Abstract
Ideal cancer gene therapies should have high tumor specificity and efficacy, and allow systemic administration to target metastases. We recently developed a bi-directional, two-step transcriptional amplification (TSTA) system driven by the tumor-specific Survivin promoter (pSurv) to amplify the correlated expression of both the reporter gene firefly luciferase (FL) and therapeutic gene tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Here, we compare the specificity and potency of an adenovirus carrying this system (Ad-pSurv-TSTA-TRAIL-FL) to a nonspecific vector (Ad-pCMV-FL) in an orthotopic hepatocellular carcinoma (HCC) rat model after systemic administration. At 24 h after injection of Ad-pCMV-FL, bioluminescence imaging revealed a trend (P=0.30) towards greater FL expression in liver versus tumor. In striking contrast, Ad-pSurv-TSTA-TRAIL-FL showed increased FL activity within the tumor compared with the liver (P<0.01), a strong trend towards reduced liver expression compared with Ad-pCMV-FL (P=0.07), and importantly, similar FL levels within tumor compared with Ad-pCMV-FL (P=0.32). Hence, this vector shows potent, tumor-specific transgene expression even after extensive liver transduction and may be of significant value in avoiding hepatotoxicity in HCC patients. Future studies will explore the benefits of tumor-specific TRAIL expression in this model, the potential to target metastases and the extension of this vector for the treatment of other Survivin-positive tumors is warranted.
Keywords: hepatocellular carcinoma, adenoviral vector, two-step transcriptional amplification, Survivin
INTRODUCTION
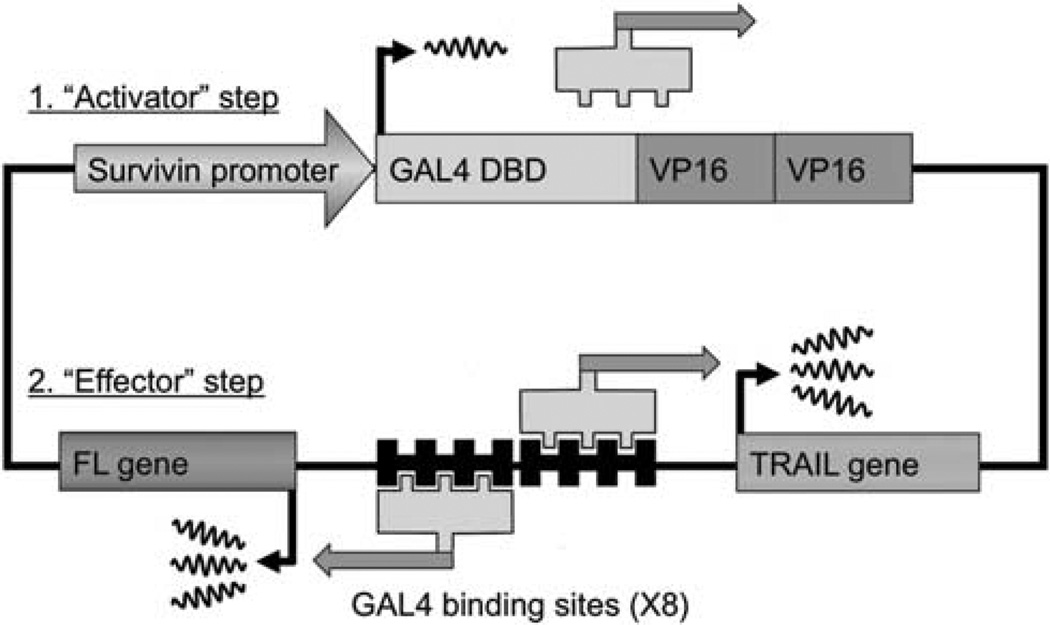
Over the past decade, different strategies have been developed to improve the tumor specificity of gene therapies to avoid toxic transgene expression in normal tissues. These strategies are particularly needed for adenoviral-based gene therapies as these vectors rapidly transduce both Kupffer cells and hepatocytes,1 potentially inducing life-threatening hepatotoxicity. In the context of hepato-cellular carcinoma (HCC), the ability to avoid expression in normal hepatocytes is particularly important owing to the impaired liver function typically seen in this patient population. Transcriptionally targeted gene therapy is a promising approach that utilizes tumor-specific promoters (TSPs) to induce therapeutic gene expression primarily in cancer cells.2 However, one challenge with this strategy is overcoming the low-level transcription induced by TSPs. Ideally, one would like to achieve levels of transgene expression in cancer cells similar to those using strong viral promoters, such as the cytomegalovirus (CMV) promoter, as this should improve therapeutic outcome. To help overcome the limitation of weak TSPs, we have previously developed and validated a two-step transcriptional amplification (TSTA) system.3,4 In the first ‘activator’ step of this system, the chosen weak promoter directly drives the expression of a fusion protein between the GAL4-DNA-binding domain and two tandem VP16 transactivation domains. In the subsequent ‘effector’ step, the GAL4-VP16-2 fusion protein promotes strong expression of a therapeutic gene under the control of multiple GAL4-binding sites and a E4TATA minimal promoter.3,4
In addition to efficient targeting, the ideal gene therapy strategy would also be capable of non-invasively monitoring the location, intensity and duration of transgene expression over time. This can be accomplished by coupling the expression of the therapeutic gene with an imaging reporter gene such as firefly luciferase (FL) for bioluminescence imaging (BLI)5 or herpes simplex virus-1 thymidine kinase for positron emission tomography (PET).6 Along these lines, we have also developed a bi-directional TSTA system, in which the GAL4-VP16-2 transactivator simultaneously promotes the expression of two genes in a highly correlated manner.7 In our latest bi-directional system (pSurv-TSTA-TRAIL-FL), we chose the tumor-specific Survivin promoter (pSurv) to drive the expression of both the pro-apoptotic gene tumor necrosis factor-α-related apoptosis-inducing ligand (TRAIL/Apo2L) and reporter gene FL (Figure 1). The tumor-targeting potential of an adenoviral serotype 5 vector carrying this system (Ad-pSurv-TSTA-TRAIL-FL) was explored in mice with colorectal cancer xenografts. Ad-pSurv-TSTA-TRAIL-FL lead to higher levels of transgene expression after intratumoral delivery compared with a one-step system where pSurv directly drove the expression of FL.5 Furthermore, the Survivin-driven TSTA system was also able to decrease FL expression in the normal liver after intravenous delivery in healthy mice compared with a CMV-promoter-driven system.5 Importantly, expressions of TRAIL and FL were highly correlated both in vitro and in vivo.5 This system has the coveted tumor-on/normal hepatocyte-off profile, leads to high levels of therapeutic gene expression and the location and level of transgene expression can be tracked with imaging. However, the ability to specifically target orthotopic tumors and avoid normal hepatocyte expression following systemic administration has not been explored and was the primary focus of this study.
