Abstract
The liver is a target of in vitro tissue engineering despite its capability to regenerate in vivo. The construction of liver tissues in vitro remains challenging. In this review, conventional 3D cultures of hepatocytes are first discussed. Recent advances in the 3D culturing of liver cells are then summarized in the context of in vitro liver tissue reconstruction at the micro- and macroscales. The application of microfluidics technology to liver tissue engineering has been introduced as a bottom-up approach performed at the microscale, whereas whole-organ bioengineering technology was introduced as a top-down approach performed at the macroscale. Mesoscale approaches are also discussed in considering the integration of micro- and macroscale approaches. Multiple parallel multiscale liver tissue engineering studies are ongoing; however, no tissue-engineered liver that is appropriate for clinical use has yet been realized. The integration of multiscale tissue engineering studies is essential for further understanding of liver reconstruction strategies.
Keywords: 3D culture, microfluidics, vascularization, tissue engineering
Introduction
It is well known that the liver has the capability to regenerate in vivo.1,2 However, on isolation and culturing in vitro, it is difficult to maintain the functions of liver cells because in vivo conditions are simply not fully reproduced by current in vitro culture techniques. Various key factors have been found to improve in vitro liver cell primary culture;3 a recent study identified small molecules for human hepatocyte expansion and differentiation.4 However, reconstructing an original three-dimensional (3D) structure by culturing hepatocytes and other liver cell types, such as endothelial cells (ECs) and hepatic stellate cells (HSCs), remains challenging.
Since the concept of tissue engineering was first reported,5 many studies have attempted to reconstruct liver tissues in vitro. The liver remains an important target of in vitro tissue engineering because liver transplantation remains the only effective therapy in cases of end-stage liver disease.6,7 In particular, the generation of microcirculation networks and their integration with engineered 3D tissues—i.e., vascularization—is of great interest. To achieve functional tissue-engineered livers, organization of the microstructure as well as large-volume tissue is important.
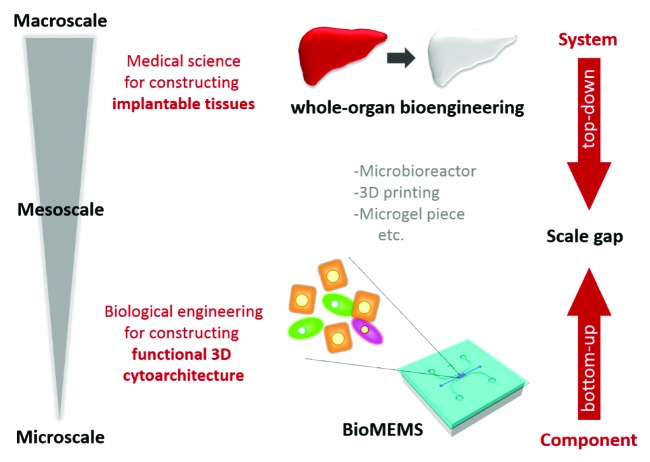
In this review, conventional 3D culture methods are first discussed. Recent advances in the 3D culturing of liver cells are then discussed in the context of liver tissue reconstruction in vitro at the micro- and macroscales. In particular, two types of complex culture models have been explored in recent tissue engineering research. One is the application of microelectromechanical system (MEMS) technology to address biological issues such as the construction of organized 3D tissues; this has been termed “bioMEMS technology.”8 BioMEMS focuses on the organization of microstructure in culture. Cellular components of the liver system are assembled in a stepwise manner to form tissue by controlling the cellular microenvironment, such as the spatial and temporal distribution of cells and biophysical and biochemical factors at the microscale. This bioMEMS approach constructs a whole system by building up cellular components, so is considered a bottom-up approach (Fig. 1). On the other hand, the second approach involves the use of decellularized whole-organ scaffolds because a whole-organ structure can be used as a scaffold to reconstruct liver tissue. This approach is performed at the macroscale and yields large-volume tissues. In this approach, the whole system resides in the culture model in which the cellular components must be organized. Becuase tissue construction is considered with directionality, from a system to the components, this whole-organ bioengineering technology is considered a top-down approach to liver tissue engineering (Fig. 1). Finally, mesoscale approaches are discussed, with consideration of the integration of micro- and macroscale approaches. Thus, recent liver tissue engineering studies have been performed at multiple scales (Fig. 1). Integration of these multiscale procedures is important for achieving the construction of functional liver tissues and organs that are appropriate for clinical use.
Figure 1. Multiscale tissue engineering. Tissue engineering research has been performed at multiscale. Bioengineers implement bioMEMS technologies to control the microenvironment of cells in culture, aiming to construct a functional 3D cytoarchitecture. These studies are performed mainly on a microscale, which can be regarded as a bottom-up approach. In contrast, medical scientists implement decellularized organs for tissue engineering, aiming to construct implantable tissues. This is performed on a macroscale, which can be regarded as a top-down approach. Although both methods aim to reconstruct liver tissues in vitro, there is a major scale gap between these approaches. Mesoscale approaches, such as microbioreactors, 3D printing technologies, and microgel piece technologies, should thus be implemented to bridge the gap.
Conventional 3D Culturing for the Construction of Liver Tissue
Spheroid culture, based on spontaneous tissue formation
It is widely recognized that culturing cells in 3D configurations better maintains their functions than culturing in 2D, because there are major differences between a flat layer of cells and a well-organized, complex 3D tissue.9,10 Hepatocytes reside in the liver, a highly organized structure, where they interact with neighboring hepatocytes, nonparenchymal cells, such as ECs and HSCs, and extracellular matrix (ECM) proteins. Thus, many 3D culture methods have been described for the construction of liver tissues in vitro.
The spheroid culture was one of the first attempts to culture hepatocytes in a 3D configuration. When seeded in a culture dish, hepatocytes usually attach and spread over the plastic surface. However, when the culture surface is treated to prevent attachment, hepatocytes instead attach to each other, resulting in the formation of multi-cellular aggregates known as hepatocyte spheroids. Although hepatocytes in two-dimensional (2D) culture rapidly lose their functions, hepatocytes in spheroids maintain their differentiated functions in long-term culture.11,12 Recent advances in microfabrication technologies have enabled formation of numerous hepatocyte spheroids with a uniform diameter.13,14
In the early stages of spheroid cultures, only hepatocytes were cultured to form 3D structures. Although spheroid culture is useful in terms of the maintenance of hepatocyte function, hepatocyte spheroids are formed by the aggregation of hepatocytes. Thus, the structure of spheroids does not reflect the well-organized liver tissue in vivo. This resulted in the extension of hepatocyte spheroid cultures by co-culturing with ECs15 and HSCs16 to better mimic physiological hepatic tissue. These co-culture spheroid models are useful for investigating heterotypic cellular interactions between hepatocytes and ECs, and hepatocytes and HSCs, respectively. However, it remains difficult to construct spheroids that reproduce the true in vivo structure formed by hepatocytes, HSCs, and ECs. Additionally, the size of hepatocyte spheroids is limited by diffusion-dependent mass transfer because of the lack of a vascular network within the 3D structures. Furthermore, tissue formation within hepatocyte spheroids depends on the spontaneous reorganization of the cells. Thus, more controllable 3D culture methods have since been developed to better mimic in vivo liver structures.
