Abstract
Imaging reporter gene expression in living subjects with various imaging modalities is a rapidly accelerating area of research. Applications of these technologies to cancer research, gene therapy, and transgenic models are rapidly expanding. We report construction and testing of several triple fusion reporter genes compatible with bioluminescence, fluorescence and positron emission tomography (PET) imaging. A triple fusion reporter vector harboring a bioluminescence synthetic Renilla luciferase (hrl) reporter gene, a reporter gene encoding the monomeric red fluorescence protein (mrfp1), and a mutant herpes simplex virus type 1 sr39 thymidine kinase [HSV1-truncated sr39tk (ttk); a PET reporter gene] was found to preserve the most activity for each protein component and was therefore investigated in detail. After validating the activities of all three proteins encoded by the fusion gene in cell culture, we imaged living mice bearing 293T cells transiently expressing the hrl-mrfp-ttk vector by microPET and using a highly sensitive cooled charge-coupled device camera compatible with both bioluminescence and fluorescence imaging. A lenti-viral vector carrying the triple fusion reporter gene was constructed and used to isolate stable expressers by fluorescence-activated cell sorting. These stable 293T cells were further used to show good correlation (R2 ~0.74–0.85) of signal from each component by imaging tumor xenografts in living mice with all three modalities. Furthermore, metastases of a human melanoma cell line (A375M) stably expressing the triple fusion were imaged by microPET and optical technologies over a 40–50-day time period in living mice. Imaging of reporter gene expression from single cells to living animals with the help of a single tri-fusion reporter gene will have the potential to accelerate translational cancer research.
INTRODUCTION
To unravel the complexity and dynamics of molecular and cellular events, it is desirable to image reporter gene expression in individual cells, living animals, and humans with the help of a single reporter gene construct. Noninvasive and repetitive imaging of molecular events within a single cell to groups of cells within a living subject using different imaging modalities should play a critical role in understanding normal physiology and disease progression (1). Fluorescent reporter genes such as green fluorescence protein (gfp), red fluorescent protein (rfp), and their various mutants have been used extensively to tag individual proteins or cells, to monitor biochemical interactions of proteins and cell trafficking, and to study cellular dynamics (2). Expression of these fluorescent proteins in living cells can be imaged in real time by confocal laser microscopy, two-photon laser microscopy, and several other advanced microscopy techniques. On the other hand, noninvasive imaging of reporter gene expression in living subjects can be accomplished by using positron emission tomography (PET), single-photon emission computed tomography, magnetic resonance imaging, and small animal optical imaging modalities with distinct reporter genes compatible with each imaging modality (1, 3, 4). All of these noninvasive imaging modalities are playing expanding roles in the biomedical sciences. Imaging of reporter gene expression by PET is being used to directly and indirectly monitor therapeutic gene expression, immune cell trafficking, and protein-protein interactions (1). Noninvasive optical imaging techniques such as bioluminescence (5) and fluorescence (6) are also being applied in many ways including imaging critical pathways involved in tumorigenesis and metastasis (1). However, each of these imaging techniques and their respective reporter genes has unique advantages and limitations (1). The optical imaging strategies have the advantages of being relatively low cost and high-throughput, but they are limited by their nontomographic nature, lack of fine spatial resolution, and inability to scale up to larger animals and humans. PET has the advantage of being applicable to all living subjects, but it has the disadvantage of requiring relatively short-lived radioactive tracers. Bioluminescence, although relatively easy to perform in small animals, is difficult to use for single cell imaging, whereas fluorescence is highly sensitive for single cell imaging.
A specific strategy for combining the different modalities, including a cell-based technique with an animal imaging technique, is to build a unified fusion gene composed of different reporter genes whose expression can be imaged with different imaging modalities in both individual cells and living subjects. This approach is useful, provided that the fusion protein could retain at least moderate levels of activity of each individual reporter protein and be stable and not broken down into its specific constituents. This approach also opens the possibility of merging PET and optical imaging techniques for applications in a single living subject. Previously, we reported construction and validation of a fusion reporter vector bearing HSV1-sr39 thymidine kinase and Renilla luciferase (tk20rl) joined by a 20-amino acid (aa)-long spacer for imaging with microPET and bioluminescence optical charge-coupled device (CCD) modalities in living mice (7). This strategy was limited by the inability to image individual cells due to the relatively low light yield from bioluminescence. In the current study, we developed and tested several triple fusion vectors bearing a bioluminescence, a fluorescence, and a PET reporter gene joined by a 14-aa-long and an 8-aa-long spacer, respectively. Among all of the vectors tested, hrl-mrfp-ttk triple fusion reporter vector containing a synthetic Renilla luciferase [hrl (8)], monomeric rfp [mrfp1 (9)], and a truncated version of sr39tk (ttk; the first 135 bp of sr39tk were deleted) could best preserve the activities of all three component proteins and was therefore pursued for additional studies. We then imaged reporter gene expression in individual living cells by inverted fluorescence microscopy and also in living mice by both microPET and a cooled CCD camera compatible with both bioluminescence and fluorescence imaging. We also developed a lentiviral vector carrying the triple fusion reporter construct to infect dividing and nondividing cells and isolated stable expressers of 293T cells by fluorescence-activated cell sorting (FACS). Finally, we used a cancer metastatic model by introducing human melanoma cells (A375M) stably expressing the triple fusion reporter gene in living mice via tail vein and imaged metastases over a period of 50 days by all three of the imaging modalities. The unique tri-fusion vector should facilitate rapid translation of approaches developed in cells to preclinical models and, eventually, clinical applications.
MATERIALS AND METHODS
Chemicals
[8-3H]Penciclovir and 14C-labeled 2′-fluoro-5-fluoro-1-β-D-arbinofuranosyluracil were obtained from Moravek Biochemicals (Brea, CA). 9-(4-[18F]Fluoro-3-hydroxymethylbutyl)guanine (FHBG) was also synthesized at University of California Los Angeles as detailed previously (10). Coelenterazine was purchased from Biotium, Inc. (Hayward, CA). The poly-clonal anti-TK antibody was a kind gift of Dr. M. Black (Washington State University, Pullman, WA), and the monoclonal anti-Renilla luciferase protein (RL) was purchased from Chemicon International (Temecula, CA).
Construction of hrl-mrfp-ttk and Other Fusion Genes
PCR amplification and standard cloning techniques were used to insert the hrl and mrfp genes from plasmid pCDNA 3.1-CMV-hrl (Promega, Madison, WI) and pCDNA3.1-CMV-mrfp1 in frame with the ttk gene into the pCDNA3.1-sr39-truncated tk (a kind gift of Dr. D. Kaufman; University of California, Los Angeles, CA). The CMV-wtk vector was obtained from Dr. M. Black and modified to truncated wtk (wttk) by deleting first 135 bp through PCR and cloned in pCDNA3.1 backbone to generate CMV-wttk plasmid. CMV-fl and CMV-egfp were purchased from Promega and BD Sciences-Clontech (Palo Alto, CA) respectively. For PCR amplifications, different 5′ and 3′ end primers were used to generate the fusion vectors. Standard cloning techniques were used to generate the lentiviral (CS-hrl-mrfp-ttk) vector as performed previously in our laboratory (11).