Figure 1.
Schematic diagram of the Survivin-targeted bi-directional TSTA system. In the first ‘activator’ step of the system, the cancer-specific Survivin promoter drives the expression of the GAL4-VP16-2 fusion protein. GAL4-VP16-2 consists of two tandem repeats of the N-terminal portion of the VP16 activation domain (aa 413–454) fused to the GAL4 DNA-binding domain (DBD; aa 1–147). In the second ‘effector’ step of the system, the GAL4-VP16-2 fusion protein binds to GAL4-responsive minimal promoter and bi-directionally drives the expression of both the reporter gene (RG) FL and therapeutic gene (TG) TRAIL. The use of the GAL4-VP16-2 fusion protein leads to the amplification of both TG and RG simultaneously, whereas the tightly coupled FL allows indirect determination of both the location and level of TRAIL expression using BLI.
RESULTS
Verification of tumor formation after implantation of McA-RH7777 cells into left hepatic lobe of buffalo rats
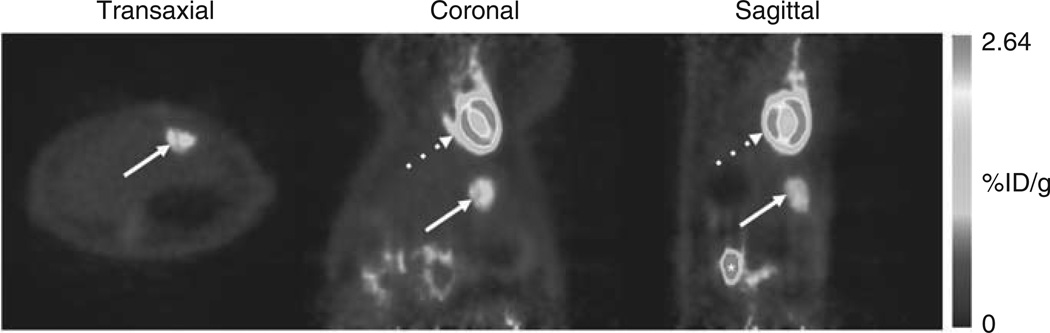
Syngeneic HCC cells (McA-RH7777 cells, 1 × 106) were surgically implanted in the left hepatic lobe of Buffalo rats (n=12) as described previously.8 At 13 days following implantation, rats were imaged with 18F-fluorodeoxyglucose (FDG) PET (Figure 2). In all rats, a clear hypermetabolic foci was detected within the liver, confirming the presence of the orthotopic HCC.
Figure 2.
18F-FDG PET imaging 2 weeks following tumor cell implantation showed glucose hypermetabolic foci in the liver, confirming successful tumor formation. Transaxial, coronal and sagittal (left to right) PET images showed evidence of hypermetabolic foci in the liver (white arrows). Dotted arrow represents tracer uptake in the normal myocardium. Asterisk indicates renal uptake.
