Abstract
Epigenetic interventions are required to induce reprogramming from one cell type to another. At present, various cellular reprogramming methods such as somatic cell nuclear transfer, cell fusion, and direct reprogramming using transcription factors have been reported. In particular, direct reprogramming from somatic cells to induced pluripotent stem cells (iPSCs) has been achieved using defined factors that play important epigenetic roles. Although the mechanisms underlying cellular reprogramming and vertebrate regeneration, including appendage regeneration, remain unknown, dedifferentiation occurs at an early phase in both the events, and both events are contrasting with regard to cell death. We compared the current status of changes in cell fate of iPSCs with that of vertebrate regeneration and suggested that substantial insights into vertebrate regeneration should be helpful for safe applications of iPSCs to medicine.
Keywords: Dedifferentiation, Direct Conversion, Pluripotency, Primed Conversion, Reprogramming, Transcription Factors, Vertebrate Regeneration, induced Pluripotent Stem Cells (iPSCs)
Introduction
Newt limbs, zebrafish fins, and xenopus tails can regenerate after amputation. The phenomena called appendage regeneration have been intensively studied; however, a gap between this regeneration and mammalian regenerative biology still remains.1-3 With regard to limb regeneration in salamanders, which is a representative example of appendage regeneration, amputated limbs are covered by the epidermis, and immature cells accumulate and proliferate beneath them, forming the “blastema.”4 The blastema, which is encased by the newly formed epidermis, comprises dedifferentiated cells that are restricted to skeletal muscle cells, chondrocytes, Schwann cells, and mesenchymal fibroblasts.5 Salamanders and zebrafish can also regenerate cardiac tissue after the resection of the ventricular apex6,7 or the genetic destruction of cardiomyocytes.8 Significant cardiomyogenesis was observed in murine hearts less than 1 wk after birth.9 The vertebrate regeneration discussed above entails a common process, dedifferentiation. However, the factors distinguishing regenerative vertebrates from nonregenerative vertebrates remain unknown.10 Thus, in regenerative biology and medicine, it is crucial to understand the molecular mechanisms underlying the processes, including growth, patterning, dedifferentiation, and redifferentiation.
Appendage Regeneration
Overview
In zebrafish fin regeneration after amputation, there are four stages, termed “regeneration epithelialization or wound healing” (0–1 d post-amputation [dpa]), “blastema formation” (1–2 dpa), “regenerative outgrowth” (2–7 dpa), and “termination.”11,12 At 1 dpa, the proximal epidermis migrates to cover the stump and form a 3–4-cell-thick layer. Inflammation then proceeds to clean the clotted plasma and cellular debris. At 2 dpa, histolysis occurs, remodeling the extracellular matrix, and cells to be liberated by histolysis begin to dedifferentiate in this period. They do not significantly activate the cell cycle and primarily form the blastema by distal migration under the wound epidermis.13 Subsequently, the accumulation blastema is achieved by a marked increase in mitosis, which is dependent on factors from the wound epidermis14 and regenerating nerve.4 Subsequent regenerative outgrowth or blastema accumulation is characterized by robust proliferation of dedifferentiated cells. An outstanding feature is the presence of rare apoptotic cells in the blastema despite the presence of avascular tissue, which could be hypoxic and possibly susceptible to apoptosis.
Proteomic analysis of the blastema in regenerating axolotl limbs revealed that dedifferentiated cells are capable of avoiding apoptosis through several mechanisms such as reduced metabolism, differential regulation of proapoptotic and antiapoptotic proteins, and initiation of an unfolded protein response.12 The tricarboxylic acid cycle and electron transport enzymes are downregulated by 7 dpa, which may be mediated via increased nitrogen oxide in the blastema. The proteasome deliberately destroys aberrant proteins that could not have been refolded by a chaperon and those that have been specifically marked by a recognition tag formed from ubiquitin. In the axolotl limb regeneration blastema, several chaperons and components of the proteasome–ubiquitin system are upregulated at an early phase, suggesting that protein quality control could play an essential role in dedifferentiation.12
Molecular mechanism
Loss-of-function experiments have revealed regulators of the regeneration process,15-17 although further elucidation is required. Till date, canonical Wnt and fibroblast growth factor (FGF) signaling pathways are the major pathways to be reported in appendage regeneration, and activin, retinoic acid, hedgehog, and noncanonical Wnt signaling is involved in the regulation of regeneration.11 Moreover, as negative regulators, noncoding microRNAs play an important role by downregulating the expression of ligands for FGF and Wnt/β-catenin signaling. Lef1, a ligand for canonical Wnt, is expressed in the newly developed epithelium over the amputated plane. Inducible DKK1, an inhibitor of canonical Wnt signaling, inhibits lef1 expression and blastema formation, and the knockdown following this stage decreases the sizes of regenerated fins.15 Although the lateral epithelial growth that wraps the amputation plane could not be prevented by DKK1 during 0–1 dpa, msxb expression, which is involved in FGF signaling, was impaired. The specific roles of canonical Wnt/β-catenin molecules for each regeneration stage and crosstalk with the FGF signaling pathway have been uncovered.