3D stacked-up culture based on controllable tissue formation
3D stacked-up culture, which is a method of 3D culture by stacking 2D cell layers, is a more controllable method of mimicking the in vivo liver structure. Because hepatic cords have a bilayer structure, 3D culture of hepatocytes has been achieved by stacking 2D cell layers. For example, small hepatocytes, a type of hepatic progenitor cell, were cultured on a pair of microporous membranes to allow them to form 2D cell layers.17 These 2D tissues with microporous membranes were stacked to form bilayer structures, because hepatocytes form such bilayer structures, known as hepatic cords, in vivo. Interestingly, small hepatocytes in the stacked structure adapted to their 3D culture microenvironment and formed bile canaliculi, highly differentiated structures, between the two cell layers. As the cells formed differentiated structures, they began to express differentiated functions—such as albumin secretion—in the 3D culture. This method was also performed using biodegradable microporous membranes composed of poly(d,l-lactide-co-glycolide).18 In this culture model, hepatocytes formed bilayer structures without membranes as the membrane was degraded during culture. Furthermore, this culture model was extended to perform tri-culturing of small hepatocytes, HSCs, and ECs.19,20 In the tri-culture model, HSC behavior was spatially and temporally controllable, providing HSC-mediated proximal layers of hepatocytes and ECs.19 Furthermore, with control of temporal hepatocyte-HSC interactions, HSCs were finally located along EC capillary-like structures stacked on hepatic organoids composed of small hepatocytes and HSCs.20
Cell sheet engineering—as proposed by Okano et al.—is a pioneering method of constructing a 3D structure by stacking 2D cell layers. In this method, cells are cultured on a temperature-responsive surface; cell sheets can then be harvested by reducing the temperature. This method can be applied to several cell types. In terms of liver tissue engineering, an EC layer has been stacked onto a hepatocyte layer.21 Hepatocytes covered by an EC layer maintained differentiated function for longer than did single hepatocytes in culture. Moreover, bile canalicular networks formed among the hepatocytes and developed well in the layered hepatocyte-EC sheets.22
Multi-layered tissues can also be constructed by the formation of a nanoscale-biocompatible polyelectrolyte scaffold, which provides a cell-adhesive surface, on top of a monolayer of cells so that a second cell type can be seeded directly onto the first layer of cells. For example, a polyelectrolyte scaffold was assembled on top of hepatocytes by sequentially depositing chitosan and DNA as the cationic and anionic polyelectrolytes.23 Second cell types, such as ECs, fibroblasts and hepatocytes, were cultured on top of the original hepatocyte layer, resulting in a layered architecture.
Magnetite nanoparticles have also been used to construct layered 3D structures in vitro;24 magnetically labeled ECs were seeded onto hepatocyte monolayers cultured under a magnetic field. Due to the magnetic field, ECs grew on the hepatocyte layer, resulting in a heterotypic, layered construct. Hepatocytes maintained a high level of albumin secretion in the heterotypic constructs.
Although these 3D stacked-up culture methods achieved relatively complex, organized tissues compared with hepatocyte spheroids, this method still failed to construct physiological liver tissues. However, the development of 3D culture techniques has enabled culturing of hepatocytes in 3D configurations in which they maintain their differentiated function. Liver tissue engineers have come to recognize that the transport of oxygen and nutrients into thick, 3D multicellular tissues is a significant issue in liver tissue engineering. Because mass transport is limited by diffusion within tissues, 3D tissues must include microvascular networks that provide oxygen and nutrients by convection.7 In this context, new approaches to achieving vascularization of 3D tissue-engineered constructs have been developed. Two such approaches using bioMEMS and decellularization technologies, based on bottom-up and top-down approaches, respectively, are described below. A method based on bioMEMS technology allowed the precise control of cellular microenvironments to construct more physiological tissues, leading to vascularization; this method is more controllable than conventional 3D culture.
Bottom-Up Approach: BioMEMS
BioMEMS: A new technology for the control of culture microenvironments
In the past two decades, microfabrication technologies have been applied to multiple life science fields, bringing new capabilities to research. The number of published papers related to microfluidics technologies has increased markedly since 2000, not only concerning liver reconstruction but also other life science fields (Fig. 2). Soft lithography is a key technology in microfluidics that is used to create microstructures by printing, molding and embossing.25 Using this technology, designed structures at a micrometer scale can be embossed on the surface of biocompatible elastomeric polymers such as poly(dimethylsiloxane) (PDMS). This technology was first used for micropatterning cell culture on a flat surface,26-29 and was then applied to create 3D culture microenvironments.30,31 PDMS with channels embossed on the surface can be bonded with a coverglass, which creates spaces for culture medium between the microfabricated PDMS and a coverglass, resulting in a microfluidic device. Such microfluidics-based cell culture devices opened new doors in life science research, particularly in cell biology32,33 and tissue engineering.34
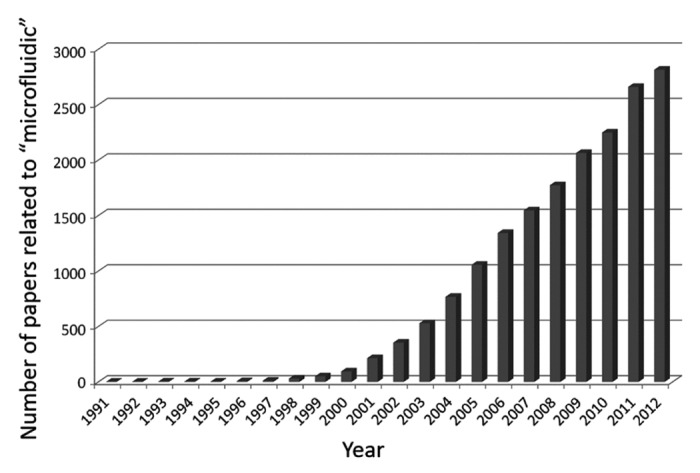
Figure 2. Increase in the number of papers related to microfluidics technologies. The number of papers from 1991–2012 in a scientific literature database, the Web of Science, was searched using the keyword “microfluidic.” Note that the number of papers increased dramatically after ~2000.