The Survivin-targeted, amplifiable vector dramatically improved specificity of transgene expression, although maintaining potency following intravenous delivery compared with a CMV-targeted vector
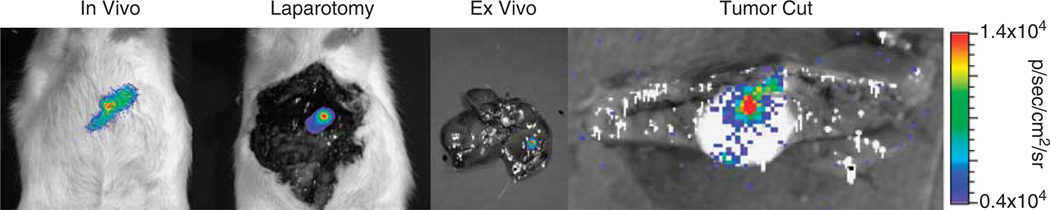
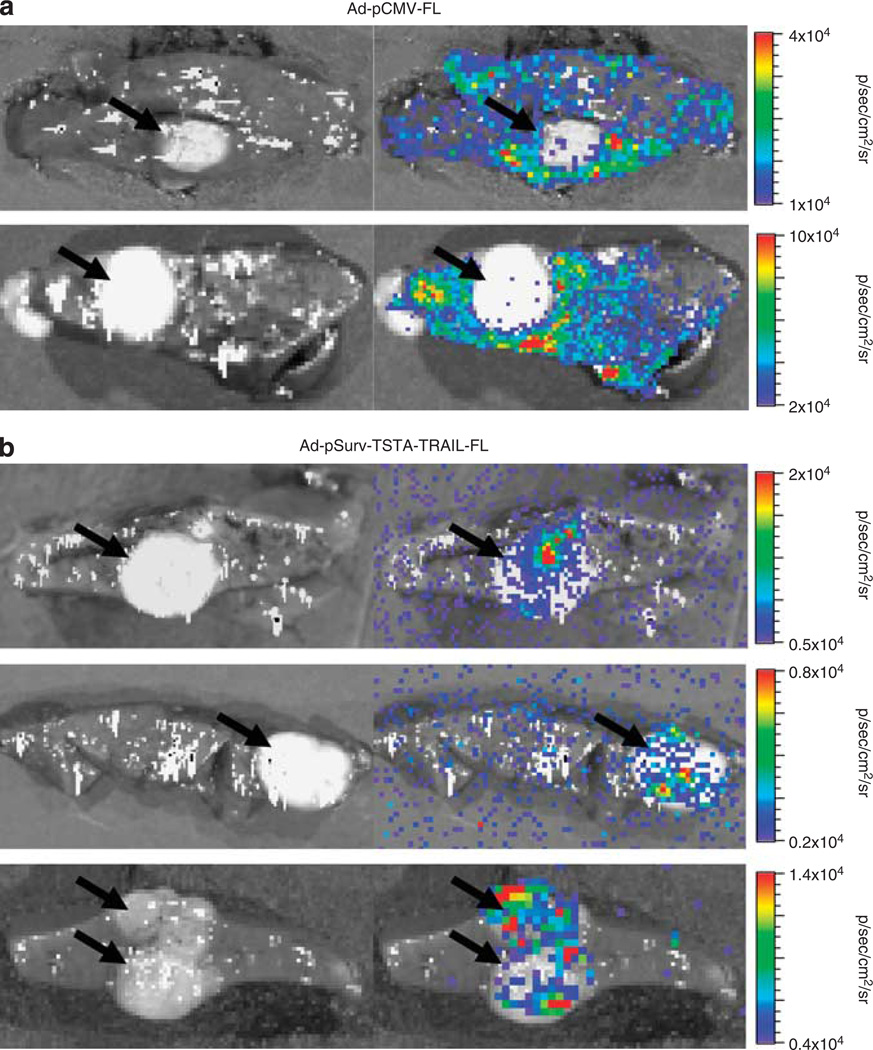
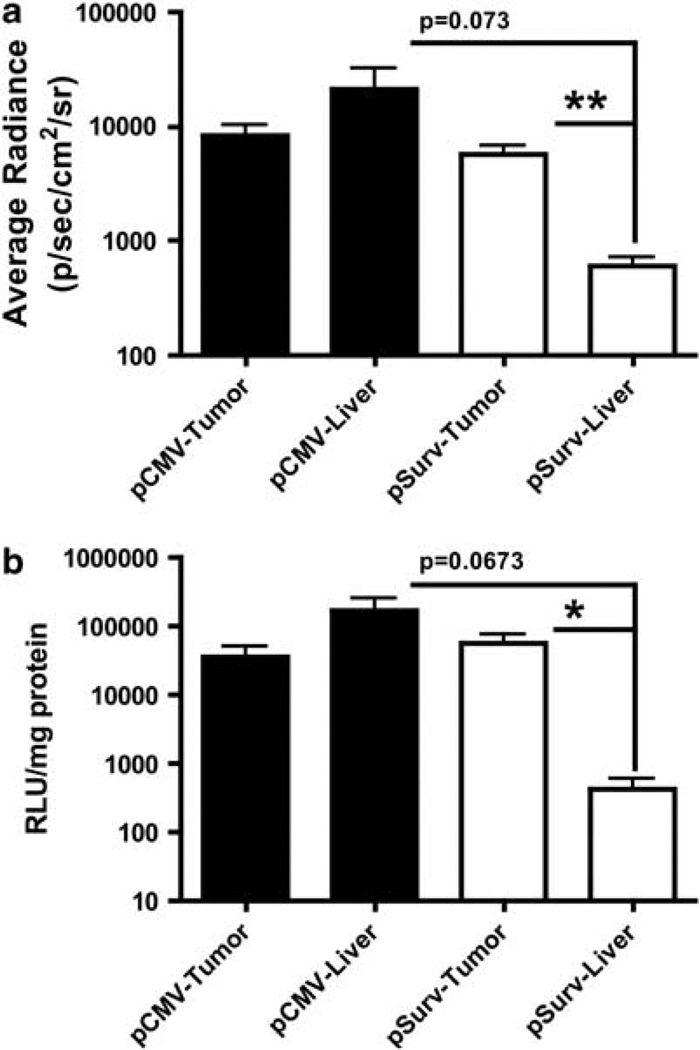
Tumor-bearing rats were administered 109 plaque-forming units (PFU) of either a CMV-targeted virus (Ad-pCMV-FL) (n=3) or Ad-pSurv-TSTA-TRAIL-FL (n=6) via the tail vein. After 48 h, a series of bioluminescent images were collected to determine the locations and levels of FL activity (Figure 3). Qualitatively, rats administered Ad-pCMV-FL showed greater FL activity within the normal liver compared with the tumor (Figure 4a). In contrast, BLI images from rats administered Ad-pSurv-TSTA-TRAIL-FL showed minimal FL activity within the normal liver and foci of high FL activity within tumors (Figure 4b). Owing to the imprecision of determining the location of FL activity in in vivo, laparotomy and ex vivo whole tumor images, quantitative information about liver and tumor FL activity was assessed from tumor cut images (Figure 5a). For two rats receiving Ad-pSurv-TSTA-TRAIL-FL, tumor cut images were not collected owing to technical difficulties; therefore, images from only four rats in the Survivin-targeted and three rats in the CMV-targeted group were analyzed. Rats administered Ad-pSurv-TSTA-TRAIL-FL showed significantly increased FL activity within tumor versus normal liver (5653±1540 versus 485.7±55.18 photons s−1 cm−2 sr−1, respectively; P<0.01). Conversely, in the rats that received Ad-pCMV-FL, there was a trend towards less FL activity in the tumor compared with the normal liver (8243±2212 versus 21 250±10 870 photons s−1 cm−2 sr−1, respectively; P=0.3062). The Survivin-driven vector showed a strong trend towards decreased liver FL expression compared with the CMV-driven vector (603±124 versus 21 250±10 870 photons s−1cm−2 sr−1; P=0.073). Importantly, FL expression within the tumor itself was not significantly different between the two vectors (P=0.3193).