Cell Lines and Transient Transfection Procedures
Neuro 2a (N2a) neuronal cell lines (a gift from Dr. Vincent Mauro; Scripps Research Institute, La Jolla, CA), 293T human embryonic kidney cells (American Type Culture Collection, Manassas, VA), and A375M human melanoma cells (a gift from Dr. M. Kolodny; University of California, Los Angeles, CA) were used. The N2a and A375M cells were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin (100 μg/ml) and streptomycin (292 μg/ml), and 293T cells were grown in MEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution. All transient transfections were carried out using the Superfect transfection reagent (Qiagen, Valencia, CA) following the protocol recommended by the manufacturer.
tTK, hRL, and β-Galactosidase (β-Gal) Activity
TK enzyme activity assays were performed as described previously (12), and β-Gal and Renilla or firefly luciferase assays were done using the β-Gal enzyme assay system and Dual-Luciferase Reporter Assay System from Promega, respectively. Each of the luciferase reactions was measured in a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA) for a period of 10 s.
Western Blot Analysis
The levels of tTK and hRL were evaluated by Western blotting with a rabbit polyclonal anti-TK antiserum and a mouse monoclonal anti-Renilla antibody using cell lysates prepared from 293T cells transfected with CMV-hrl-mrfp-ttk, CMV-ttk, and CMV-hrl plasmids (12).
Lentiviral Production
Lentivirus was developed and used to infect 293T and A375M cells as described previously (11).
Fluorescence Microscopy, CCD Imaging, and FACS
Expression of mRFP1 was observed under a Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss Microimaging Inc., Thornwood, NY) with DsRed filter setting (λex, 546 nm; λem, 605 nm) and analyzed with MetaMorph software (University Imaging Corp., Downingtown, PA). For quantification of the expression level of mRFP1 present in the CMV-hrl-mrfp-ttk and CMV-mrfp1, 1 × 104 and 1 × 105 of 293T, A375M, or N2a cells expressing the vectors were seeded in black-bottomem clear 96-well plates and imaged in the Xenogen IVIS optical imaging system (Xenogen Corp., Almeda, CA) with an excitation filter at 500–550 nm and an emission filter at 575–650 nm. Regions of interest (ROIs) were drawn over the cell area and quantified by using Living Image Software version 2.20. For FACS, 1 × 106 of CS-hrl-mrfp-ttk infected 293T and A375M cells were sorted by using a Becton Dickinson FACSvantage SE cell sorter.
MicroPET Imaging of Mice
Animal care and euthanasia were performed with the approval of the University of California Animal Research Committee. Male 12–14-week-old nude mice (nu/nu) received s.c. injection with ~10 × 106 293T cells transiently expressing the CMV-hrl-mrfp-ttk fusion, CMV-ttk, CMV-hrl, and CMV-mrfp1 on the ventral side, and mice (n = 4) were scanned the next day using a microPET as described previously (12). Additionally, 10 × 106 of each of four differentially expressing clones of 293T cells stably expressing hrl-mrfp-ttk gene were implanted in three mice and scanned in the microPET ~24 h later. The microPET images were reconstructed by using three-dimensional filtered back projection and an iterative maximum a posteriori algorithm (13). ROIs were drawn over the tumor area. The ROI counts were converted to percentage of injected dose/g (ID/g) using filtered back projection as described previously (12), and images shown were reconstructed with maximum a posteriori algorithm.
Bioluminescence and Fluorescence Imaging of mRFP1 and RL Expression in Living Mice
For in vivo fluorescence imaging, mice implanted with the cells described above were anesthetized, and each mouse was placed in a light tight chamber equipped with a halogen light source, and whole body image was acquired for 1 s using the Xenogen IVIS optical imaging system with an excitation filter at 500–550 nm and an emission filter at 575–650 nm. ROIs were drawn over implanted cell area and quantified by using Living Image Software version 2.20. For bioluminescence imaging, each mouse next received injection with 10 μl (2 μg/μl dissolved in methanol) of coelenterazine diluted in 90 μl of PBS (pH 7) via tail vein. Each animal was then placed supine in the same light tight chamber, and whole body images were obtained and quantified as described previously (8). Both bioluminescence and fluorescence signals were recorded as maximum [photons/second/centimeter2/steradian (photons/s/cm2/sr)].
Multimodality Imaging of Cancer Metastasis in Living Mice Using a Human Melanoma Cell Line (A375M) Stably Expressing the Triple Fusion Vector
Three 8-week-old Beige severe combined immunodeficient mice received injection with 7 × 105 A375M cells stably expressing the hrl-mrfp-ttk gene via tail vein and were imaged repeatedly with fluorescence, bioluminescence, and microPET. At day 40, the mice were first imaged with microPET and bioluminescence (as described above) and then sacrificed and imaged; the chest was cut open with Illuminatool Tunable lighting system using the 540 nm excitation filter and RFP viewing glass (Lightools Research). Fluorescence imaging and light photograph of mice were digitally captured with a Nikon camera for 2 s.
RESULTS
A Multimodality Fusion Vector Harboring the hrl Gene (Bioluminescence), Gene Encoding for mRFP1 (Fluorescence), and a Mutant Truncated HSV1-sr39 Thymidine Kinase (PET) Reporter Gene Maintains hRL, mRFP1, and tTK Activity in Several Cell Lines
We first constructed a fusion gene vector (hrl-ttk) carrying hrl (hrl, gene; hRL, enzyme) and ttk (ttk, gene; tTK, enzyme) reporter genes separated by a 22-aa-long spacer (LENSHASAGYQACGTAG-PGSTG) and then inserted the PCR-amplified mrfp1 gene fragment in the middle of the spacer (at the position of Cys-Gly) to generate a hrl-mrfp-ttk triple fusion reporter gene. The PCR-amplified hrl gene fragments from pCMV-hrl vector were inserted in frame with ttk gene (the first 45 aa of sr39tk gene were truncated to delete the nuclear localization signal of tk gene) cloned in pCDNA3.1+ separated by the above-mentioned spacer under the control of a CMV promoter. The resultant vector was then digested with HindIII and SacII and ligated in frame to PCR-amplified and HindIII/SacII-digested mrfp1 fragments (without stop codon) from pRSETB vector to generate the hrl-mrfp-ttk fusion vector. The order of the three different reporter genes and spacers in this triple fusion vector is as follows: hrl-spacer (LENSHASAGYQAST)-mrfp-spacer (TAGPGSAT)-ttk gene. The final vector was fully verified by sequencing.