An important advantage of microfluidics-based cell culture devices is that cells can be cultured under flow conditions.35 Microfluidic perfusion culture allows more physiologically relevant culture environments compared with conventional culture devices such as culture dishes and flasks, in which cells are cultured on a flat surface under static conditions. Specifically, the delivery and removal of soluble biochemical factors as well as mechanical force exerted via fluid flow can be controlled precisely in a microfluidic culture device.
Microfluidic devices have additional advantages compared with conventional culture methods. For example, cells can be cultured under a growth factor concentration gradient. Cellular distribution can also be controlled in culture, as the microchannel pattern can be designed. In addition to spatial control of cellular distribution, its temporal control is also possible because cells can be added during culture to designated regions as required via a microchannel. Microfluidic devices also provide suitable environments for cell culture because of the larger surface: volume ratio, more similar to that in vivo, which enhances cell–cell interactions via concentrated soluble factors. Microfluidic devices also have several limitations. For example, protein expression analysis, such as by western blotting, is difficult due to the limited number of cells harvested from a microfluidic device. Evaporation of culture medium easily affects osmolality, which can damage cells. Surface coating of the microfluidic channels is required to facilitate cell adhesion. However, the advantages of microfluidic devices are critical in the context of tissue engineering because functional tissue can be constructed using well-controlled culture conditions, optimized for the formation of organized tissue.
Microfluidics-based culture systems to mimic liver structures and microenvironments
One of the earlier attempts to culture liver cells in a microfluidic device was reported by Leclerc et al.36 The study focused on the establishment of a cell culture in a microfluidic device, such as sterilization steps and collagen pre-coating, rather than on constructing physiological liver tissues. They developed a device with a 3D microfluidic structure composed of two stacked layers of PDMS incorporated in a continuous fluid perfusion circuit to enhance 3D cell arrangement. HepG2 cells were cultured in the device and their functions were monitored over 10 d. This device was then modified to a microfluidic PDMS bioreactor for large-scale culture of hepatocytes.37 The design involved a combination of microchannels for cell culture and an oxygen chamber separated vertically by thin PDMS membranes. The high gas permeability of PDMS allows supply of oxygen to the cells cultured in microchannels. Consequently, a large number of cells were cultured successfully by stacking the PDMS layers. A cell density of ~3–4 × 107/cm3 was achieved, consistent with other macroscale bioreactors.
Carraro et al.38 developed a microfluidics-based bilayer device with a discrete parenchymal chamber modeled on the liver architecture. Microfluidic channels were designed to provide appropriate flow behavior based on physiological data from human microvasculature. Hepatocytes were cultured in a parenchymal chamber separated by a nanoporous membrane that allows transport of metabolites and small proteins while protecting hepatocytes from the effects of shear stress. Hepatocyte functions—such as albumin secretion and urea production—were maintained at significantly higher levels for at least 7 d in a perfused microfluidic device compared with in static culture dishes. Although this study reported that hepatocytes maintained differentiated functions in a microfluidic device, hepatocytes failed to form physiological structures similar to those in vivo.
Because the microfluidic device allows precise control of the culture microenvironment, the technology can be used for constructing physiological liver tissues by building the individual cellular components of the liver. Based on this bottom-up approach, a microfluidic device has been used to construct the smallest unit of liver structure, the hepatic cord. Lee et al.39 developed a microfluidic device with a design inspired by the liver microstructure. Hepatocytes were cultured in a constrained space of 50 × 30 × 500 μm, resulting in the formation of hepatic cord-like structures. They also designed a microfluidic structure to mimic the natural endothelial barrier layer in the liver sinusoid, which exhibited mass transport properties similar to the liver in vivo, defined tissue and fluid transport regions, and continuous nutrient exchange. Hepatocytes were maintained in the microfluidic device for at least 7 d. A similar microfluidic device was developed for high-density culture of hepatocytes.40 In this microfluidic device, a high density of hepatocytes is in close contact with the microcirculation, which mimics the liver tissue configuration in vivo. These microfluidic devices were modified such that the hepatocytes grew in two lines, resulting in the formation of linear bile canaliculi along the hepatic-cord-like structure.41 The control of bile canalicular formation has the potential to enable collection of bile secreted by hepatocytes in culture. Although the hepatocytes constructed physiological structures, such as bile canaliculi, within this culture model, the system was a 2D culture. Thus, the 2D microfluidic culture system needed to be developed into a 3D culture system to construct more complex 3D tissues, based on this bottom-up approach.
Microfluidics-based 3D culture systems for hepatocytes
Microfluidics-based 2D culture models were further extended to 3D in the context of liver tissue engineering. Microfluidic devices have enabled us to generate 3D structures composed of hepatocytes under controlled culture environments. Toh et al.42 developed a 3D-cell-perfusion culture system using microchannels. In this device, hepatocytes were cultured in a microchannel with an array of micropillars that immobilize and support cells. Consequently, cells were cultured in a physiological microenvironment with 3D cell–cell and cell–matrix interactions. Cell viability changed significantly according to the flow conditions. Flow rates were optimized to achieve the greatest hepatocyte viability. Hepatocytes seeded in this device showed aggregation within 1 d. Cells were then remodeled into 3D aggregates with smooth surfaces, similar to hepatocyte spheroids. Hepatocytes maintained cell–cell interactions in the 3D culture, as confirmed by cortical localization of actin filaments between hepatocytes. Furthermore, a metabolic function of hepatocytes, UDP-glucuronyltransferase activity, was maintained at a significantly higher level in the 3D aggregates than in 2D monolayer cultures.
This microfluidic device was further modified to have multiplexed culture channels for drug toxicity testing.43 The modified device has parallel cell culture channels that are addressed independently by the outputs of a concentration gradient generator. The dose-dependent hepatotoxicity of model drugs was investigated. Hepatocytes in the microfluidic device were more sensitive to the hepatotoxic effects of most of the drugs tested than those in multi-well plate cultures, demonstrating the potential of this device for in vitro drug testing in a biologically relevant and efficient manner.
Goral et al.44 developed a similar perfusion-based microfluidic device for human hepatocyte culture, with some modifications. Cryopreserved primary human hepatocytes were cultured in the device, resulting in the formation of bile canalicular networks within 3D hepatocyte tissue-like structures under perfusion conditions, whereas most were dead within 2 wk under static conditions. Continuous delivery of nutrients and waste removal were achieved under perfusion conditions, which seemed to mimic in vivo blood perfusion. Furthermore, hepatocytes forming bile canaliculi exhibited a transport function from the cytoplasm to the luminal space of bile canaliculi via multi-drug resistance associated protein 2. This microfluidics-based culture system allowed the construction of 3D hepatocyte tissues. However, this 3D tissue was composed of hepatocytes alone. Thus, this culturing technology needed to be developed into a co-culture system to construct more complex tissues, based on the bottom-up approach.
