Abstract
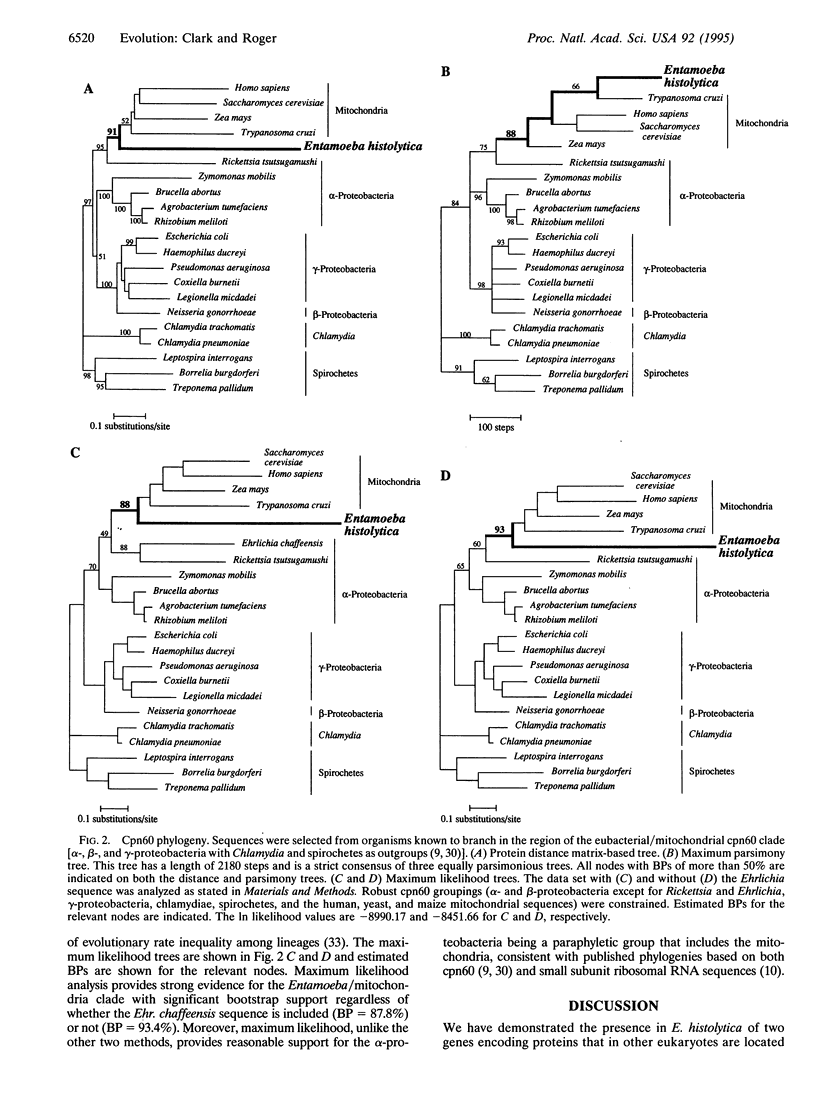
Archezoan protists are though to represent lineages that diverged from other eukaryotes before acquisition of the mitochondrion and other organelles. The parasite Entamoeba histolytica was originally included in this group. Ribosomal RNA based phylogenies, however, place E. histolytica on a comparatively recent branch of the eukaryotic tree, implying that its ancestors had these structures. In this study, direct evidence for secondary loss of mitochondrial function was obtained by isolating two E. histolytica genes encoding proteins that in other eukaryotes are localized in the mitochondrion: the enzyme pyridine nucleotide transhydrogenase and the chaperonin cpn60. Phylogenetic analysis of the E. histolytica homolog of cpn60 confirmed that it is specifically related to the mitochondrial lineage. The data suggest that a mitochondrial relic may persist in this organism. Similar studies are needed in archezoan protists to ascertain which, if any, eukaryotic lineages primitively lack mitochondria.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakker-Grunwald T., Wöstmann C. Entamoeba histolytica as a model for the primitive eukaryotic cell. Parasitol Today. 1993 Jan;9(1):27–31. doi: 10.1016/0169-4758(93)90161-8. [DOI] [PubMed] [Google Scholar]
- Bruchhaus I., Leippe M., Lioutas C., Tannich E. Unusual gene organization in the protozoan parasite Entamoeba histolytica. DNA Cell Biol. 1993 Dec;12(10):925–933. doi: 10.1089/dna.1993.12.925. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993 Dec;57(4):953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D. M., Loo T. W., Gillam S., Bragg P. D. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur J Biochem. 1986 Aug 1;158(3):647–653. doi: 10.1111/j.1432-1033.1986.tb09802.x. [DOI] [PubMed] [Google Scholar]
- Diamond L. S., Harlow D. R., Cunnick C. C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72(4):431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Ellis J. E., Setchell K. D., Kaneshiro E. S. Detection of ubiquinone in parasitic and free-living protozoa, including species devoid of mitochondria. Mol Biochem Parasitol. 1994 Jun;65(2):213–224. doi: 10.1016/0166-6851(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Harlow D. R., Weinbach E. C., Diamond L. S. Nicotinamide nucleotide transhydrogenase in Entamoeba histolytica, a protozoan lacking mitochondria. Comp Biochem Physiol B. 1976;53(2):141–144. doi: 10.1016/0305-0491(76)90024-9. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Fujiwara M. Relative efficiencies of the maximum likelihood, maximum parsimony, and neighbor-joining methods for estimating protein phylogeny. Mol Phylogenet Evol. 1993 Mar;2(1):1–5. doi: 10.1006/mpev.1993.1001. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Hashimoto T., Adachi J., Iwabe N., Miyata T. Early branchings in the evolution of eukaryotes: ancient divergence of entamoeba that lacks mitochondria revealed by protein sequence data. J Mol Evol. 1993 Apr;36(4):380–388. doi: 10.1007/BF00182185. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Huelsenbeck J. P., Cunningham C. W. Application and accuracy of molecular phylogenies. Science. 1994 Apr 29;264(5159):671–677. doi: 10.1126/science.8171318. [DOI] [PubMed] [Google Scholar]
- Jackson J. B. The proton-translocating nicotinamide adenine dinucleotide transhydrogenase. J Bioenerg Biomembr. 1991 Oct;23(5):715–741. doi: 10.1007/BF00785998. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992 Jun;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kramer R. A., Tomchak L. A., McAndrew S. J., Becker K., Hug D., Pasamontes L., Hümbelin M. An Eimeria tenella gene encoding a protein with homology to the nucleotide transhydrogenases of Escherichia coli and bovine mitochondria. Mol Biochem Parasitol. 1993 Aug;60(2):327–331. doi: 10.1016/0166-6851(93)90144-m. [DOI] [PubMed] [Google Scholar]