Figure 3.
Sequence of BLI. At 48h after tail-vein administration of Ad-pSurv-TSTA-TRAIL-FL (shown) or Ad-pCMV-FL, tumor-bearing rats were imaged intact and following laparotomy, and livers were imaged ex vivo following whole dissection and after a single incision through the tumor (tumor cut). Scale bar corresponds to average radiance values in tumor cut images.
Figure 4.
Systemic administration of the Survivin-driven TSTA virus (Ad-pSurv-TSTA-TRAIL-FL) results in improved cancer specificity of FL activity in orthotopic hepatocellular carcinomas compared with CMV-targeted virus (Ad-pCMV-FL). (a) Representative white light image alone (left) and white light image with overlying BLI image (right) of tumor (black arrow) and surrounding the normal liver parenchyma from two rats infected with Ad-pCMV-FL. Note the extensive FL activity from surrounding the normal liver compared with the tumor. (b) Similar images of tumor-bearing livers from three rats infected with Ad-pSurv-TSTA-TRAIL-FL 24h before imaging. Significantly improved tumor specificity of FL expression (black arrow) is seen compared with the images in (a) owing to much lower signal from the normal liver. Note the scale differences between the different sets of images from individual rats.
Figure 5.
Analysis of BLI imaging and luminometer measurements of tissue lysates. (a) Analysis of cut tumor images revealed a nonsignificant trend towards increased FL activity in the normal liver compared with the tumor in rats infected with Ad-pCMV-FL (mean±s.e.m.; n=3). In contrast, the Survivin-targeted virus (n=4) significantly improved the tumor specificity of transgene expression, and showed a strong trend towards abrogating FL activity in the normal liver compared with the CMV-targeted virus. Importantly, administration of either the Survivin- or CMV-targeted viruses resulted in similar levels of FL activity within tumors. (b) Luminometer readings of FL activity from tumor or normal liver lysates from animals infected with either virus corroborated the imaging results. *P<0.05 and **P<0.01 as determined via a two-tailed, unpaired t-tests.
After BLI, luciferase activity was measured in tissue lysates using a luminometer assay (Figure 5b). Results from this assay corroborated our imaging results. In rats receiving the CMV-driven vector, normal liver parenchyma showed 4.75-fold higher luciferase activity than tumor, but these differences were not statistically significant (167 900±85 970 versus 35 390±15 390 relative light units per mg protein, respectively; P=0.2037). Rats receiving the Survivin-driven vector showed 135-fold higher luciferase activity in the tumor than in the normal liver (57 860±15 900 versus 430±187 relative light units per mg protein, respectively; P<0.05). Finally, no difference in FL activity within the tumor was found comparing the two vectors (P=0.3697) and a nonsignificant trend towards decreased liver FL expression using the Survivin-driven vector was noted (P=0.0673).
Intratumoral delivery of the Survivin-targeted vector dramatically increased tumor-to-liver transgene expression versus systemic delivery
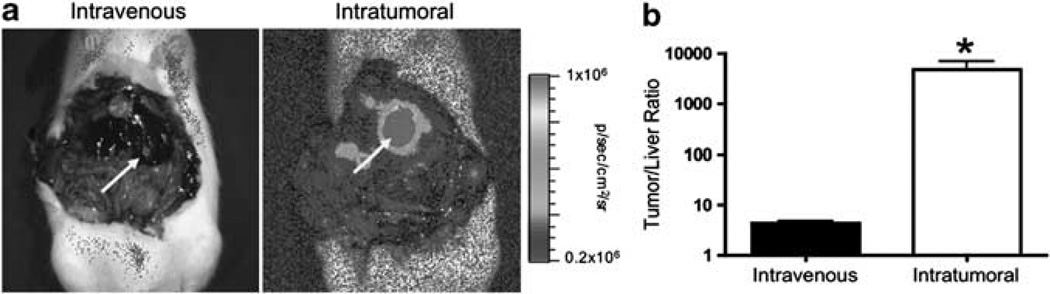
We also performed direct intratumoral injection of Ad-pSurv-TSTATRAIL-FL with open abdominal surgery (n=3). As expected, qualitatively higher light output was detected within the tumor following intratumoral versus intravenous delivery (Figure 6a). This corresponded to an approximately 1100-fold increase in tumor-to-liver signal ratio using intratumoral versus intravenous delivery (P<0.05; Figure 6b). Finally, for one rat we delivered 10-fold higher dose of the Survivin-targeted virus (1010 PFU) via the tail vein and obtained 90-fold higher FL signal intensity from tumor compared with rats receiving the 109 PFU dose intravenously (data not shown).
Figure 6.