Microfluidics-based 3D culture systems for liver cell co-culture
In addition to the hepatocyte culture, the establishment of EC culture is important in achieving vascularization of tissue-engineered constructs in a microfluidic device. Hydrogel is an important scaffold that provides a 3D environment for the formation of microvascular networks by ECs. This led to the development of a hydrogel-incorporating microfluidic device. Although hepatocytes were also cultured in a microfluidic device incorporating collagen gel, the hydrogel-containing device was first used to monitor the dynamics of individual cells during 3D angiogenesis.45 This device has a great advantage in terms of high-resolution live-cell imaging. An in vitro 3D angiogenesis model has been used widely to investigate vascular sprouting and capillary extension and development. In this model, ECs were cultured on collagen gel formed at the bottom of a culture dish. Because vascular sprouts extended perpendicular to the microscopic viewing plane, z-stack images could be acquired by a confocal laser-scanning microscope and reconstructed as a 3D projection image for investigation of 3D capillary morphogenesis. However, this microfluidic device provides a window for monitoring sprouting angiogenesis, due to the unique geometry of the device. Sprouting angiogenesis can be monitored using a phase-contrast microscope because vascular sprouts grow predominantly in the microscopic viewing plane. This device was then modified to investigate 3D migration of various cell types, including ECs and cancer cells, and the effect of co-culture with other cell types.46-48
The hydrogel-containing device was then applied to the co-culture of hepatocytes and ECs.49 As vascularization of tissue-engineered constructs is challenging, it is important to investigate the interaction between hepatocytes and ECs. In particular, the interaction between epithelial tissues and capillaries should be clarified. Although many studies on the co-culture of hepatocytes and ECs have been conducted, little is known about the interaction of hepatocytes, which form 3D tissues, and ECs, which form capillary structures. In this device, hepatocytes were first cultured under interstitial flow conditions to enhance the formation of 3D tissue structures. The presence of interstitial flow promoted significant formation of 3D tissue structures compared with static conditions. This hepatocyte culture model was coupled with the 3D angiogenesis model. Hepatocyte-EC co-culture in the microfluidic device revealed that the presence of hepatocytes promoted significant capillary formation by ECs. This culture model demonstrated interactions between hepatocyte-tissues and endothelial capillaries, suggesting the potential for creating vascularized hepatocyte tissues.
The formation of hydrogel fibers based on microfluidics technologies can also be applied to liver tissue engineering. Because hepatic cords consist of aligned single or coupled hepatocytes, this aligned structure can be mimicked by hydrogel microfibers. Yamada et al.50 reported formation of such hepatocyte structures in alginate gel microfibers. Fibroblasts were also embedded in the alginate gel to enhance hepatocyte differentiation via cell−cell communications. Heterotypic and homotypic cell−cell interactions in this culture system enhanced hepatocyte differentiation in long-term culture for more than 30 d.
Microfluidics-based 3D culture systems allowed the construction of increasingly more complex tissues, as described above. Because microfluidics technologies can control the culture microenvironment, reorganization of hepatocyte tissues to include other cell types, such as ECs, can be achieved. Such culture models are useful for constructing physiological 3D tissues similar to the in vivo situation. However, these culture models can only be performed at a microscale. The resulting micro-physiological hepatic tissues can be used for the study of further tissue organization, liver cell biology, physiology, and drug testing. The critical limitation of these microfluidics-based systems is the limited volume of the tissue-engineered constructs, which is not adequate for clinical use.
In terms of tissue engineering and regenerative medicine, achieving a large volume of tissue-engineered construct(s) is essential. Thus, a top-down approach, performed at the macroscale, is also required. Whole-organ bioengineering is such an approach to construction of liver tissue at the macroscale.
Top-Down Approach: Whole-Organ Bioengineering
Decellularization and recellularization of the liver
The most important advantage of decellularized whole organs is that they comprise the intact original 3D architecture and specific ECM proteins of the organ. In particular, vascular networks retain the original 3D structures, allowing connection to the recipient’s circulation for rapid oxygen and nutrient delivery after transplantation. Therefore, transplantation of reconstructed organs is possible upon the successful repopulation of the decellularized organ. In this context, a decellularized liver scaffold is a practical platform for reconstructing liver tissues in vitro because it has a macroscopic structure and contains decellularized vascular networks that can be surgically connected to the recipient’s vascular networks when transplanted.
Decellularization is a technique for removing all cells from a tissue/organ by physical, enzymatic, and/or chemical treatments, which has advanced dramatically in recent years (Fig. 3). The selection of decellularization agents among non-ionic, ionic, and zwitterionic detergents, enzymatic agents, physical agents, and the direct application of force, along with the development of new protocols, are critical steps for successful decellularization.51-53 It is important to remove all cellular material without adversely affecting the composition, mechanical integrity, or the eventual biological activity of the remaining ECM, and original structures, including vascular and lymphatic networks, which must be optimized for each tissue/organ.54
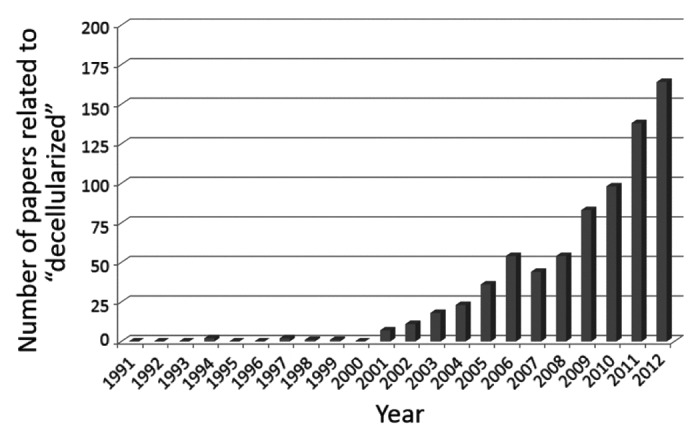
Figure 3. Increase in the number of papers related to decellularization technologies. The number of papers from 1991–2012 in a scientific literature database, the Web of Science, was searched using the keyword “decellularized.” Note that the number of papers increased dramatically after ~2000.