- Lahti C. J., Bradley P. J., Johnson P. J. Molecular characterization of the alpha-subunit of Trichomonas vaginalis hydrogenosomal succinyl CoA synthetase. Mol Biochem Parasitol. 1994 Aug;66(2):309–318. doi: 10.1016/0166-6851(94)90157-0. [DOI] [PubMed] [Google Scholar]
- Länge S., Rozario C., Müller M. Primary structure of the hydrogenosomal adenylate kinase of Trichomonas vaginalis and its phylogenetic relationships. Mol Biochem Parasitol. 1994 Aug;66(2):297–308. doi: 10.1016/0166-6851(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Müller M. Energy metabolism of protozoa without mitochondria. Annu Rev Microbiol. 1988;42:465–488. doi: 10.1146/annurev.mi.42.100188.002341. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAY H. N., SEN GUPTA P. C. A cytochemical study of entamoeba histolytica. J Indian Med Assoc. 1954 Sep;23(12):529–533. [PubMed] [Google Scholar]
- Reeves R. E. Metabolism of Entamoeba histolytica Schaudinn, 1903. Adv Parasitol. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- Reiner D. S., Shinnick T. M., Ardeshir F., Gillin F. D. Encystation of Giardia lamblia leads to expression of antigens recognized by antibodies against conserved heat shock proteins. Infect Immun. 1992 Dec;60(12):5312–5315. doi: 10.1128/iai.60.12.5312-5315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. W., Feng D. F., Doolittle R. F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992 Dec;17(12):489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Sogin M. L. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991 Dec;1(4):457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Soltys B. J., Gupta R. S. Presence and cellular distribution of a 60-kDa protein related to mitochondrial hsp60 in Giardia lamblia. J Parasitol. 1994 Aug;80(4):580–590. [PubMed] [Google Scholar]
- Vermeulen A. N., Kok J. J., van den Boogaart P., Dijkema R., Claessens J. A. Eimeria refractile body proteins contain two potentially functional characteristics: transhydrogenase and carbohydrate transport. FEMS Microbiol Lett. 1993 Jun 15;110(2):223–229. doi: 10.1111/j.1574-6968.1993.tb06324.x. [DOI] [PubMed] [Google Scholar]
- Viale A. M., Arakaki A. K., Soncini F. C., Ferreyra R. G. Evolutionary relationships among eubacterial groups as inferred from GroEL (chaperonin) sequence comparisons. Int J Syst Bacteriol. 1994 Jul;44(3):527–533. doi: 10.1099/00207713-44-3-527. [DOI] [PubMed] [Google Scholar]
- Viale A. M., Arakaki A. K. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 1994 Mar 21;341(2-3):146–151. doi: 10.1016/0014-5793(94)80446-x. [DOI] [PubMed] [Google Scholar]
- Weinbach E. C., Takeuchi T., Elwood Claggett C., Inohue F., Kon H., Diamond L. S. Role of iron-sulfur proteins in the electron transport system of Entamoeba histolytica. Arch Invest Med (Mex) 1980;11(1 Suppl):75–81. [PubMed] [Google Scholar]
- Williams R., Cotton N. P., Thomas C. M., Jackson J. B. Cloning and sequencing of the genes for the proton-translocating nicotinamide nucleotide transhydrogenase from Rhodospirillum rubrum and the implications for the domain structure of the enzyme. Microbiology. 1994 Jul;140(Pt 7):1595–1604. doi: 10.1099/13500872-140-7-1595. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Hatefi Y., Trach K., Hoch J. A. Amino acid sequence of the signal peptide of mitochondrial nicotinamide nucleotide transhydrogenase as determined from the sequence of its messenger RNA. Biochem Biophys Res Commun. 1988 Nov 30;157(1):24–29. doi: 10.1016/s0006-291x(88)80005-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Hatefi Y., Trach K., Hoch J. A. The primary structure of the mitochondrial energy-linked nicotinamide nucleotide transhydrogenase deduced from the sequence of cDNA clones. J Biol Chem. 1988 Feb 25;263(6):2761–2767. [PubMed] [Google Scholar]
- Yu Y., Samuelson J. Primary structure of an Entamoeba histolytica nicotinamide nucleotide transhydrogenase. Mol Biochem Parasitol. 1994 Dec;68(2):323–328. doi: 10.1016/0166-6851(94)90178-3. [DOI] [PubMed] [Google Scholar]