In contrast, Lef1 expression in the developed epithelium leads to fgfr1 expression in mesenchymal cells of the blastema. Chemical impairment of fgfr1 prevented blastema formation, msxb expression, and consequently cell proliferation.16 Moreover, blastema formation was blocked in fgf20a null mutants.17 One example of the negative impact to Wnt/β-catenin signaling is Wnt5b, a noncanonical Wnt.15 In addition, miR-203 represses lef1 expression as a mediator of Wnt/β-catenin.18 In contrast, miR-133 is involved in the FGF signaling pathway.19 An array of noncoding RNAs should form a sophisticated regulatory network for appendage regeneration, which may share features with the regulatory network for carcinogenesis.
Comprehensive transcriptional profiling20 and RNA sequencing21 during limb regeneration revealed significant upregulation of c-Myc (myelocytomatosis oncogene) and Krüppel-like factors 4 (Klf4). In contrast Oct4 (POU domain, class 5, transcriptional factor 1), Sox2 (sex-determining region Y-box 2), and Nanog were not upregulated. SALL4, which is involved in the maintenance of pluripotency, was overexpressed during blastema formation. In contrast, SALL1 and SALL3 were only gradually expressed during the patterning phase. During epithelialization and blastema formation, the expression of many oncogenes such as ATF3, JUN3, EGR1, NR4A2, and FOS increased; however, these genes were then downregulated during the patterning process. Proteomic analysis of the blastema in regenerating axolotl limbs showed upregulation of LIN28, which is related to cellular reprogramming. Also, antiapoptotic mechanisms, such as reduced metabolism and initiation of an unfolded protein response were activated.12
Cell sources
Determination of the origin of blastema cells has been one of the main concerns of regenerative biology for a long time.1 Genetic lineage tracing elucidated the origin and differentiation capability of blastema cells in amputated axolotl limbs and zebrafish fins. Transplantation experiment using green fluorescent protein (GFP) cells from various tissues of axolotol limbs showed that grafted cells dedifferentiate, proliferate, and redifferentiate into cells that are restricted to the origin.5 Cre/loxP-based genetic marking to track osteoblasts in zebrafish fin regeneration clearly demonstrated the dedifferentiation of pre-existing osteoblasts and redifferentiation to osteoblasts.22 However, it is possible that resident stem cells are involved in appendage regeneration,23 particularly in the case of skeletal muscles, which are accompanied by a population of stem cells called satellite cells.24 Moreover, genetic ablation of all skeletal osteoblasts in zebrafish fins resulted in de novo osteogenetic process, rather than through the dedifferentiation and redifferentiation process.25
Vertebrate Regeneration in the Heart
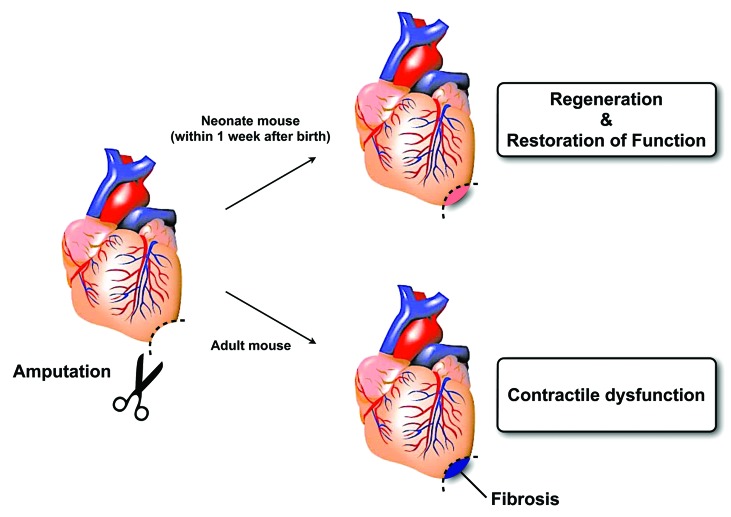