Plasmid DNA prepared from four to five clones of CMV-hrl-mrfp-ttk triple fusion were transiently transfected in 293T cells, and the cells were first observed in a fluorescence microscope for mRFP1 activity and further assayed for hRL and tTK activity. The plasmid clone exhibiting the highest mRFP1, hRL, and tTK activities was selected for additional studies. To extend our study to different variants of bioluminescence/fluorescence/PET reporter genes, we also generated several functionally active multimodality reporter fusion vectors, i.e., fl-mrfp-ttk [by replacing hrl with firefly luciferase (fl)], fl/hrl-egfp/rfp-ttk (mrfp1 is replaced with egfp or tetrameric rfp known as DsRed2), and fl/hrl-rfp-wttk [by replacing the truncated HSV1-sr39tk (ttk) with wild-type HSV1-truncated thymidine kinase (wttk)]. The nature and the order of the spacers for all these constructs were equivalent to CMV-hrl-mrfp-ttk vector described earlier. The ttk, wttk, fl, hrl, rfp, gfp, and mrfp1 genes were also cloned in pCDNA3.1+ backbone to generate positive control plasmids to directly compare the results of each fusion. All these fusion vectors were functionally active with respect to each individual protein, however the level of activity varied for each construct (Table 1). Overall, the hrl-mrfp-ttk fusion construct showed the highest activity for all three of the component proteins in comparison with other vectors and thus was further studied for multimodality imaging.
Table 1.
Fluorescent, bioluminescent and PETa reporter gene expressions exhibited by different triple fusion constructs in comparison to the respective positive controls assayed from transiently transfected 293T cells (tTK and hRL activities are normalized with cotransfected β-Gal activity)
| Constructs | % TK activity | % wTK activity | % hRL activity | % FL activity | RFP/eGFP/mRFP (fluorescence activity by microscopy) |
|---|---|---|---|---|---|
| hrl-rfp-ttk | 39.7 | 27.8 | Medium | ||
| fl-rfp-ttk | 29.6 | 22.1 | Low | ||
| hrl-egfp-ttk | 50 | 33 | High | ||
| fl-egfp-ttk | 43 | 20 | High | ||
| hrl-mrfp-ttk | 149 | 54 | High | ||
| fl-mrfp-ttk | 100 | 53.6 | Medium | ||
| hrl-rfp-wttk | 76 | 44.7 | Medium | ||
| fl-rfp-wttk | 61.6 | 62.6 | Low | ||
| ttk | 100 | ||||
| wttk | 100 | ||||
| hrl | 100 | ||||
| fl | 100 | ||||
| mrfpl | Very high |
PET, positron emission tomography; eGFP, enhanced green fluorescence protein; β-Gal, β-galactosidase.
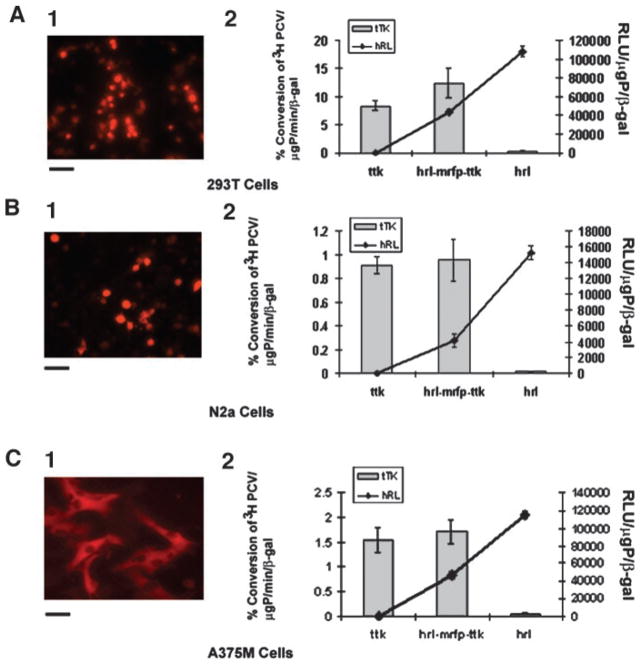
To compare the levels of reporter gene expression of each of the components of the CMV-hrl-mrfp-ttk plasmid, three different cell lines [293T, N2a, and A375M (Fig. 1, A–C) were transiently transfected with the triple fusion plasmid along with proper positive controls (pCDNA3.1-ttk, pCDNA3.1-mrfp1, pCDNA3.1-hrl) and negative controls (mock-transfected control cells). Each cell line was also cotransfected with the CMV-β-gal reporter gene to normalize for transfection efficiency. After 24 h, the expression of mrfp1 was observed in the inverted fluorescence microscope, and then activity of the other three reporter genes was assayed from the same cell lysates, and tTK and hRL activities were normalized to β-Gal activity. In all three of the different cell lines, CMV-hrl-mrfp-ttk showed equal or slightly higher tTK activity (not statistically significant) compared with the positive control (pCDNA3.1-ttk) but had a lower hRL activity [33% (A375M), 27.4% (N2a), and 54% (293T)] compared with that of the positive control pCDNA3.1-hrl plasmid (P < 0.05). The expression level of mrfp1 of this triple fusion vector in all of the cell lines was ~60 –70% of the positive control pCDNA3.1-mrfp1 vector, as determined by the fluorescence signal using the CCD camera.
Fig. 1.
mRFP1, hRL, and tTK activity exhibited by 293T, N2a, and A375M cell lines transiently transfected with the hrl-mrfp-ttk fusion construct. 293T (A), N2a (B), and A375M (C) cells were transiently cotransfected with CMV-β-gal and either CMV-hrl-mrfp-ttk, CMV-ttk, CMV-hrl, or CMV-mrfp1; harvested 24 h later; and assayed for mRFP1 expression (A.1, B.1, and C.1; by fluorescence microscopy) and tTK/hRL enzyme activities (A.2, B.2, and C.2). Bars for the fluorescence micrographs represent 100 μm. Values for tTK and hRL activity were normalized with β-galactosidase activity for each transfection. The tTK activity is expressed as (percentage of conversion of [8-3H]penciclovir to its phosphorylated form)/μg protein/min. The hRL activity is expressed as relative light units (RLU)/μg protein. Error bars represent SE for triplicate measurements. The slightly higher tTK activity exhibited by the hRL-mRFP-tTK fusion protein in comparison with the positive tTK protein is not statistically significant; however, the hRL activity of the hRL-mRFP-tTK fusion protein is significantly lower (P < 0.05) than the positive hRL protein.