Decellularization techniques have been applied for parts of tissues/organs, such as heart valves,55,56 vessel walls,57,58 the skin,59,60 and the trachea.61 Recent advances in this technique have facilitated the preparation of decellularized whole-organ scaffolds. Specifically, it has been shown that perfusing a detergent solution through the vascular network, rather than relying on agitation and diffusion alone, allows decellularization of entire organs.51-53 Ott et al.62 reported the decellularization of a heart by coronary perfusion with detergents. The decellularized heart, a 3D cardiac ECM scaffold with native vascular channels, was repopulated with cardiac cells or ECs, resulting in macroscopic contractions by day 4, and pump function by day 8. This decellularization technique based on detergent perfusion has since been modified and adopted for the liver,63 lung,64 and kidney.65
3D architecture and complex mixtures of native ECM in the decellularized liver
The ECM as well as the 3D structure of a decellularized liver is important for constructing liver tissues in vitro. While only one component or a few ECM proteins, such as collagen, laminin, and fibronectin, have been used to create in vitro culture models, the ECM in vivo is a complex mixture of functional proteins. These mixtures of ECM proteins, which are unable to be replicated in vitro, play an important role in the maintenance of tissue structures and functions.66 Decellularized ECM scaffolds, especially those derived from urinary bladder and small intestinal submucosa, have been well-characterized.54 The specific composition and distribution of ECM are important for the maintenance of cells in each tissue/organ because decellularized ECM contains proteins secreted by the resident cells of the tissue/organ from which they are prepared.
In the case of liver ECM, even in the absence of the 3D structure of an intact liver, decellularized liver-derived ECM facilitated differentiated functions of primary rat hepatocytes in culture.67 Liver-derived ECM was useful for culture of not only hepatocytes but also sinusoidal endothelial cells (SEC), the liver-specific ECs. In particular, the ECM source was important because liver-derived ECM maintained the greatest degree of SEC differentiation compared with ECM derived from other organs, such as the bladder and small intestinal submucosa.68
Application of repopulated liver grafts
Decellularization of the liver was first reported by Uygun et al.,63 who applied the perfusion-based decellularization technique to the rat liver, and showed that ECM proteins, such as collagen type I, collagen type IV, fibronectin and laminin, were retained similarly to the native liver. Importantly, the vascular tree, including the microvasculature, remained intact in the decellularized liver. They further demonstrated that the decellularized liver could be repopulated with primary rat hepatocytes. Hepatocytes injected through the portal vein remained in and around the vessels after 4 h of in vitro perfusion culture and were distributed throughout the decellularized liver matrix within a few days. Albumin and urea production by the repopulated hepatocytes was also quantified. In particular, the cumulative urea levels in the recellularized liver system were significantly higher during the 5 d of the culture period than in control hepatocytes cultured in a collagen gel sandwich configuration. Furthermore, expression levels of Adh1, encoding alcohol dehydrogenase-1, and Cyp3a18, encoding cytochrome P450, family 3, subfamily a, polypeptide 18, were higher in recellularized liver than in the sandwich control culture, although they remained much lower than in a normal liver. In addition to hepatocytes, ECs were also introduced to the hepatocyte-repopulated liver graft; this is essential for transplantation of recellularized liver grafts. The recellularized liver graft was finally transplanted to a rat and maintained in vivo for 8 h. A remaining challenge is improving recellularization for engineering of an entire liver because the liver is composed not only of hepatocytes, but also nonparenchymal cells. Specifically, recellularization should be performed with optimized seeding strategies, and temporally controlled seeding of hepatocytes, with the addition of nonparenchymal cells, such as SECs, HSCs, biliary epithelial cells, and Kupffer cells.
Baptista et al.69 further investigated liver recellularization techniques. Both human fetal liver cells and ECs were seeded into decellularized whole liver scaffolds from ferrets, resulting in hepatocyte localization in the parenchyma, biliary epithelial cells in biliary tubular structures, and ECs around the vascular structures. In particular, they showed that simultaneous cell seeding, through the vena cava and the portal vein, was useful for achieving ECs covering the entire length of the vascular network, including the pericentral and periportal regions, which is critical for preventing blood clotting. In this study, endothelialized liver scaffolds showed significant reduction in the presence and adhesion of platelets, compared with the decellularized liver scaffold without EC seeding. However, further improvement, to achieve complete endothelialization, is required for successful transplantation. Recently, the decellularization and recellularization of liver, which were first performed using rats, were also adopted for porcine livers—which are of a relevant size.70,71 These results of recellularization using primary human cells and the generation of a human-sized organ suggest that whole-organ bioengineering can be applied clinically.
Decellularization and recellularization techniques have great potential for liver tissue engineering. Because they comprise the architecture of the entire organ including an intact vascular tree, they can be transplanted readily. Bao et al.72 demonstrated the transplantation of recellularized rat liver to hepatectomized rats, resulting in improvement of liver function and prolonged survival from 16 to 72 h. Although this top-down approach is promising in terms of the reconstruction of liver tissues at the macroscale, it is necessary to refine the microarchitecture of the liver tissue. In particular, endothelialization of the entire liver graft is essential to completely prevent hemorrhage and thrombosis after the recirculation of blood on transplantation. The lack of bile ducts is another issue in terms of the construction of the bile-drainage system, another critical function of the liver. Also, a clear limitation of decellularization concept is the limited number of decellularized livers vs. the larger numbers of required donor organs, although with decellularization, livers can be obtained from donated organs that are discarded due to damage or that are otherwise deemed unsuitable for transplantation. Thus, microscale-engineered liver construction should be scaled up and conducted at the macroscale to be appropriate for clinical use. In this context, mesoscale approaches are also needed to bridge the micro- and macroscale approaches.
Mesoscale Approaches to Bridge the Micro- and Macro-scale Approaches
Microbioreactor systems
The group of Griffith and colleagues reported pioneering works in the context of microbioreactor systems for perfused 3D culture of liver cells; their system has the potential to be scaled up, and is thus a mesoscale approach with the potential to bridge the gap between micro- and macoscale approaches. This microbioreactor was designed to have a flow rate sufficient for oxygen supply while providing fluid shear stress in a physiological range.73 Hepatocytes formed 3D tissues and showed well-maintained metabolic functions in the microbioreactor.74 Co-culture of hepatocytes and liver nonparenchymal cells was also performed in the microbioreactor; SECs persisted for 13 d in culture in the absence of VEGF and serum.75 Furthermore, this microbioreactor system was modified to integrate it with a multiwell plate format.76 Liver cells can be cultured under constant microperfusion in the microbioreactor equipped with microfabricated polystyrene and polycarbonate scaffolds coated with collagen, micropumps, and oxygen sensors. Because the system uses a 24-well format, it has a higher throughput capability, which is beneficial for conducting liver toxicology and metabolism assays.