The zebrafish heart has been intensively investigated for its regenerative capacity, and amputation experiments have recently provided convincing proof of the dedifferentiation model in vertebrate regeneration using genetic fate mapping.7,26 Genetic cell ablation by inducible diphtheria toxin expression in zebrafish hearts has strengthened the evidence that newly developed cardiomyocytes are derived from pre-existing cardiomyocytes through a dedifferentiation process.8 Msp1, which is a mitotic checkpoint kinase, and GATA4, which is a transcription factor (TF) of early cardiac development, are essential for zebrafish cardiac regeneration.7,27 Similar to limb regeneration, three phases of cardiac apex regeneration have been distinguished as “inflammatory,” “reparative,” and “regenerative,” and these correspond to wound healing, blastema formation, and outgrowth and termination phases in limb regeneration, respectively.28 With dedifferentiation following apex amputation, epicardiac cells proceed into the epithelial–mesenchymal transition (EMT) in response to FGF and PDGF.29,30 Thereafter, cardiomyocytes with disorganized sarcomeres are similar to immature cardiomyocytes that have been derived from induced pluripotent stem cells (iPSCs) using current cardiac differentiation protocols as monolayers or using embryonic body methods. These detach from one another and launch proliferation with the expression of positive cell cycle regulators such as polo-like kinase 1 (plk1) and cdc2.26 Within 1 wk of birth, neonatal mice repopulate amputated cardiac apexes with newly developed cardiomyocytes,9 which are formed through dedifferentiation and redifferentiation, a phenomenon similar to that observed in zebrafish hearts. This ability was found to be lost by 1 wk of age, and injured apexes were then filled with fibrotic tissues (Fig. 1).
Figure 1. Vertebrate regeneration in mouse neonates hearts.
Reprogramming into Pluripotency
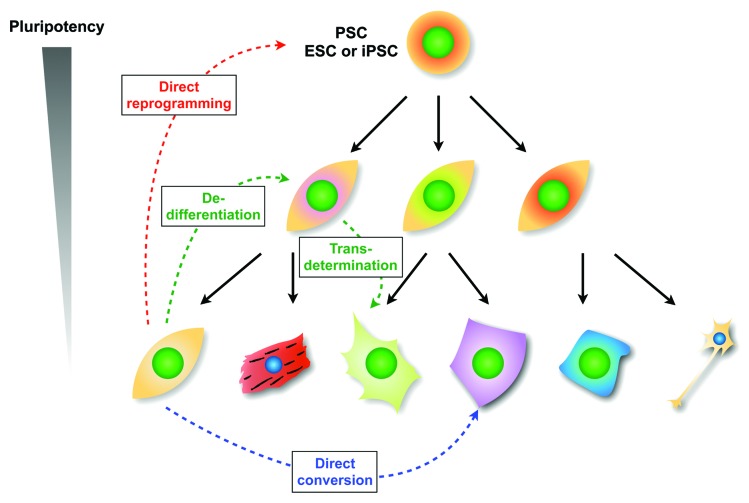
During development, gene expression is regulated by TF and epigenomic networks. One method of determining whether the gene regulatory mechanisms can be reversed is somatic cell nuclear transfer (SCNT). The concept of nuclear transfer31 originates from proposals by Hans Spemann in 1936, who was motivated to test Weismann’s theory that cell differentiation is the consequence of unequal nuclear division during embryonic development. In 1952, Briggs and King successfully accomplished nuclear transplantation of a cell from the blastula into frog eggs for the first time.32 In 1958, John Gurdon demonstrated that nuclei from intestinal epithelial cells could be developed to produce offspring when transferred into enucleated eggs.33 In 1997, Ian Willmut verified that SCNT was successful in sheep.34 Shinya Yamanaka revealed a set of genes to drive reprogramming to pluripotency in 2006.35 Until the discovery of iPSCs, the differentiation process was considered to be one-way, with the exception of the fertilization process. This process of pluripotent stem cell (PSC) differentiation has been shown to be regulated and maintained by complex transcriptional and epigenetic networks.36-38 According to the classical view of cell fate hierarchy based on the Waddington epigenetic landscape,39 PSCs reside at the top of the hierarchy above differentiated somatic cells (Fig. 2). This model indicates a natural restriction of the cell differentiation potential during normal cellular development along each lineage.