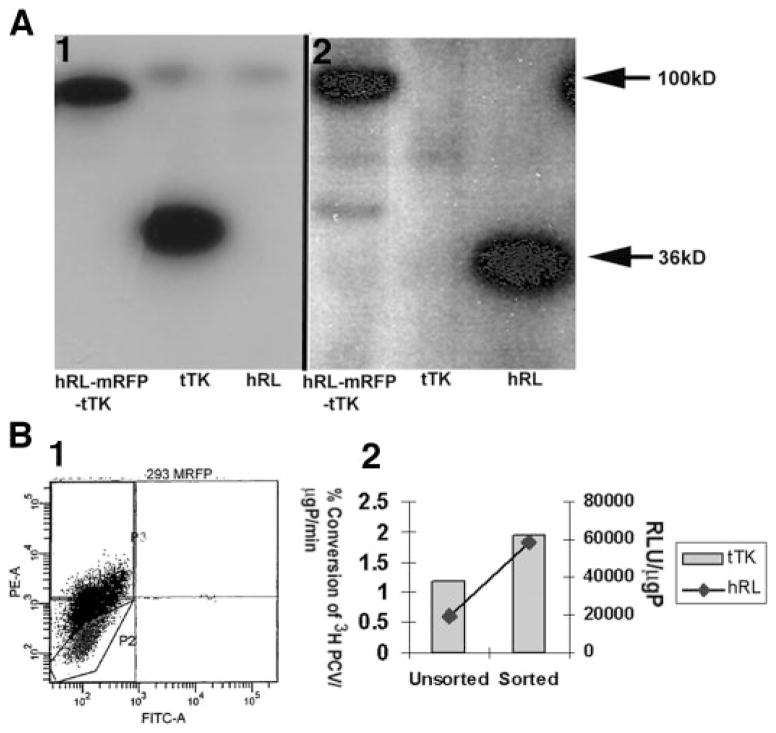
Western Blot Analysis of Extracts from Cells Transiently Transfected with hrl-mrfp-ttk Probed with Anti-TK and Anti-RL Antibodies Reveals the Presence of a 100-kDa Fragment, the Predicted Size of the Protein Encoded by the hrl-mrfp-ttk Fusion
To investigate hrl-mrfp-ttk fusion reporter gene expression at the protein level, cell lysates from hrl-mrfp-ttk-, ttk-, and hrl-transfected 293T cells were resolved by 10% SDS-PAGE and analyzed on Western blots by using antibodies specific for TK and RL (Fig. 2A). The predominant band in the triple fusion sample recognized by anti-TK (Fig. 2A.1) and anti-RL (Fig. 2.A.2) antibodies is about 100 kDa, the expected size of the hRL-mRFP-tTK fusion protein. The tTK and the hRL proteins were recognized at about 36 kDa band by their specific antibodies. Similar results were also obtained from N2a and A375M cell extracts (data not shown).
Fig. 2.
Biochemical and flow cytometric characterization of hrl-mrfp-ttk fusion reporter gene expression. A, Western blot analysis of hRL-mRFP-tTK fusion protein. Twenty μg of total cellular protein obtained from the cell lysates of transiently transfected 293T cells with hrl-mrfp-ttk, ttk, and hrl plasmids were resolved in a 10% SDS poly-acrylamide gel and transferred and probed with anti-TK (A.1) and anti-RL (A.2) antibodies. A 100-kDa band was specifically recognized by the antibodies only from the hRL-mRFP-tTK fusion samples. The polyclonal anti-TK and the monoclonal anti-RL antibody recognize tTK (second lane in A.1) and hRL (third lane in A.2) at about 36 kDa, respectively. B, flow cytometry plot and enzymatic activities of the positive expressers of CS-hrl-mrfp-ttk (lentiviral vector)-infected 293T cells. One million 293T cells were infected with CS-hrl-mrfp-ttk vector and sorted with fluorescence-activated cell sorting with a filter at 585 ± 42 nm band setting. Highly fluorescing cells (~33%) that migrated to the P3 sector (B.1) were collected and further tested for tTK and hRL enzyme activities (B.2). The sorted fraction showed higher tTK and hRL activities than the unsorted population. The TK activity is expressed as (percentage of conversion of [8-3H]penciclovir to its phosphorylated form)/μg protein/min. The RL activity is expressed as relative light units (RLU)/μg protein.
A Lentiviral Vector Carrying the hrl-mrfp-ttk Triple Fusion Is Able to Stably Transfect 293T and A375M Cells, and the Positive Expressers Can Be Sorted by FACS Analysis
One potential use of a lentiviral vector is to deliver the genes of interest to any type of dividing or nondividing cell lines or target tissues in living animals. Therefore, the full-length fusion gene cassette was cloned in the NheI and XhoI site of a second-generation lentiviral vector (11), and viruses carrying the triple fusion reporter gene were used to transduce 293T cells. Five million 293T cells infected with lentivirus were sorted by FACS using a 585 ± 42 nm filter setting, and 33% positive expressers were collected (Fig. 2B.1). The sorted and unsorted cells were then assayed for tTK and hRL expression (Fig. 2B.2) and clearly show a significant (P < 0.05) gain in expression in the sorted cell population. Similarly, the A375M cells were transduced with the lentivirus carrying the triple fusion reporter gene, and positive expressers were selected by two rounds of FACS sorting and verifed to have significant hRL and tTK activities (data not shown).
293T Cells Transiently Expressing the hrl-mrfp-ttk Fusion Reporter Gene Can Be Imaged in Living Mice with the MicroPET and Optical Cooled CCD Imaging Systems
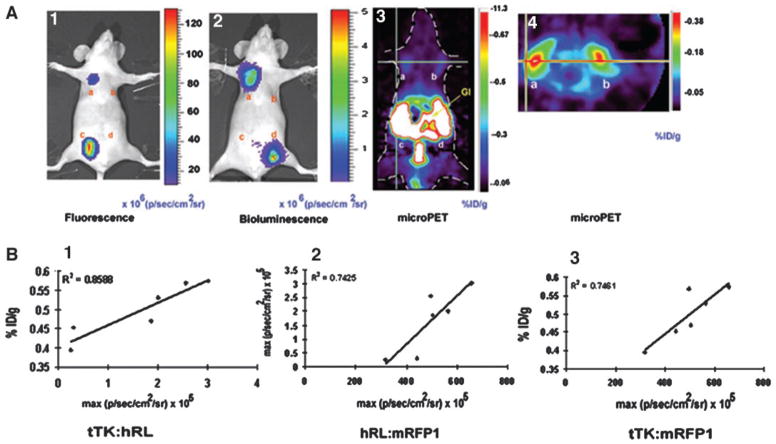
Our aim of building a fusion reporter vector was to test its efficacy for simultaneous imaging of reporter gene expression quantitatively and repeatedly in living subjects using different modalities. We therefore injected 10 × 106 293T cells transiently transfected with either CMV-hrl-mrfp-ttk, CMV-ttk, CMV-hrl, or CMV-mrfp1 vectors s.c. at four different sites on the ventral sides of four 12–14-week-old nu/nu nude mice. The mice were first scanned using the cooled CCD camera for fluorescence followed by a bioluminescence scan after injection of 20 μg of coelenterazine via tail vein. Fluorescence imaging of these mice reveals that the cells expressing the hrl-mrfp-ttk fusion (Fig. 3A.1, a) show lower fluorescence [~11.03 ± 6.03 × 108 maximum (p/s/cm2/sr)] in comparison with the cells expressing mrfp1 vector (Fig. 3A.1, c) alone [~65.8 ± 32.29 × 108 maximum (p/s/cm2/sr); Fig. 3A.1]. No significant signal is observed from the other two sites of implantation carrying the CMV-ttk (Fig. 3A.1, b)- and CMV-hrl (Fig. 3A.1, d)-expressing cells. A bioluminescence scan of the mice shows a signal of 5.8 ± 3.7 × 106 maximum (p/s/cm2/sr) from the cells expressing the fusion reporter gene (Fig. 3A.2, a) and about 7.37 ± 4 × 106 maximum (p/s/cm2/sr) from the CMV-hrl-expressing cells (Fig. 3A.2, d). Similar to fluorescence imaging, the other two sites carrying CMV-mrfp1 (Fig. 3A.2, c)- and CMV-ttk (Fig. 3A.2, b)-expressing cells did not show any significant bioluminescence signal. Because the FHBG mass used for PET imaging is 1000-fold lower (due to the presence of radioactive isotope) than the coelenterazine mass used for bioluminescence imaging, PET imaging is not as sensitive as bioluminescence at superficial depths (1). Furthermore, PET imaging benefits from well-vascularized