Comparison of FL activity in both tumor and normal liver after either systemic or intratumoral injection of pSurv-TSTA-TRAIL-FL virus. (a) Although both systemic and intratumoral injection resulted in notable tumor specificity using the Survivin-targeted system, as expected direct intratumoral delivery (right image) resulted in visibly greater FL activity within tumors (white arrow) compared with intravenous delivery. The scale bar is the same for both images to highlight differences in expression levels. (b) Analysis of BLI images revealed significantly improved tumor/liver ratio of FL activity (mean±s.e.m.) following intratumoral (n=3) versus systemic (n=5) viral administration. *P<0.05 as determined via two-tailed, unpaired t-test.
DISCUSSION
HCC is a primary cancer of the liver and annually accounts for approximately 600 000 deaths worldwide.9 HCC is particularly prevalent in developing countries where risk factors for HCC such as hepatitis B/C infection are also prevalent, but HCC incidence in developed countries is rising. The prognosis of HCC is extremely poor—primarily because HCC is typically diagnosed at advanced stages; and secondarily, because the best curative options, tumor resection or liver transplantation, are not feasible for the majority of patients (>80%). Even those patients selected for surgical interventions only have a 30–40% 5-year survival rate owing to recurrence or metastasis.10 Hence, novel therapies are urgently needed, and ideally should be both useful for the majority of patients and aimed at targeting both primary HCC and distant metastasis. Systemically administered gene therapies may fulfill this need, but specificity, safety and efficacy of these therapies must be determined.
Effective gene therapy of HCC and its distant metastases will require vectors that can be administered systemically, promote strong tumor-specific transgene expression and be able to avoid expression in normal tissues to prevent unwanted side effects. In this study, we have assessed the ability to target tumors in an orthotopic HCC rat model following systemic intravenous administration of adenoviral vector carrying a TSP driving a bi-directional TSTA system5 to produce the expression of a therapeutic gene, TRAIL, and a reporter gene, FL.11
Adenoviral vectors have several features that are highly advantageous for successful gene therapy including: an ability to infect a wide range of human cells; an ability to yield high levels of gene transfer compared with other vectors; a low pathogenicity in human beings; a tolerance to large recombinant inserts (up to ~7.5 kb); an ability to infect non-dividing cells; and a high tendency to avoid genomic integration, thereby minimizing the chances of insertional mutagenesis.12 Despite all these advantages, there are several limitations of adenoviral vectors. Firstly, they are notoriously hepatotropic and if a strong ubiquitous promoter (for example, pCMV) controls expression, this can lead to significant levels of normal hepatocyte transgene expression and possible hepatotoxicity. We have also shown the hepatotropic nature of such vectors in this study, as intravenous delivery of the Ad-pCMV-FL vector lead to abundant FL activity throughout the normal liver. A second limitation of adenoviruses is the inability to administer effective multiple doses of virus over time owing to the production of neutralizing antibodies following the first dose. Therefore, prevention of transgene expression in the liver while maintaining high tumor expression following a single dose is a challenge with adenoviral vectors. Several strategies have been designed to try to overcome these limitations. The first and most obvious is to introduce the viruses directly into the tumor. As shown in Figure 6, this results in significantly higher tumor expression compared with systemic administration. However, this strategy is not without its limitations. First, systemic leakage is difficult to avoid, resulting in extra-tumoral, particularly hepatic, transduction. Second, intratumoral injection is not always feasible in all patients, particularly those with multiple tumor foci. Finally, this type of strategy has limited therapeutic benefit for treating small, distant metastases. To better target all tumor cells, including those in single large primary tumors, multiple small tumors or even micrometastases and single metastasized cells, a systemically administered therapy is required. However, with this strategy the risk of non-tumor transcription is increased and tissue-specific control of therapeutic protein production is needed. To achieve this control, one common strategy is called transcriptional targeting. This involves the use of tissue-specific promoter or TSP to drive transgene expression. Therefore, even though non-tumor tissues are transduced with the adenoviral vector, gene transcription is limited to tumor cells. Numerous TSPs have been used for a variety of cancers, including human α-fetoprotein for HCC,13,14 human carcinoembryonic antigen for colorectal carcinoma,15 human α-lactalbumin and ovine β-lactoglobulin for breast cancer16 and human telomerase reverse transcriptase for TERT-positive cancers.17,18 However, one major limitation of TSPs is the low levels of transcription they induce. Therefore, even though they provide the wanted specificity, the efficacy of these vectors may be suboptimal. Previous work from our lab has shown that this limitation may be overcome through the combined use of the TSTA system with a cell-specific promoter such as variants of the prostate-specific antigen enhancer/promoter,3,19–21 the vascular endothelial growth factor (VEGF) promoter22 and the mucin-1 promoter.23 Therefore, the TSTA strategy has broad applications for augmenting the activity of a weak promoter. In the vector used here, we employed the promoter for Survivin (pSurv). Survivin, a member of the inhibitor of apoptosis family, is involved in controlling mitotic progression and preventing cell death, and is overexpressed in many cancers (HCC, lung, breast, colon, ovarian, skin and so on), but not in normal adult tissues.24–28 Therefore, pSurv is well suited for transcriptional targeting systems, has been utilized for this purpose in models of lung, breast, glioma and colon cancer,5,29–31 and our Survivin-driven TSTA system may have broad applicability for effective cancer therapy in numerous tumor types. Following systemic administration of our Survivin-targeted vector, transgene expression was primarily restricted to tumor cells even though the normal liver was transduced extensively, as shown using the CMV-targeted vector. However, the use of transcriptional targeting with pSurv in combination with the amplification capability of the GAL4-VP16-2 fusion protein in our TSTA system, we were not only able to improve tumor specificity, but also attained similar levels of transgene expression within the tumor compared with the CMV-targeted vector, one of the strongest and most commonly used promoters in clinical trials.