Figure 2. Scheme of cell fate changes based on Waddington’s epigenetic landscape. Direct reprogramming is the reversion of terminally differentiated cells such as fibroblasts to a pluripotent state. Direct conversion is the alteration from one cell type to another, such as fibroblasts to cardiomyocytes. Dedifferentiation is defined as a reversion of specialized phenotypes into an undifferentiated state. Transdetermination is the switching of somatic stem/progenitor cells from one determined state to another closely related state.110,111
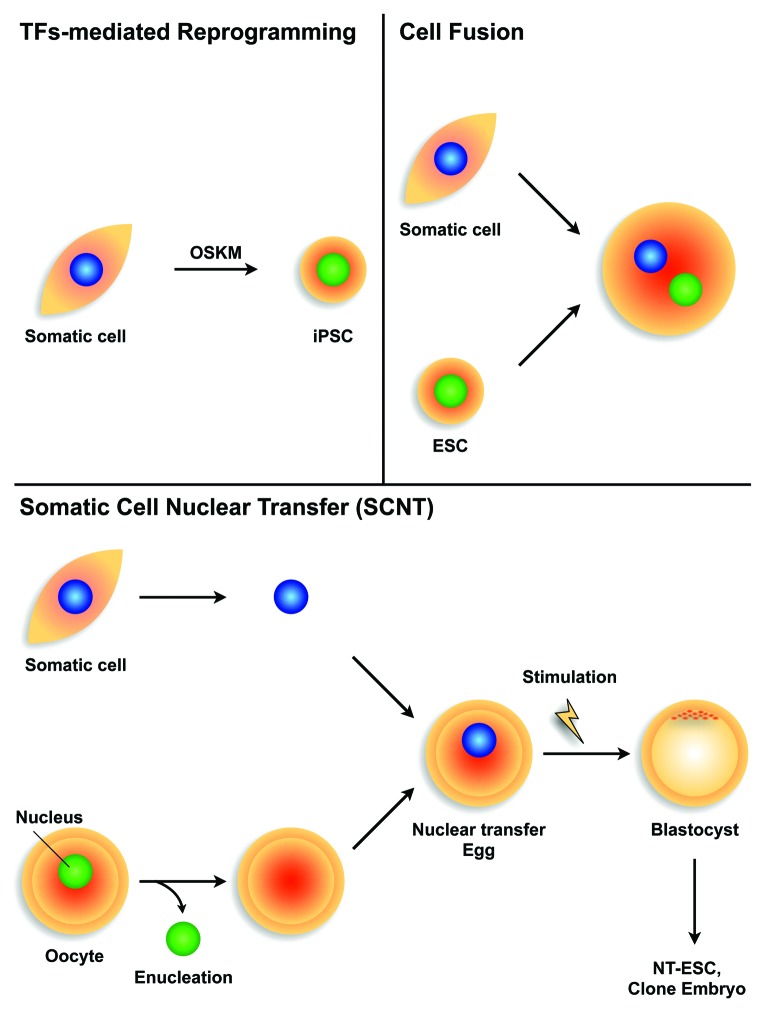
Three cellular reprogramming methods for converting somatic cells to PSCs have been reported, including SCNT, cell fusion, and direct reprogramming using TFs (Fig. 3). Briggs and King29 reported the first cellular reprogramming by transplanting intact nuclei from amphibian blastocysts into enucleated oocytes using SCNT. The transferred oocyte nuclei were activated to zygotes, which developed into tadpoles. In addition, John Gurdon, Nobel laureate in 2012, challenged SCNT using more differentiated nuclei from a tadpole intestinal cell and succeeded in producing offspring.33 These results indicate that genomic DNA from differentiated cells remains unchanged and retains the capacity to develop reproductive organisms. Although the efficiency of successful cloning is only 1–2%,40 some reports demonstrate cloning of mammals such as sheep41 and mice42 from nuclei of adult cells. Recently, Tachibana et al.43 reported the reprogramming of human somatic cells to pluripotent cells using SCNT. However, the requirement of skillful handling, ethical issues, limited oocyte availability, and low success rates hamper this approach for studying reprogramming mechanisms.
Figure 3. Study design of nuclear reprogramming Transcriptional factor (TF)-mediated reprogramming (upper left); reprogramming TFs such as Oct4, Sox2, Klf4, and c-Myc (OSKM) are introduced into iPSCs from somatic cells. Cell fusion (upper right); two or more different types of cells are fused using methods such as electrical cell fusion, polyethylene glycol cell fusion, or Sendai virus-induced cell fusion. Somatic cell nuclear transfer (SCNT; lower); nuclei from donor cells are transferred into enucleated oocytes. Blastocysts derived from SCNT-oocytes can then be cultured as nuclear transfer-embryonic stem cells (NT-ESCs) or can be implanted into pseudopregnant mice to produce offspring.