tissues with relatively high levels of reporter gene expression. We therefore implanted the cells expressing the fusion gene and ttk gene in the right and left axillary region of the mice. We quantified the signal from each of the sites expressing the CMV-hrl-mrfp-ttk and CMV-ttk directly from the microPET images to determine the percentage of ID/g tumor for FHBG. This percentage of ID/g is a measure of the amount of tracer accumulated in a given tissue site normalized to the injected amount and to the mass of the tissue examined. The mean percentage of ID/g for FHBG accumulation in the CMV-hrl-mrfp-ttk-expressing cells (0.303 ± 0.09; Fig. 3A.3, a and Fig. 3A.4, a) did not differ significantly from that of the CMV-ttk-expressing cells (0.313 ± 0.09; Fig 3A.3, b and Fig. 3A.4, b) for the four mice (Fig. 3A.3 and 3A.4). Preservation of a high level of tTK activity and moderate levels of hRL and mRFP1 activities by this tri-fusion vector thus allows simultaneous imaging of transient expression of all three of the reporter genes in living mice with all three of the imaging techniques. Repetitive imaging of the same mouse over a 10-day period produced signals for all three of the reporter genes that increased with time as the tumor burden increased (data not shown).
Fig. 3.
Results of imaging living mice. A, fluorescence, bioluminescence, and micro-positron emission tomography (PET) imaging of hrl-mrfp-ttk expression in the same living nude mouse. Ten million 293T cells transiently expressing the CMV-hrl-mrfp-ttk, CMV-ttk, CMV-mrfp1, and CMV-hrl plasmids were implanted s.c. at four sites on the ventral side of a nude mouse and imaged the next day for fluorescence/bioluminescence and PET using a cooled charge-coupled device (CCD) camera and microPET, respectively. Fluorescence imaging was performed by placing the mouse in a CCD camera for 1 s, and a fluorescence image was acquired with a excitation filter at 500–550 nm and an emission filter at 575– 650 nm. Cells expressing the fusion (A.1, a) and mrfp1 (A.1, c) genes showed fluorescence, and the signal is recorded as maximum photons/sec/cm2/sr (A.1). The same mouse was then scanned in the CCD camera for bioluminescence after injection of coelenterazine via tail vein, and bioluminescence signal was found in cells expressing the fusion (A.2, a) and hrl (A.2, d) and recorded as maximum photons/sec/cm2/sr (A.2). After the optical scan, the same mouse was imaged by microPET using 9-(4-[18F]fluoro-3-hydroxymethylbutyl)guanine (FHBG). Cells expressing the fusion reporter gene (A.3, a and A.4, a) and ttk gene (A.3, b and A.4, b) showed FHBG accumulation in coronal section (A.3) and trans-axial section (A.4). Nonspecific accumulation of tracer was found in the gastrointestinal tracts and bladder (attributable to clearance of FHBG; A.3). B, in vivo correlation of hrl, mrfp1, and ttk gene expression exhibited by four clones of 293T cells stably but differentially expressing the hrl-mrfp-ttk fusion. Ten million cells of each clone were implanted on the axillary regions of the ventral side of three nude mice (two clones in each mouse), and after 24 h, mice were imaged by the cooled CCD camera and microPET. Plots of percentage of ID/g versus bioluminescence [expressed as maximum photons/second/centimeter2/steradian (p/s/cm2/sr); B.1], bioluminescence versus fluorescence (both expressed as maximum p/s/cm2/sr; B.2), and percentage of ID/g versus fluorescence (expressed as maximum p/s/cm2/sr; B.3) were obtained from the regions of interest drawn over the regions of cell implantation. Each of the six data points of each plot represents region of interest data from the fluorescence, bioluminescence, and microPET images of the same living mouse, with a total of three mice (2 points/mouse)
The lentivirus-infected 293T cells stably expressing the hrl-mrfp-ttk fusion gene described in the previous section were diluted to single cells, and four clones with differential expression of all three genes were selected. In cell culture, these four clones exhibited good correlation between tTK and hRL (R2 = 0.96), hRL and mRFP1 (R2 = 0.94), and tTK and mRFP1 (R2 = 0.86) activities (data not shown). For quantification of mRFP1 activity, 3.5 × 105 cells of each clone and 293T cells were seeded in black, clear-bottomed 96-well plates in triplicate and imaged in the CCD camera with DsRed2 excitation and emission filter options. ROIs were drawn on each well, and fluorescence was measured quantitatively for each group. The basal fluorescence exhibited by 293T cells was subtracted from the fluorescence of each well of each clone, and the mean of the three wells of each clone was taken as absolute fluorescence activity, expressed as maximum (p/s/cm2/sr) for each clone. tTK and hRL assays were performed as described earlier. To measure the correlation between the expression of the three reporter genes across three different imaging modalities, 10 × 106 cells of each clone were implanted on the two axillary regions on the ventral side of three nude mice (two clones in each mouse), and mice were imaged for fluorescence, bioluminescence, and microPET on the same day as described above. The fluorescence and bioluminescence signals and the percentage of ID/g values for FHBG of the hrl-mrfp-ttk expressing clones across the three mice were all well correlated: R2 = 0.85 (hRL:tTK; Fig. 3B.1); R2 = 0.74 (hRL:mRFP1; Fig. 3.B.2); and R2 = 0.74 (tTK:mRFP1; Fig. 3B.3).