It should be mentioned that another level of safety in this vector is the use of TRAIL as the pro-apoptotic gene. TRAIL is a transmembrane protein, member of the tumor necrosis factor family and rapidly induces apoptosis in a wide variety of cancer cells upon interaction with the death receptors DR4/TRAIL-R1 and DR5/TRAIL-R2.11 Notably, the pro-apoptotic effects of TRAIL are primarily limited to cancer cells as normal cells express decoy receptors (DcR1/TRAIL-R3 and DcR2/TRAIL-R4), which competitively bind TRAIL, but are incapable of activating the extrinsic apoptotic cascade.11 Hence, TRAIL is a promising agent for selective cancer treatment32–34 and a good candidate for successful gene therapy as shown by intratumoral injection of a replication-competent Ad5-TRAIL adenovirus.35,36 TRAIL is also known to induce apoptosis in adjacent, non-transduced cells by the ‘bystander effect’.36–38 Armeanu et al.39 showed that adenoviral gene transfer of pCMV-TRAIL was superior to soluble TRAIL for inducing cell death in human HCC lines. Unfortunately, because a nonspecific CMV promoter was used, TRAIL also induced a strong apoptotic response in 53% of normal hepatocytes. Our system couples TRAIL production with the cancer-specific pSurv to limit TRAIL expression in HCC to HCC cells, with relative sparing of hepatocytes. Therefore, any off-tumor expression should not result in significant apoptosis of normal cells. Ongoing studies are exploring the effects of TRAIL expression in both tumor and normal tissue.
One limitation of this study is that transduction of Ad-pSurv-TSTA-TRAIL-FL induces expression of both therapeutic TRAIL and reporter FL protein, which can decrease FL activity following tumor cell damage by TRAIL-induced apoptosis. Therefore, cancer specificity of pSurv as measured by FL reporter gene expression may be underestimated. Another limitation is that the model we employed does not allow us to provide survival data to show the beneficial effects of our gene therapy strategy (death is not an allowable end point for this model at our institution). Currently, we are exploring other Survivin-positive tumor models that will allow us to test the effects of animal survival.
In summary, this study explores the ability to target orthotopically administered HCC cells with an adenoviral vector (Ad-pSurv-TSTA-TRAIL-FL) carrying both a therapeutic gene and reporter gene following systemic administration. Our system has several important generalizable features amenable for successful gene therapy: (1) it utilizes transcriptional targeting to restrict transgene expression to tumor cells; (2) the TSTA system overcomes the weak transcriptional strength of TSPs leading to potent transgene expression; and (3) the reporter gene FL can be used to track therapeutic gene expression as its expression is highly correlated to the expression of the therapeutic gene.5 In the future, we can couple the therapeutic gene to a PET reporter gene such as herpes simplex virus thymidine kinase,6 the sodium iodide transporter40 or a multimodality reporter gene41 for clinical applications.
MATERIALS AND METHODS
Cell line and media
Morris hepatoma cells (McA-RH7777) were purchased from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 1% penicillin/streptomycin. All cells were maintained in a humidified incubator with 5% CO2 in the atmosphere at 37 °C.
Orthotopic HCC rat model
Animal experiments were carried out in accordance with the institutional guidelines. Buffalo rats were purchased from Charles Rivers Laboratory (Wilmington, MA, USA). Rats were anesthetized with isoflurane inhalation: for induction, 5% isoflurane was mixed in 1 l O2 per min, and for maintenance, 2–4% isoflurane was mixed in 1 l O2 per min. Analgesia was provided by both a subcutaneous injection of buprenorphine (0.01–0.05mg kg−1) and an intramuscular injection of Flunixin meglumine (2.5mg kg−1). For tumor implantation, a subxiphoid incision was made and the liver was mobilized to expose the left lateral segment. A total of 106 syngeneic McA-RH7777 cells suspended in 100 µl of phosphate-buffered saline were injected slowly (typically over 30–60 s) under the capsule of the left lateral lobe as described previously.8 A cotton applicator was applied for 2–3min over the needle insertion site, followed by the application of ~100 µl of 70% ethanol to the peritoneal cavity to prevent extra-hepatic cell spillage. The incision was then closed in layers with suture.
PET imaging
At 13 days following cell implantation, 18F-FDG PET imaging was performed to confirm tumor formation in the liver. Rats were anesthetized using isoflurane inhalation as above, intravenously administered ~500 µCi of 18F-FDG and imaged with a Concorde R4 microPET system (Siemens AG, Malvern, PA, USA). A 5-min static scan with an approximate resolution of 2mm in each axial direction was obtained in all animals. Images were reconstructed with the OSEM (ordered-subsets expectation maximization) algorithm.
Adenovirus construction
All adenoviruses are first-generation E1, E3 deleted. The bi-directional TSTA adenovirus, Ad-pSurv-TSTA-TRAIL-FL, was cloned by Vector Biolabs (Philadelphia, PA, USA), as described previously5. The control virus, Ad-pCMV-FL, was constructed using the AdEasy kit (Agilent Technologies: Stratagene Products, Santa Clara, CA, USA) according to the manufacturer’s guidelines. In short, all gene sequences were inserted into the pShuttle intermediate plasmid and then recombined into the adenoviral backbone in bacteria. Viruses were later packaged and amplified in HEK-293 cells and titers were determined using an adenovirus titer immunoassay kit (QuickTiter, Cell Biolabs Inc., San Diego, CA, USA).
Adenoviral administration
The next day after verification of tumor formation with 18F-FDG PET, 109 PFU of adenovirus (Ad-pSurv-TSTA-TRAIL-FL or Ad-pCMV-FL) in 300 µl of phosphate-buffered saline were administered into the rats either by intravenous (tail vein) or intratumoral injection. For direct intratumoral delivery, the abdominal wall of the animal was reopened, viruses were injected into 3–5 foci within the tumor and the incision was closed in layers with suture.