3D printing technologies
The combination of 3D printing technology and a 3D sacrificial molding also has the potential to overcome the scale gap between the micro- and macroscale approaches in tissue engineering. Miller et al.77 fabricated 3D interconnected filament networks of carbohydrate glass using 3D printing technology; these were then used as a cytocompatible sacrificial mold. After casting of hydrogel materials with/without cells around the mold, they were placed in medium to dissolve the carbohydrate glass lattice, resulting in a hydrogel scaffold with perfusable microchannel networks. The microchannel can be lined by ECs, and use of such a gel with channels rescues protein expression in the gel core, most dramatically around each perfused channel. Because this type of scaffold can be fabricated at a millimeter scale with predefined, multiscale, and reproducible patterns, this technology seems useful for scaling up tissue-engineered constructs. Thus, 3D printing represents a mesoscale approach.
Microfabricated hydrogel pieces
Hydrogel fibers and blocks also have potential to be scaled up to construct 3D tissues of a practical size. For example, hydrogel blocks were synthesized by photolithography and transferred into a culture dish. The microgel blocks were assembled into linear, branched, random and offset structures on a millimeter scale.78 Because cells can be encapsulated into the microgel blocks, the assembled blocks encapsulating various cell types have the potential to organize into complex 3D tissues. Hydrogel fibers also have the potential to be scaled up because fiber-shaped materials can be assembled into complex 3D tissues by folding, bundling, reeling and weaving. Onoe et al.79 reported meter-long hydrogel fibers with encapsulated viable cells such as cardiomyocytes, ECs, cortical cells, HepG2 cells, and primary islet cells. These fibers could be assembled into macroscopic cellular structures with various spatial patterns, which represents a mesoscale approach.
Concluding Remarks
BioMEMS technology has great potential for the precise control of microenvironments in culture, such as spatial and temporal cellular configurations, concentration gradients of soluble factors, and flow conditions around cells. The physiological microscale culture models can be useful for investigating the formation of functional and vascularized tissues. This bottom-up approach based on bioMEMS technologies has allowed us to investigate construction of functional 3D tissues from individual cells in a step-by-step manner; consequently, increasingly complex tissues can be constructed. Furthermore, recent advances in bioMEMS studies created a new field called ‘organs-on-a-chip’ or ‘human-on-a-chip,’ in which interconnected organ models, which include liver and other organs, that better mimic organ-level functions in vivo are constructed.80-82
Advances in bioMEMS technologies have facilitated construction of complex tissues and organs. However, the resulting tissues are on a micrometer scale. To be useful for regenerative medicine, these tissues must be scaled up. The top-down approach, liver decellularization, has enabled construction of liver tissues on a macroscale. This technique has a significant advantage in terms of a more clinically relevant scale compared with the liver tissues reconstructed by bioMEMS technologies. After functional tissues have been reconstructed in the decellularized liver scaffold, they can be transplanted in vivo. However, the construction of macroscale liver tissues with a refined microarchitecture remains challenging. Specifically, the establishment of a functional microcirculation is essential.
Mesoscale culture models, such as microreactors fabricated at the millimeter scale, have also been reported. It is possible to culture a mass of hepatocytes and create relatively large liver organoids, which has potential for scaling up of microscale models to the macroscale. However, precise control of the culture microenvironment and optimizing tissue organization are problematic due to difficulties with visualization of the process of 3D tissue organization.
In conclusion, parallel studies on micro-, meso-, and macroscale liver tissue engineering are ongoing (Fig. 1). Each scale has its own advantages and limitations, and gaps exist among the models; these gaps must be bridged to achieve large-scale tissue organization with functional microarchitectures. The knowledge or basic principles of constructing liver tissues from one scale model should be applied to other scale models to complement liver tissue engineering studies at each scale. Furthermore, the integration of multiscale tissue-engineering studies, including both bottom-up and top-down approaches, is essential for construction of functional liver tissues in vitro.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (25282135, 25560208, 20246041) from the Japan Society for Promotion of Science.
Glossary
Abbreviations:
- 2D
two-dimensional
- 3D
three-dimensional
- EC
endothelial cells
- ECM
extracellular matrix
- HSC
hepatic stellate cell
- MEMS
microelectromechanical system
- PDMS
poly(dimethylsiloxane)
- SEC
sinusoidal endothelial cells
References
- 1.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–6. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitaka T. The current status of primary hepatocyte culture. Int J Exp Pathol. 1998;79:393–409. doi: 10.1046/j.1365-2613.1998.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–20. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 6.Griffith LG, Naughton G. Tissue engineering--current challenges and expanding opportunities. Science. 2002;295:1009–14. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 7.Khademhosseini A, Vacanti JP, Langer R. Progress in tissue engineering. Sci Am. 2009;300:64–71. doi: 10.1038/scientificamerican0509-64. [DOI] [PubMed] [Google Scholar]
- 8.Borenstein JT, Vunjak-Novakovic G. Engineering tissue with BioMEMS. IEEE Pulse. 2011;2:28–34. doi: 10.1109/MPUL.2011.942764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbott A. Cell culture: biology’s new dimension. Nature. 2003;424:870–2. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 10.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 11.Landry J, Bernier D, Ouellet C, Goyette R, Marceau N. Spheroidal aggregate culture of rat liver cells: histotypic reorganization, biomatrix deposition, and maintenance of functional activities. J Cell Biol. 1985;101:914–23. doi: 10.1083/jcb.101.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koide N, Sakaguchi K, Koide Y, Asano K, Kawaguchi M, Matsushima H, Takenami T, Shinji T, Mori M, Tsuji T. Formation of multicellular spheroids composed of adult rat hepatocytes in dishes with positively charged surfaces and under other nonadherent environments. Exp Cell Res. 1990;186:227–35. doi: 10.1016/0014-4827(90)90300-Y. [DOI] [PubMed] [Google Scholar]