Metastasis of A375M Human Melanoma Cells Expressing the hrl-mrfp-ttk Reporter Gene Can Be Imaged by MicroPET and an Optical CCD Camera in Living Mice
To apply the tri-fusion strategy to a relevant preclinical cancer study, we used a melanoma metastatic model. A375M human melanoma cells are known to metastasize to other organs once injected in the animal i.v. and form pulmonary and brain metastases with some rare occurrence of bone metastases (14, 15). A375M cells (7 × 105) stably expressing the hrl-mrfp-ttk reporter gene were injected in three 8-week-old Beige severe combined immunodeficient mice via tail vein. On the first day of cell injection, bioluminescence signal was detectable from the lungs (the primary route of cell migration; Fig. 4A), but not from microPET images (data not shown). The mice were then subsequently imaged over time every 6–7 days for a period of 40–50 days. At day 40, moderate microPET signal (~0.35% ID/g) from the lungs and strong signal (~0.78% ID/g) from the chest region are detected from one of the three mice (Fig. 4C). A corresponding bioluminescence signal (2 × 105 p/s/cm2/sr) is detected from the lungs of the same mouse on the same day (Fig. 4B). The relatively high PET signal from the chest region (Fig. 4C) is not evident with bioluminescence imaging (Fig. 4C), likely due to relatively poor penetration of light produced by Renilla luciferase from greater depths. A faint bioluminescence signal (5 × 103 p/s/cm2/sr) was also seen from the pelvic region of the mouse that was undetectable in microPET, likely due to hindrance by the nonspecific signal due to FHBG tracer clearance from the kidneys and gastrointestinal tract. In vivo fluorescence imaging of metastases did not produce good images due to significant autofluorescence caused by the presence of hair. However, when the mouse was sacrificed, and internal organs were exposed, several small metastatic tumors were found with fluorescence (Fig. 4E). Among the other two mice, one showed bioluminescence signal in the abdominal region at day 48; however we could not detect specific microPET signal, likely due to the presence of moderate levels of nonspecific signal resulting from the clearence of FHBG through kidneys and the gastrointestinal tract.
Fig. 4.

Multimodality imaging of metastasis of A375M cells stably expressing the hrl-mrfp-ttk fusion reporter gene in living mice. A, bioluminescence imaging of a SCID mouse injected with A375M cells expressing the hrl-mrfp-ttk vector at day 0. 7 × 105 A375M cells stably expressing the triple fusion were injected via tail-vein in a SCID mouse and two hours later imaged for bioluminescence signal following tail-vein injection of coelenterazine. Prominent bioluminescence signal was found from the region of both the lungs [1.3–1.5 × 105 max (p/sec/cm2/sr)]. B, bioluminescence imaging of the same SCID mouse at day 40. At day 40, the same mouse was imaged and relatively high bioluminescence signal [2 × 105 max (p/sec/cm2/sr)] was found from the left lung region and moderate signal from the right lung region. A faint bioluminescence signal (5 × 103 p/sec/cm2/sr) was also present from the right pelvic region. C, microPET imaging of the same SCID mouse at day 40. Following a bioluminescence scan, the mouse was imaged in microPET using FHBG. Shown is a thin coronal slice of ~1-mm thickness. A strong signal (~0.78% ID/g) was present from the chest region (Ch) with lower signal (0.35% ID/g) from the lung region. The stronger PET signal was found to be from a metastatic tumor present deep inside the body, as evident from the fluorescence photograph (4.E). Note the gallbladder (GB) retains FHBG so background signal from the GB is also seen in the microPET images. D, light photograph of the same SCID mouse after sacrifice and organ exposure (image has been modified by using Adobe Photoshop version 6). E, whole body fluorescence imaging of the same SCID mouse. Fluorescing metastatic tumors were found in lung and chest regions that correspond with the bioluminescence and PET images.
DISCUSSION
In this study, we report the construction of several novel triple fusion reporter genes including one harboring a bioluminescence reporter gene (synthetic Renilla luciferase), a fluorescence reporter gene (monomeric red fluorescence protein), and a PET reporter gene (truncated version of HSV1-sr39 thymidine kinase) and validate its application in living cells (cell microscopy and FACS) and in living mice using three different small animal imaging technologies (in vivo fluorescence, in vivo bioluminescence, and microPET). Use of bi-fusion reporter genes for molecular imaging has been validated previously by us (7) and by other investigators (16, 17). These previous approaches have been limited by the inability to image single cells (7) or to take advantage of the low background signal with bioluminescence (16) or the advantage of tomographic imaging with PET (17). However, the ability to have a fluorescence, bioluminescence, and PET signal provides the full spectrum of coverage needed for many reporter gene applications. One can use this tri-fusion reporter gene to sort cells and to image in small living animals using either fluorescence or bioluminescence and in larger subjects, including humans, using PET. The ability to move between imaging technologies without having to use a different reporter gene for each application will greatly simplify various biological models including transgenics, cell trafficking, anticancer pharmaceutical research, and gene therapy.
Although our previous bi-fusion reporter construct (tk20rl) showed well-correlated expression of PET and bioluminescence imaging modalities (7), it was somewhat limited by decreased TK activity and was susceptible to enzymatic cleavage into its component proteins. By changing the orientation of the fusion partner in the current vector, we could gain a significant amount of TK activity, indicating that the COOH-terminal end of thymidine kinase protein may be crucial for ensuring TK enzyme activity. In contrast, the hRL activity of the current construct showed a decrease in enzyme activity as opposed to our previous vector, which showed a gain in RL activity. However, this new synthetic version of Renilla luciferase (hRL) is 40–50-fold more active than original Renilla luciferase (18), and therefore a drop in RL activity did not affect the efficacy of this vector significantly. Moreover, the tk gene in this triple fusion vector has a deletion of the first 135 bp that contains a nuclear localization signal and a cryptic testis-specific transcriptional start point (19, 20). Thus, this deletion leads to more cytoplasmic localization of TK enzyme, likely resulting in more TK activity (21) due to the availability of greater amount of substrate (FHBG). This deletion mutant will also likely overcome the problem of male sterility in transgenic mice carrying the thymidine kinase gene due to production of a shorter transcript in testis from a cryptic transcriptional initiation site (20) present in the first 135 bp of the gene. Another added advantage of this vector over our previous one and other vectors reported in the literature is that it can retain its integrity as a fusion protein when expressed, so that signal from each component of the tri-fusion protein will not be susceptible to problems related to cleavage. The absence of cleavage of the triple fusion vector is likely due to change of certain amino acids (Cys-Gly to Ser-Thr) present in the spacer in contrast to the previously reported 20-aa spacer of the tk20rl vector.
In the process of building a better multimodality vector, we constructed several other fusion vectors (see Table 1). Most of these vectors had lower tTK, luciferase, and RFP activity, probably due to the inherent nature of RFP (DsRed2) of forming obligate tetramer for proper maturation of the flurophores (22) present in the fusion genes. The tetrameric nature of RFP present in hrl/fl-rfp-ttk fusions might impose structural and functional constrains on the other partner proteins resulting in decreased TK and luciferase activity. Our hrl/fl-egfp-ttk vectors did show a better TK and luciferase activity than hrl/fl-rfp-ttk vectors, due to the monomeric nature of eGFP proteins, but did not show better activity than the hrl/fl-mrfp-ttk fusion vectors. Moreover, the excitation and emission spectra of GFP (λex, 489 nm; λem, 508 nm) is not as favorable for fluorescence imaging in living subjects as compared with RFP and mRFP because of the better penetration of red and near-infrared light in tissues (1). We also consistently observed a drop in tTK and RFP activity of the triple fusions with firefly luciferase in comparison with the fusions bearing Renilla luciferase. Fusion reporter vectors bearing truncated wild-type thymidine kinase also preserved a better wild-type thymidine kinase and luciferase activity (with both firefly and Renilla), and these vectors should be useful in the future when using other substrates (e.g., 2′-fluoro-5-fluoro-1-β-D-arbinofuranosyluracil/2′-deoxy-2′-fluoro-5-fluoro-1-β-D-arbinofuranosyluracil) that are more sensitive when used with wild-type thymidine kinase (23). It is likely that one tri-fusion will not serve the needs for all applications, and investigators will need to choose from a library of tri-fusions for a given application.