Bioluminescence imaging
At 2 days after viral delivery, BLI of rats was performed using an IVIS Spectrum imaging system (Xenogen, Alameda, CA, USA). Rats were anesthetized as above and 5 min before imaging d-luciferin was injected intraperitoneally (45mg per rat; Xenogen, Alameda, CA, USA). Animals were placed in a light-tight chamber and photons emitted were collected with a cooled charge-coupled device camera. Immediately after in vivo imaging, a laparotomy was performed and open abdominal images of each rat were collected. Immediately following this, livers were excised and ex vivo images were acquired of both the whole liver and a portion of the liver following an incision through the middle of the tumor. All images were acquired with an integration time of 60 s and regions of interest of equal size were drawn within the tumor and adjacent normal liver parenchyma in tumor cut images to measure average radiance (expressed as photons s−1 cm−2 sr−1).
Bioluminescence luminometer assay
Following BLI, FL activity in tumor and liver was determined with a Luciferase Assay kit (Promega, Sunnyvale, CA, USA) in a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA, USA). An integration time of 10 s was used for all measurements. The protein content of tissue lysates was determined using a Bio-Rad protein assay system (Bio-Rad, Hercules, CA, USA) in a Beckman DU-50 spectrophotometer (Beckman Instruments, Fullerton, CA, USA). Luminescence results were reported as relative light units per mg protein.
Statistical testing
Statistical comparisons between treatments were performed with a two-tailed, unpaired t-test. Nominal P-values <0.05 were considered statistically significant.
ACKNOWLEDGEMENTS
We would like to acknowledge grant support from NCI ICMIC P50CA114747 (SSG) and NCI RO1 CA082214 (SSG). We would also like to thank Tim Doyle (Stanford Center for Innovation in In Vivo Imaging, Stanford University) for help with small animal imaging. JAR is supported by the Canadian Institutes of Health Research Postdoctoral Fellowship. B-CA received financial support from the Nuclear Infra Construction of Nuclear R&D program, Korean ministry of Science and Technology.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Akiyama M, Thorne S, Kirn D, Roelvink PW, Einfeld DA, King CR, et al. Ablating CAR and integrin binding in adenovirus vectors reduces nontarget organ transduction and permits sustained bloodstream persistence following intraperitoneal administration. Mol Ther. 2004;9:218–230. doi: 10.1016/j.ymthe.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Lo H-W, Day C-P, Hung M-C. Cancer-specific gene therapy. Adv Genet. 2005;54:235–255. doi: 10.1016/S0065-2660(05)54010-0. [DOI] [PubMed] [Google Scholar]
- 3.Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir SS. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci USA. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Adams JY, Billick E, Ilagan R, Iyer M, Le K, et al. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- 5.Ray S, Paulmurugan R, Patel MR, Ahn BC, Wu L, Carey M, et al. Noninvasive imaging of therapeutic gene expression using a bidirectional transcriptional amplification strategy. Mol Ther. 2008;16:1848–1856. doi: 10.1038/mt.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambhir SS, Herschman HR, Cherry SR, Barrio JR, Satyamurthy N, Toyokuni T, et al. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray S, Paulmurugan R, Hildebrandt I, Iyer M, Wu L, Carey M, et al. Novel bidirectional vector strategy for amplification of therapeutic and reporter gene expression. Hum Gene Ther. 2004;15:681–690. doi: 10.1089/1043034041361271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van den Bosch JJ, Higgins L, Hwang G, Katzenberg RH, Willmann JK, Paulmurugan R, et al. Novel rat hepatocellular carcinoma model designed forin vivo evaluation of image-guided therapies. J Vasc Interv Radiol. 2008;19:S136. [Google Scholar]
- 9.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 10.Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 12.Vorburger SA, Hunt KK. Adenoviral gene therapy. Oncologist. 2002;7:46–59. doi: 10.1634/theoncologist.7-1-46. [DOI] [PubMed] [Google Scholar]
- 13.Nakaya H, Ishizu A, Ikeda H, Tahara M, Shindo J, Itoh R, et al. In vitro model of suicide gene therapy for alpha-fetoprotein-producing gastric cancer. Anticancer Res. 2003;23:3795–3800. [PubMed] [Google Scholar]
- 14.Ye X, Liang M, Meng X, Ren X, Chen H, Li Z-Y, et al. Insulation from viral transcriptional regulatory elements enables improvement to hepatoma-specific gene expression from adenovirus vectors. Biochem Biophys Res Commun. 2003;307:759–764. doi: 10.1016/s0006-291x(03)01251-8. [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, Li S, Nyati MK, DeRemer S, Parsels J, Rehemtulla A, et al. Regional delivery and selective expression of a high-activity yeast cytosine deaminase in an intrahepatic colon cancer model. Cancer Res. 2003;63:658–663. [PubMed] [Google Scholar]
- 16.Anderson LM, Krotz S, Weitzman SA, Thimmapaya B. Breast cancer-specific expression of the Candida albicans cytosine deaminase gene using a transcriptional targeting approach. Cancer Gene Ther. 2000;7:845–852. doi: 10.1038/sj.cgt.7700191. [DOI] [PubMed] [Google Scholar]