- 13.Nakazawa K, Izumi Y, Fukuda J, Yasuda T. Hepatocyte spheroid culture on a polydimethylsiloxane chip having microcavities. J Biomater Sci Polym Ed. 2006;17:859–73. doi: 10.1163/156856206777996853. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda J, Nakazawa K. Hepatocyte spheroid arrays inside microwells connected with microchannels. Biomicrofluidics. 2011;5:22205. doi: 10.1063/1.3576905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamori M, Mizumoto H, Kajiwara T. An approach for formation of vascularized liver tissue by endothelial cell-covered hepatocyte spheroid integration. Tissue Eng Part A. 2009;15:2029–37. doi: 10.1089/ten.tea.2008.0403. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Absi SF, Hansen LK, Hu WS. Three-dimensional co-culture of hepatocytes and stellate cells. Cytotechnology. 2004;45:125–40. doi: 10.1007/s10616-004-7996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudo R, Mitaka T, Ikeda M, Tanishita K. Reconstruction of 3D stacked-up structures by rat small hepatocytes on microporous membranes. FASEB J. 2005;19:1695–7. doi: 10.1096/fj.04-3269fje. [DOI] [PubMed] [Google Scholar]
- 18.Kasuya J, Sudo R, Tamogami R, Masuda G, Mitaka T, Ikeda M, Tanishita K. Reconstruction of 3D stacked hepatocyte tissues using degradable, microporous poly(d,l-lactide-co-glycolide) membranes. Biomaterials. 2012;33:2693–700. doi: 10.1016/j.biomaterials.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Kasuya J, Sudo R, Mitaka T, Ikeda M, Tanishita K. Spatio-temporal control of hepatic stellate cell-endothelial cell interactions for reconstruction of liver sinusoids in vitro. Tissue Eng Part A. 2012;18:1045–56. doi: 10.1089/ten.tea.2011.0351. [DOI] [PubMed] [Google Scholar]
- 20.Kasuya J, Sudo R, Masuda G, Mitaka T, Ikeda M, Tanishita K. Reconstruction of hepatic stellate cell-incorporated liver capillary structures in small hepatocyte tri-culture using microporous membranes. J Tissue Eng Regen Med. 2012 doi: 10.1002/term.1630. In press. [DOI] [PubMed] [Google Scholar]
- 21.Harimoto M, Yamato M, Hirose M, Takahashi C, Isoi Y, Kikuchi A, Okano T. Novel approach for achieving double-layered cell sheets co-culture: overlaying endothelial cell sheets onto monolayer hepatocytes utilizing temperature-responsive culture dishes. J Biomed Mater Res. 2002;62:464–70. doi: 10.1002/jbm.10228. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Ohashi K, Utoh R, Kano K, Okano T. Preserved liver-specific functions of hepatocytes in 3D co-culture with endothelial cell sheets. Biomaterials. 2012;33:1406–13. doi: 10.1016/j.biomaterials.2011.10.084. [DOI] [PubMed] [Google Scholar]
- 23.Rajagopalan P, Shen CJ, Berthiaume F, Tilles AW, Toner M, Yarmush ML. Polyelectrolyte nano-scaffolds for the design of layered cellular architectures. Tissue Eng. 2006;12:1553–63. doi: 10.1089/ten.2006.12.1553. [DOI] [PubMed] [Google Scholar]
- 24.Ito A, Takizawa Y, Honda H, Hata K, Kagami H, Ueda M, Kobayashi T. Tissue engineering using magnetite nanoparticles and magnetic force: heterotypic layers of cocultured hepatocytes and endothelial cells. Tissue Eng. 2004;10:833–40. doi: 10.1089/1076327041348301. [DOI] [PubMed] [Google Scholar]
- 25.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–73. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 26.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 27.Folch A, Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227–56. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 28.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104:5722–6. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–6. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 30.Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR, Vacanti JP. Microfabrication technology for vascularized tissue engineering. Biomed Microdevices. 2002;4:167–75. doi: 10.1023/A:1016040212127. [DOI] [Google Scholar]
- 31.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–11. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 32.Paguirigan AL, Beebe DJ. Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. Bioessays. 2008;30:811–21. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni M, Tong WH, Choudhury D, Rahim NA, Iliescu C, Yu H. Cell culture on MEMS platforms: a review. Int J Mol Sci. 2009;10:5411–41. doi: 10.3390/ijms10125411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puleo CM, Yeh HC, Wang TH. Applications of MEMS technologies in tissue engineering. Tissue Eng. 2007;13:2839–54. doi: 10.1089/ten.2007.0214. [DOI] [PubMed] [Google Scholar]
- 35.Kim L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip. 2007;7:681–94. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 36.Leclerc E, Sakai Y, Fujii T. Cell culture in 3-dimensional microfluidic structure of PDMS (polydimethylsiloxane) Biomed Microdevices. 2003;5:109–14. doi: 10.1023/A:1024583026925. [DOI] [Google Scholar]
- 37.Leclerc E, Sakai Y, Fujii T. Microfluidic PDMS (polydimethylsiloxane) bioreactor for large-scale culture of hepatocytes. Biotechnol Prog. 2004;20:750–5. doi: 10.1021/bp0300568. [DOI] [PubMed] [Google Scholar]
- 38.Carraro A, Hsu WM, Kulig KM, Cheung WS, Miller ML, Weinberg EJ, Swart EF, Kaazempur-Mofrad M, Borenstein JT, Vacanti JP, et al. In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed Microdevices. 2008;10:795–805. doi: 10.1007/s10544-008-9194-3. [DOI] [PubMed] [Google Scholar]
- 39.Lee PJ, Hung PJ, Lee LP. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007;97:1340–6. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 40.Zhang MY, Lee PJ, Hung PJ, Johnson T, Lee LP, Mofrad MR. Microfluidic environment for high density hepatocyte culture. Biomed Microdevices. 2008;10:117–21. doi: 10.1007/s10544-007-9116-9. [DOI] [PubMed] [Google Scholar]
- 41.Nakao Y, Kimura H, Sakai Y, Fujii T. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics. 2011;5:22212. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toh YC, Zhang C, Zhang J, Khong YM, Chang S, Samper VD, van Noort D, Hutmacher DW, Yu H. A novel 3D mammalian cell perfusion-culture system in microfluidic channels. Lab Chip. 2007;7:302–9. doi: 10.1039/b614872g. [DOI] [PubMed] [Google Scholar]
- 43.Toh YC, Lim TC, Tai D, Xiao G, van Noort D, Yu H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9:2026–35. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 44.Goral VN, Hsieh YC, Petzold ON, Clark JS, Yuen PK, Faris RA. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip. 2010;10:3380–6. doi: 10.1039/c0lc00135j. [DOI] [PubMed] [Google Scholar]
- 45.Vickerman V, Blundo J, Chung S, Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–77. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–75. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 47.Chung S, Sudo R, Vickerman V, Zervantonakis IK, Kamm RD. Microfluidic platforms for studies of angiogenesis, cell migration, and cell-cell interactions. Ann Biomed Eng. 2010;38:1164–77. doi: 10.1007/s10439-010-9899-3. [DOI] [PubMed] [Google Scholar]