One of the potential uses of the multimodality reporter vectors in gene therapy is to target any type of cell line or tissues and then follow gene expression using a multimodality approach. Viral vectors, especially the lentiviral ones, are among the most standardized and widely used vectors to deliver any gene of interest to target tissues or an organism and to isolate cells, particularly nondividing cells stably expressing the gene. Recently, a bicistronic lentiviral vector carrying tk and fl reporter genes has been successfully used for PET and bioluminescence imaging in our laboratory (11). The new lentiviral construct carrying the triple fusion gene reported in this work has been used successfully with FACS analysis to isolate lentiviral infected 293T and A375M cells stably expressing the triple fusion reporter. This lentiviral construct should have tremendous potential in wide variety of research applications. Our preliminary data with the A375M metastatic melanoma model further confirm the usefulness of this lentiviral vector carrying triple fusion reporter gene to follow progression of cancer metastases by molecular imaging. Extensions of this study with drug treatment are currently in progress (24).
Use of light is probably the oldest method of analyzing tissues in biomedical science (25). The various optical imaging approaches including fluorescence microscopy (at the cellular level), diffuse optical tomography, and intravital microscopy (for deeper structures at the organism level) are commonly used (26, 27). However, intrinsic absorption and scattering of light through the tissues and autofluorescence properties of biological molecules (e.g., tryptophan, collagen, elastin, nicotineamide adenine dinucleotide, hemoglobin, oxyhemoglobin, and so forth) impose certain restrictions for using fluorescence as an imaging tool in small living subjects. However, both light attenuation and autofluorescence decline as wavelength increases, especially in the red to near-infrared region (>600 nm). A fluorescence protein/fluorochrome with excitation and emission toward red (560 nm onward) has better penetrability through the tissues than that with excitation and emission in the blue or green region. Moreover, hemoglobin and water, which are responsible for the highest absorption of light among all other biological molecules, have their lowest coefficient of absorption in the red and near-infrared region. Therefore, in vivo fluorescence imaging is more suitable in the red and near-infrared region than in the green or yellow region. Optical imaging in living subjects at the near-infrared region (650–900 nm) has therefore been used extensively by applying different fluorochromes that emit light at near-infrared region spectra (28) and in combination with other imaging modalities (29). However, synthesis and attachment of these fluorochromes to proteins/copolymers require complex chemical procedures and are more difficult to generalize. Also, these strategies are not directly applicable to genetically encoded reporters. The mRFP1 protein used in this study has an excitation and emission range in the far-red region (584–607 nm) and thus is one of the better reporter gene choices for fluorescence imaging in living subjects (9). The monomeric nature of this protein also confers a better functional preservation as fusion partner as compared with RFP or DsRed2 (tetramer) and HcRed (dimer). However, we still observed a significant amount of autofluorescence from the mouse relative to bioluminescence, which is not limited by autofluorescence, and therefore bioluminescence produced a better signal:background ratio in living animals. However, bioluminescence imaging of gene expression in a single cell is not easily possible due to generation of relatively low amounts of light. Therefore, the current results would support using the fluorescence component for cell imaging/sorting with limited in vivo imaging, bioluminescence for small animal imaging even with a very few number of cells, and PET for tomographically imaging living subjects including larger animals and humans.
Our cancer metastatic model shows that metastases can be imaged by microPET and bioluminescence in living mice using this triple fusion reporter gene, with certain limitations for each technique. The bioluminescence signal from Renilla luciferase is not detectable from metastases present at greater depths but is easily detectable from superficial metastases from any region of the body. On the other hand, microPET reveals metastases from deep inside the body, but signals from metastases in the abdomen/pelvis are somewhat obscured by the nonspecific signal in the gastrointestinal tract and urinary collecting system due to tracer clearance. Finally, autofluorescing properties of biological molecules limit detection of metastases by in vivo fluorescence imaging in living animals. However, metastases can be easily visualized in sacrificed animals with exposed tissues in situ using whole body fluorescence imaging. It is likely that the fluorescence signal to background can be improved by using an excitation source under the animal and imaging with a camera above the animal to help minimize autofluorescence. The bioluminescence signal can also be markedly improved by injecting higher doses of substrate (coelenterazine), as we have demonstrated recently (18). As red-shifted bioluminescent reporters with high substrate utilization capacity are developed, this will also likely aid in helping to use bioluminescence-based reporter fusions. In addition, the microPET signal can be improved by using tracers with longer half-lives (e.g., 124I-labeled 2′-fluoro-5-fluoro-1-β-D-arbinofuranosyluracil) to allow the background signal from the gastrointestinal tract and renal collecting system to be reduced by waiting longer after tracer injection before imaging animals. With continued refinement in reporter genes, substrates for reporter proteins, and physical instrumentation, it is likely that higher spatial resolution imaging with greater sensitivity for detecting smaller numbers of cells will eventually be possible.
Additional studies quantitatively comparing fluorescence, bioluminescence, and PET in small living animals should also help to better define the potential roles of each modality in specific applications. Cancer research, including imaging of preclinical models of tumors and metastases, immune cell trafficking, transgenic models, gene therapy, and monitoring therapy in general should all benefit from the strategies developed in the current work.
Acknowledgments
Grant support: Grant DE-FG03-01ER63276 (to R. Y. Tsien); NIH Grants P50 CA86306 (to S. S. Gambhir), R01 CA82214 (to S. S. Gambhir), and SAIRP R24 CA92865 (to S. S. Gambhir); and Department of Energy Contracts DE-FC03-87ER60615 and DE-FG02-03ER63687 (to S. S. Gambhir).
We thank W. Ladno, J. Edwards, X. Lewis, and D. Stout for technical assistance. We appreciate the kind help of Ian Chen and Shariar Yaghoubi. The members of the FACS core facility of University of California Los Angeles-Jonsson Comprehensive Cancer Center are duly acknowledged for their help.