- 17.Bilsland AE, Anderson CJ, Fletcher-Monaghan AJ, McGregor F, Evans TRJ, Ganly I, et al. Selective ablation of human cancer cells by telomerase-specific adenoviral suicide gene therapy vectors expressing bacterial nitroreductase. Oncogene. 2003;22:370–380. doi: 10.1038/sj.onc.1206168. [DOI] [PubMed] [Google Scholar]
- 18.Katz MH, Spivack DE, Takimoto S, Fang B, Burton DW, Moossa AR, et al. Gene therapy of pancreatic cancer with green fluorescent protein and tumor necrosis factor-related apoptosis-inducing ligand fusion gene expression driven by a human telomerase reverse transcriptase promoter. Ann Surg Oncol. 2003;10:762–772. doi: 10.1245/aso.2003.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Iyer M, Salazar FB, Lewis X, Zhang L, Carey M, Wu L, et al. Noninvasive imaging of enhanced prostate-specific gene expression using a two-step transcriptional amplification-based lentivirus vector. Mol Ther. 2004;10:545–552. doi: 10.1016/j.ymthe.2004.06.118. [DOI] [PubMed] [Google Scholar]
- 20.Iyer M, Salazar FB, Lewis X, Zhang L, Wu L, Carey M, et al. Non-invasive imaging of a transgenic mouse model using a prostate-specific two-step transcriptional amplification strategy. Transgenic Res. 2005;14:47–55. doi: 10.1007/s11248-004-2836-1. [DOI] [PubMed] [Google Scholar]
- 21.Iyer M, Salazar FB, Wu L, Carey M, Gambhir SS. Bioluminescence imaging of systemic tumor targeting using a prostate-specific lentiviral vector. Hum Gene Ther. 2006;17:125–132. doi: 10.1089/hum.2006.17.125. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Iyer M, Annala A, Wu L, Carey M, Gambhir SS. Noninvasive indirect imaging of vascular endothelial growth factor gene expression using bioluminescence imaging in living transgenic mice. Physiol Genomics. 2006;24:173–180. doi: 10.1152/physiolgenomics.00308.2004. [DOI] [PubMed] [Google Scholar]
- 23.Huyn ST, Burton JB, Sato M, Carey M, Gambhir SS, Wu L. A potent, imaging adenoviral vector driven by the cancer-selective mucin-1 promoter that targets breast cancer metastasis. Clin Cancer Res. 2009;15:3126–3134. doi: 10.1158/1078-0432.CCR-08-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080–1085. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- 25.Lu B, Makhija SK, Nettelbeck DM, Rivera AA, Wang M, Komarova S, et al. Evaluation of tumor-specific promoter activities in melanoma. Gene Ther. 2005;12:330–338. doi: 10.1038/sj.gt.3302385. [DOI] [PubMed] [Google Scholar]
- 26.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 27.Chen J-S, Liu J-C, Shen L, Rau K-M, Kuo H-P, Li YM, et al. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004;11:740–747. doi: 10.1038/sj.cgt.7700752. [DOI] [PubMed] [Google Scholar]
- 28.Chen W-C, Liu Q, Fu J-X, Kang S-Y. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol. 2004;10:2886–2889. doi: 10.3748/wjg.v10.i19.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Liu X, Fan J, Qi R, Bo L, Gu J, et al. A survivin-mediated oncolytic adenovirus induces non-apoptotic cell death in lung cancer cells and shows antitumoral potential in vivo. J Gene Med. 2006;8:1232–1242. doi: 10.1002/jgm.953. [DOI] [PubMed] [Google Scholar]
- 30.Van Houdt WJ, Haviv YS, Lu B, Wang M, Rivera AA, Ulasov IV, et al. The human survivin promoter: a novel transcriptional targeting strategy for treatment of glioma. J Neurosurg. 2006;104:583–592. doi: 10.3171/jns.2006.104.4.583. [DOI] [PubMed] [Google Scholar]
- 31.Zhu ZB, Makhija SK, Lu B, Wang M, Rivera AA, Kim-Park S, et al. Incorporating the survivin promoter in an infectivity enhanced CRAd-analysis of oncolysis and anti-tumor effects in vitro and in vivo. Int J Oncol. 2005;27:237–246. [PubMed] [Google Scholar]
- 32.Oikonomou E, Kothonidis K, Taoufik E, Probert E, Zografos G, Nasioulas G, et al. Newly established tumourigenic primary human colon cancer cell lines are sensitive to TRAILinduced apoptosis in vitro and in vivo. Br J Cancer. 2007;97:73–84. doi: 10.1038/sj.bjc.6603835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley SK, Harris LA, Xie D, Deforge L, Totpal K, Bussiere J, et al. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther. 2001;299:31–38. [PubMed] [Google Scholar]
- 35.Griffith TS, Anderson RD, Davidson BL, Williams RD, Ratliff TL. Adenoviral-mediated transfer of the TNF-related apoptosis-inducing ligand/Apo-2 ligand gene induces tumor cell apoptosis. J Immunol. 2000;165:2886–2894. doi: 10.4049/jimmunol.165.5.2886. [DOI] [PubMed] [Google Scholar]
- 36.Kagawa S, He C, Gu J, Koch P, Rha SJ, Roth JA, et al. Antitumor activity and bystander effects of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene. Cancer Res. 2001;61:3330–3338. [PubMed] [Google Scholar]
- 37.Wu GS. TRAIL as a target in anti-cancer therapy. Cancer Lett. 2009;285:1–5. doi: 10.1016/j.canlet.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 38.Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armeanu S, Lauer UM, Smirnow I, Schenk M, Weiss TS, Gregor M, et al. Adenoviral gene transfer of tumor necrosis factor-related apoptosis-inducing ligand overcomes an impaired response of hepatoma cells but causes severe apoptosis in primary human hepatocytes. Cancer Res. 2003;63:2369–2372. [PubMed] [Google Scholar]
- 40.Niu G, Gaut AW, Ponto LLB, Hichwa RD, Madsen MT, Graham MM, et al. Multimodality noninvasive imaging of gene transfer using the human sodium iodide symporter. J Nucl Med. 2004;45:445–449. [PubMed] [Google Scholar]
- 41.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]