the study of homodimeric interactions in living environments.
Development of novel drugs based on inhibition of active protein assemblies
The implementation of molecular imaging approaches in the drug discovery process offers the strong advantage of being able to meaningfully study a potential drug, labeled for imaging in an animal model, often before phenotypic changes become obvious. This can facilitate a more rapid progression into human studies. It is likely that preclinical studies can be accelerated to rule out homodimerization inhibitors with unfavorable biodistribution and/or pharmacokinetics before human studies (21). This would likely be useful in the design of inhibitors preventing receptor-mediated signal transduction, as well as other classes of compounds, such as antibiotic, antiviral (especially those targeting HIV-1 protease), and antiparasitic drugs (34). Ultimately, we foresee innovative molecular imaging tools, such as the one we present, enhancing our appreciation of entire biological pathway systems and their pharmacological regulation, and accelerating the achievement of a ‘systems biology’ understanding of biological complexity.
Acknowledgments
We thank Z. Walls for his useful comments. This work was supported by grants from the National Institutes of Health (T.F.M., S.S.G.) (2 RO1 CA82214-05), The Health Foundation (T.F.M.), General Electric Medical Systems (T.F.M.), Palgrave Brown Foundation (T.F.M.), Cancer Prevention Research Trust (T.F.M.), Royal College of Radiologists (T.F.M.), Steel Charitable Trust (T.F.M.), Sir Samuel Scott of Yews Trust (T.F.M.), and Department of Energy (S.S.G.) (DE-FG02-03ER63687).
References
- 1.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 2.Valdar WS, Thornton JM. Protein–protein interfaces: Analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–124. [PubMed] [Google Scholar]
- 3.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 4.Williams NE. Immunoprecipitation procedures. Methods Cell Biol. 2000;62:449–453. [PubMed] [Google Scholar]
- 5.Bollag DM. Gel-filtration chromatography. Methods Mol Biol. 1994;36:1–9. doi: 10.1385/0-89603-274-4:1. [DOI] [PubMed] [Google Scholar]
- 6.Hansen JC, Lebowitz J, Demeler B. Analytical ultracentrifugation of complex macromolecular systems. Biochemistry. 1994;33:13,155–13,163. doi: 10.1021/bi00249a001. [DOI] [PubMed] [Google Scholar]
- 7.Doyle ML. Characterization of binding interactions by isothermal titration calorimetry. Curr Opin Biotechnol. 1997;8:31–35. doi: 10.1016/s0958-1669(97)80154-1. [DOI] [PubMed] [Google Scholar]
- 8.Lakey JH, Raggett EM. Measuring protein-protein interactions. Curr Opin Struct Biol. 1998;8:119–123. doi: 10.1016/s0959-440x(98)80019-5. [DOI] [PubMed] [Google Scholar]
- 9.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 11.Heyduk T, Ma Y, Tang H, Ebright RH. Fluorescence anisotropy: Rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 1996;274:492–503. doi: 10.1016/s0076-6879(96)74039-9. [DOI] [PubMed] [Google Scholar]
- 12.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 13.Blakely BT, Rossi FM, Tillotson B, Palmer M, Estelles A, Blau HM. Epidermal growth factor receptor dimerization monitored in live cells. Nat Biotechnol. 2000;18:218–222. doi: 10.1038/72686. [DOI] [PubMed] [Google Scholar]
- 14.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc Natl Acad Sci USA. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocheville M, Lange DC, Kumar U, Sasi R, Patel RC, Patel YC. Subtypes of the somatostatin receptor assemble as functional homo- and heterodimers. J Biol Chem. 2000;275:7862–7869. doi: 10.1074/jbc.275.11.7862. [DOI] [PubMed] [Google Scholar]
- 16.McVey M, Ramsay D, Kellett E, Rees S, Wilson S, Pope AJ, Milligan G. Monitoring receptor oligomerization using time-resolved fluorescence resonance energy transfer and bioluminescence resonance energy transfer. The human delta -opioid receptor displays constitutive oligomerization at the cell surface, which is not regulated by receptor occupancy. J Biol Chem. 2001;276:14,092–14,099. doi: 10.1074/jbc.M008902200. [DOI] [PubMed] [Google Scholar]
- 17.Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 18.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massoud TF, Gambhir SS. Molecular imaging in living subjects: Seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 20.Wurth C, Thomas RM, Folkers G, Scapozza L. Folding and self-assembly of herpes simplex virus type 1 thymidine kinase. J Mol Biol. 2001;313:657–670. doi: 10.1006/jmbi.2001.5060. [DOI] [PubMed] [Google Scholar]
- 21.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR, Iyer M, Namavari M, Phelps ME, Herschman HR. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci USA. 2000;97:2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 24.Ray P, Pimenta H, Paulmurugan R, Berger F, Phelps ME, Iyer M, Gambhir SS. Noninvasive quantitative imaging of protein–protein interactions in living subjects. Proc Natl Acad Sci USA. 2002;99:3105–3110. doi: 10.1073/pnas.052710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99:15,608–15,613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wild K, Bohner T, Folkers G, Schulz GE. The structures of thymidine kinase from herpes simplex virus type 1 in complex with substrates and a substrate analogue. Protein Sci. 1997;6:2097–2106. doi: 10.1002/pro.5560061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautier I, Tramier M, Durieux C, Coppey J, Pansu RB, Nicolas JC, Kemnitz K, Coppey-Moisan M. Homo-FRET microscopy in living cells to measure monomer–dimer transition of GFP-tagged proteins. Biophys J. 2001;80:3000–3008. doi: 10.1016/S0006-3495(01)76265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saijo M, Suzutani T, Niikura M, Morikawa S, Kurane I. Importance of C-terminus of herpes simplex virus type 1 thymidine kinase for maintaining thymidine kinase and acyclovir-phosphorylation activities. J Med Virol. 2002;66:388–393. doi: 10.1002/jmv.2157. [DOI] [PubMed] [Google Scholar]
- 29.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 30.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 31.Tavare JM, Fletcher LM, Welsh GI. Using green fluorescent protein to study intracellular signalling. J Endocrinol. 2001;170:297–306. doi: 10.1677/joe.0.1700297. [DOI] [PubMed] [Google Scholar]
- 32.Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 33.Bardsley B, Cho YR, Westwell MS, Williams DH. Induction of asymmetry into homodimers. Chirality. 1998;10:14–23. [PubMed] [Google Scholar]
- 34.Veselovsky AV, Ivanov YD, Ivanov AS, Archakov AI, Lewi P, Janssen P. Protein-protein interactions: mechanisms and modification by drugs. J Mol Recognit. 2002;15:405–422. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]