- 48.Zervantonakis IK, Kothapalli CR, Chung S, Sudo R, Kamm RD. Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics. 2011;5:13406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudo R, Chung S, Zervantonakis IK, Vickerman V, Toshimitsu Y, Griffith LG, Kamm RD. Transport-mediated angiogenesis in 3D epithelial coculture. FASEB J. 2009;23:2155–64. doi: 10.1096/fj.08-122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada M, Utoh R, Ohashi K, Tatsumi K, Yamato M, Okano T, Seki M. Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions. Biomaterials. 2012;33:8304–15. doi: 10.1016/j.biomaterials.2012.07.068. [DOI] [PubMed] [Google Scholar]
- 51.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arenas-Herrera JE, Ko IK, Atala A, Yoo JJ. Decellularization for whole organ bioengineering. Biomed Mater. 2013;8:014106. doi: 10.1088/1748-6041/8/1/014106. [DOI] [PubMed] [Google Scholar]
- 53.Yagi H, Soto-Gutierrez A, Kitagawa Y. Whole-organ re-engineering: a regenerative medicine approach to digestive organ replacement. Surg Today. 2013;43:587–94. doi: 10.1007/s00595-012-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Bader A, Schilling T, Teebken OE, Brandes G, Herden T, Steinhoff G, Haverich A. Tissue engineering of heart valves--human endothelial cell seeding of detergent acellularized porcine valves. Eur J Cardiothorac Surg. 1998;14:279–84. doi: 10.1016/S1010-7940(98)00171-7. [DOI] [PubMed] [Google Scholar]
- 56.Cebotari S, Lichtenberg A, Tudorache I, Hilfiker A, Mertsching H, Leyh R, Breymann T, Kallenbach K, Maniuc L, Batrinac A, et al. Clinical application of tissue engineered human heart valves using autologous progenitor cells. Circulation. 2006;114(Suppl):I132–7. doi: 10.1161/CIRCULATIONAHA.105.001065. [DOI] [PubMed] [Google Scholar]
- 57.Conklin BS, Richter ER, Kreutziger KL, Zhong DS, Chen C. Development and evaluation of a novel decellularized vascular xenograft. Med Eng Phys. 2002;24:173–83. doi: 10.1016/S1350-4533(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 58.Zehr KJ, Yagubyan M, Connolly HM, Nelson SM, Schaff HV. Aortic root replacement with a novel decellularized cryopreserved aortic homograft: postoperative immunoreactivity and early results. J Thorac Cardiovasc Surg. 2005;130:1010–5. doi: 10.1016/j.jtcvs.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 59.Chen RN, Ho HO, Tsai YT, Sheu MT. Process development of an acellular dermal matrix (ADM) for biomedical applications. Biomaterials. 2004;25:2679–86. doi: 10.1016/j.biomaterials.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 60.Schechner JS, Crane SK, Wang F, Szeglin AM, Tellides G, Lorber MI, Bothwell AL, Pober JS. Engraftment of a vascularized human skin equivalent. FASEB J. 2003;17:2250–6. doi: 10.1096/fj.03-0257com. [DOI] [PubMed] [Google Scholar]
- 61.Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto PP, et al. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023–30. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 62.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 63.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, Milwid J, Kobayashi N, Tilles A, Berthiaume F, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 65.Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19:646–51. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Badylak SF. The extracellular matrix as a scaffold for tissue reconstruction. Semin Cell Dev Biol. 2002;13:377–83. doi: 10.1016/S1084952102000940. [DOI] [PubMed] [Google Scholar]
- 67.Lin P, Chan WC, Badylak SF, Bhatia SN. Assessing porcine liver-derived biomatrix for hepatic tissue engineering. Tissue Eng. 2004;10:1046–53. doi: 10.1089/ten.2004.10.1046. [DOI] [PubMed] [Google Scholar]
- 68.Sellaro TL, Ravindra AK, Stolz DB, Badylak SF. Maintenance of hepatic sinusoidal endothelial cell phenotype in vitro using organ-specific extracellular matrix scaffolds. Tissue Eng. 2007;13:2301–10. doi: 10.1089/ten.2006.0437. [DOI] [PubMed] [Google Scholar]
- 69.Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–17. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 70.Barakat O, Abbasi S, Rodriguez G, Rios J, Wood RP, Ozaki C, Holley LS, Gauthier PK. Use of decellularized porcine liver for engineering humanized liver organ. J Surg Res. 2012;173:e11–25. doi: 10.1016/j.jss.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 71.Yagi H, Fukumitsu K, Fukuda K, Kitago M, Shinoda M, Obara H, Itano O, Kawachi S, Tanabe M, Coudriet GM, et al. Human-scale whole-organ bioengineering for liver transplantation: a regenerative medicine approach. Cell Transplant. 2013;22:231–42. doi: 10.3727/096368912X654939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bao J, Shi Y, Sun H, Yin X, Yang R, Li L, Chen X, Bu H. Construction of a portal implantable functional tissue-engineered liver using perfusion-decellularized matrix and hepatocytes in rats. Cell Transplant. 2011;20:753–66. doi: 10.3727/096368910X536572. [DOI] [PubMed] [Google Scholar]
- 73.Powers MJ, Domansky K, Kaazempur-Mofrad MR, Kalezi A, Capitano A, Upadhyaya A, Kurzawski P, Wack KE, Stolz DB, Kamm R, et al. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol Bioeng. 2002;78:257–69. doi: 10.1002/bit.10143. a. [DOI] [PubMed] [Google Scholar]
- 74.Powers MJ, Janigian DM, Wack KE, Baker CS, Beer Stolz D, Griffith LG. Functional behavior of primary rat liver cells in a three-dimensional perfused microarray bioreactor. Tissue Eng. 2002;8:499–513. doi: 10.1089/107632702760184745. b. [DOI] [PubMed] [Google Scholar]
- 75.Hwa AJ, Fry RC, Sivaraman A, So PT, Samson LD, Stolz DB, Griffith LG. Rat liver sinusoidal endothelial cells survive without exogenous VEGF in 3D perfused co-cultures with hepatocytes. FASEB J. 2007;21:2564–79. doi: 10.1096/fj.06-7473com. [DOI] [PubMed] [Google Scholar]
- 76.Domansky K, Inman W, Serdy J, Dash A, Lim MH, Griffith LG. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–8. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11:768–74. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du Y, Lo E, Ali S, Khademhosseini A. Directed assembly of cell-laden microgels for fabrication of 3D tissue constructs. Proc Natl Acad Sci U S A. 2008;105:9522–7. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, et al. Metre-long cell-laden microfibres exhibit tissue morphologies and functions. Nat Mater. 2013;12:584–90. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 80.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–92. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 81.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–54. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van de Stolpe A, den Toonder J. Workshop meeting report Organs-on-Chips: human disease models. Lab Chip. 2013;13:3449–70. doi: 10.1039/c3lc50248a. [DOI] [PubMed] [Google Scholar]