References
- 1.Massoud T, Gambhir S. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Van Roessel P, Brand A. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat Cell Biol. 2002;4:E15–E20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 3.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 4.Bremer C, Weissleder R. In vivo imaging of gene expression. Acad Radiol. 2001;8:15–23. doi: 10.1016/s1076-6332(03)80739-0. [DOI] [PubMed] [Google Scholar]
- 5.Vooijs M, Jonkers J, Lyons S, Berns A. Noninvasive imaging of spontaneous retinoblastoma pathway-dependent tumors in mice. Cancer Res. 2002;62:1862–1867. [PubMed] [Google Scholar]
- 6.Bouvet M, Wang J, Nardin SR, Nassirpour R, Yang Meng, Baranov E, Jiang P, Moossa AR, Hoffman RM. Real-time optical imaging of primary tumor growth and multiple metastatic events in a pancreatic cancer orthotopic model. Cancer Res. 2002;62:1534–1540. [PubMed] [Google Scholar]
- 7.Ray P, Wu A, Gambhir S. Optical bioluminescence and positron emission tomography imaging of a novel fusion reporter gene in tumor xenografts of living mice. Cancer Res. 2003;63:1160–1165. [PubMed] [Google Scholar]
- 8.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell RE, Tour O, Palmer AE, Steinbach PA, Geoffrey S, Baird GS, David AZ, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaghoubi S, Barrio JR, Dahlbom M, Iyer M, Namavari M, Satyamurthy N, Goldman R, Herschman HR, Phelps ME, Gambhir SS. Human pharmacokinetic and dosimetry studies of [18F]FHBG: a reporter probe for imaging herpes simplex virus type-1 thymidine kinase reporter gene expression. J Nucl Med. 2001;42:1225–1234. [PubMed] [Google Scholar]
- 11.De A, Lewis XH, Gambhir SS. Noninvasive imaging of lentiviral-mediated reporter gene expression in living mice. Mol Ther. 2003;7:681–691. doi: 10.1016/s1525-0016(03)00070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR, Iyer M, Namavari M, Phelps ME, Herschman HR. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci USA. 2000;97:2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi JY, Leahy RM, Cherry SR, Chatziioannou A, Farquhar TH. High-resolution 3D Bayesian image reconstruction using the microPET small-animal scanner. Phys Med Biol. 1998;43:1001–1013. doi: 10.1088/0031-9155/43/4/027. [DOI] [PubMed] [Google Scholar]
- 14.Seftor RE, Seftor EA, Hendrix MJ. Molecular role(s) for integrins in human melanoma invasion. Cancer Metastasis Rev. 1999;18:359–375. doi: 10.1023/a:1006317125454. [DOI] [PubMed] [Google Scholar]
- 15.Giavazzi R, Garofalo A, Bani MR, Abbate M, Ghezzi P, Boraschi P, Mantovani A, Dejana E. Interleukin-1 induced augmentation of experimental metastases from a human melanoma in nude mice. Cancer Res. 1990;50:4771–4775. [PubMed] [Google Scholar]
- 16.Jacobs A, Dubrovin M, Hewett J, Sena-Esteves M, Tan CW, Slack M, Sadelain M, Breakefield XO, Tjuvajev JG. Functional coexpression of HSV-1 thymidine kinase and green fluorescent protein: implications for noninvasive imaging of transgene expression. Neoplasia. 1999;1:154–161. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Yu Y, Shabahang S, Wang G, Szalay A. Renilla luciferase-Aequorea GFP (Ruc-GFP) fusion protein, a novel dual reporter for real-time imaging of gene expression in cell cultures and in live animals. Mol Genet Genomics. 2002;268:160–168. doi: 10.1007/s00438-002-0751-9. [DOI] [PubMed] [Google Scholar]
- 18.Bhaumik S, Lewis XZ, Gambhir SS. Optical imaging of synthetic Renilla luciferase reporter gene expression in living mice. J Biomed Optics. 2004 doi: 10.1117/1.1647546. in press. [DOI] [PubMed] [Google Scholar]
- 19.Degreve B, Johansson M, Declercq E, Karlsson A, Balzarini J. Differential intracellular compartmentalization of herpetic thymidine kinases (TKS) in TK gene-transfected tumor cells. Molecular characterization of the nuclear localization signal of the herpes simplex virus type 1 TK. J Virol. 1998;72:9535–9543. doi: 10.1128/jvi.72.12.9535-9543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen JL, Boyer O, Salomon B, Onclercq R, Depetris D, Lejeune L, Dubus-Bonnet V, Bruel S, Charlotte F, Mattei MG. Fertile homozygous transgenic mice expressing a functional truncated herpes simplex thymidine kinase ΔTK gene. Transgenic Res. 1998;7:321–330. doi: 10.1023/a:1008893206208. [DOI] [PubMed] [Google Scholar]
- 21.Luker GD, Sharma V, Pica CM, Dahlheimer JL, Li W, Ochesky J, Ryan CE, Piwnica-Worms H, Piwnica-Worms D. Noninvasive imaging of protein-protein interactions in living animals. Proc Natl Acad Sci USA. 2002;99:6961–6966. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacchetti A, Subramaniam V, Jovin T, Alberti S. Oligomerization of DsRed is required for the generation of a functional red fluorescent chromophore. FEBS Lett. 2002;525:13–19. doi: 10.1016/s0014-5793(02)02874-0. [DOI] [PubMed] [Google Scholar]
- 23.Tjuvajev JG, Doubrovin M, Akhurst T, Cai S, Balatoni J, Alauddin MM, Finn R, Bornmann W, Thaler H, Conti PS, Blasberg RG. Comparison of radiolabeled nucleoside probes (FIAU, FHBG, and FHPG) for PET imaging of HSV1-tk gene expression. J Nucl Med. 2002;43:1072–1083. [PubMed] [Google Scholar]
- 24.De A, Collison E, Kolodney M, Gambhir S. Lentiviral reporter gene delivery as a novel way of studying therapeutic effects on cancer metastasis by noninvasive imaging. Mol Ther. 2003;7:S137. [Google Scholar]
- 25.Anderssonengels S, Afklinteberg C, Svanberg K, Svanberg S. In vivo fluorescence imaging for tissue diagnostics. Phys Med Biol. 1997;42:815–824. doi: 10.1088/0031-9155/42/5/006. [DOI] [PubMed] [Google Scholar]
- 26.Boas DA, Brooks DH, Miller EL, DiMarzio CA, Kilmer M, Gaudette RJ, Zhang Q. Imaging the body with diffuse optical tomography. IEEE Signal Processing Magazine. 2001;18:57–75. [Google Scholar]
- 27.Jain RK, Munn LL, Fukumura D. Dissecting tumour pathophysiology using intravital microscopy. Nat Rev Cancer. 2002;2:266–276. doi: 10.1038/nrc778. [DOI] [PubMed] [Google Scholar]
- 28.Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol. 2002;13:195–208. doi: 10.1007/s00330-002-1524-x. [DOI] [PubMed] [Google Scholar]
- 29.Zaheer A, Lenkinski RE, Mahmood A, Jones AG, Cantley LC, Frangioni JV. In vivo near-infrared fluorescence imaging of osteoblastic activity. Nat Biotechnol. 2001;19:1148–1154. doi: 10.1038/nbt1201-1148. [DOI] [PubMed] [Google Scholar]