Another cellular reprogramming method uses human ESCs to reprogram myeloid precursors following cell–cell fusion.44 Several methods promote cell fusion, including the use of polyethylene glycol, Sendai viruses, and electric pulses. Cell fusion using human or mouse ESCs offers an inexpensive and accessible system, in which the sequence of remodeling events results in the successful conversion of somatic cells to an ES-like state.45-49 This approach provided essential insights into the mechanisms underlying the reversion of cell fate, including epigenetic plasticity. However, cell fusions of two or more different somatic cells led to the formation of heterokaryons, in which parental nuclei remain discrete within the same cytoplasm and are therefore unsuitable for medical applications such as in regenerative medicine.
The hypothesis that reprogramming factors exist in the cytoplasm, as suggested by SCNT,32,33,43 ESC fusion experiments,47,49,50 and myogenic differentiation using overexpressed MyoD,51 open a door into the frontier of TF-mediated reprogramming. Yamanaka et al.43,44 generated PSCs from somatic cells using TFs.35,52 The four TFs, Oct4, Sox2, Klf4, and c-Myc (referred to as OSKM in this review), were sufficient to reprogram somatic cells into pluripotent cells known as iPSCs.53-56 At present, many studies have reported alternative factors such as TFs,55 small molecules,57,58 microRNAs,59,60 proteins,61 mRNAs,62 and plasmids63,64 that can accelerate and increase the reprogramming state.
Mechanisms in Reprogramming during the Initiation Phase
To obtain mechanistic insights into reprogramming, various approaches, including proteomics, comprehensive microarray analysis, metabolomics, and single cell technologies, have been applied to the process of TF-mediated reprogramming to iPSCs. The reprogramming process is delineated into three phases on the basis of the gene expression status. These are known as “initiation,” “maturation,” and “stabilization.”65 Changes in both mRNA and microRNA expression as well as histone modification result in two big waves during the initiation phase and at the start of the stabilization phase, known as the first and second waves, respectively.54
During the initiation phase, donor cells promptly initiate MET,55,66 robust cell proliferation, metabolic changes, and alternative histone modifications. Immediately after exogenous OSKM expression, MK (c-Myc, Klf4) proteins attach to the promoters of genes with open chromatin and proceed to either activate or repress depending on the type of the downstream gene, and OSK (Oct4, Sox2, Klf4) proteins bind multiple distal enhancer regions of genes, except those involved in maintaining pluripotency, which they do not occupy as seen in the case of PSCs.67 Therefore, OSKs are known as promiscuous pioneer factors.68 Such promiscuous binding facilitates the de novo accumulation of histone H3 that is dimethylated at lysine 4 (H3K4me2) on early genes of the reprogramming process such as F-box only protein 15 (Fbxo15), Fgf4, and Sall4; somatic genes such as those involved in the cell cycle (Cdc20 and Cdc25c); and metabolic genes such as phosphofructokinase liver B-type (pfkl) and glucose phosphate isomerase (Gpi). In contrast, binding of MK proteins to the promoter regions of fibroblast-specific genes, such as thymus cell antigen 1 (Thy-1) and collagen type V α 2 (Col5a2), represses transcription and causes loss of pre-existing H3K4me2. The first wave of changes in gene expression (both increase and decrease) is primarily regulated by c-MYC, and the leading part of the second wave is mediated by OCT4 and SOX2. Single cell quantitative analysis unveiled this heterogeneity of expression in a subset of pluripotency genes.55 However, because of the stochastic nature of reprogramming, no predictable specific marker was found for cells that were poised to become bona fide (truly reprogrammed iPSCs) iPSCs prior to the maturation phase.69
Evolving Artificial Reprograming of Cell Fate
Dedifferentiation and Redifferentiation
A newly developed strategy for cell fate switching is similar to a physiological process of appendage regeneration, which was attained by sequential treatments using ectopic OSKM expression for short periods; inhibition of JAK/STAT signaling, involving the maintenance of pluripotency; and exposure to the cardiac differentiation culture condition.70 OSKM was putatively assumed to induce the dedifferentiation of donor cells into the intermediate state. Thereafter, the cells were intended to be driven into cardiac lineages rather than being launched into the deterministic phase for reprogramming to pluripotency. Because this “primed conversion”71 operates on a scheme similar to appendage regeneration, with initial dedifferentiation and subsequent redifferentiation, it may be attractive to develop a medical treatment in humans.
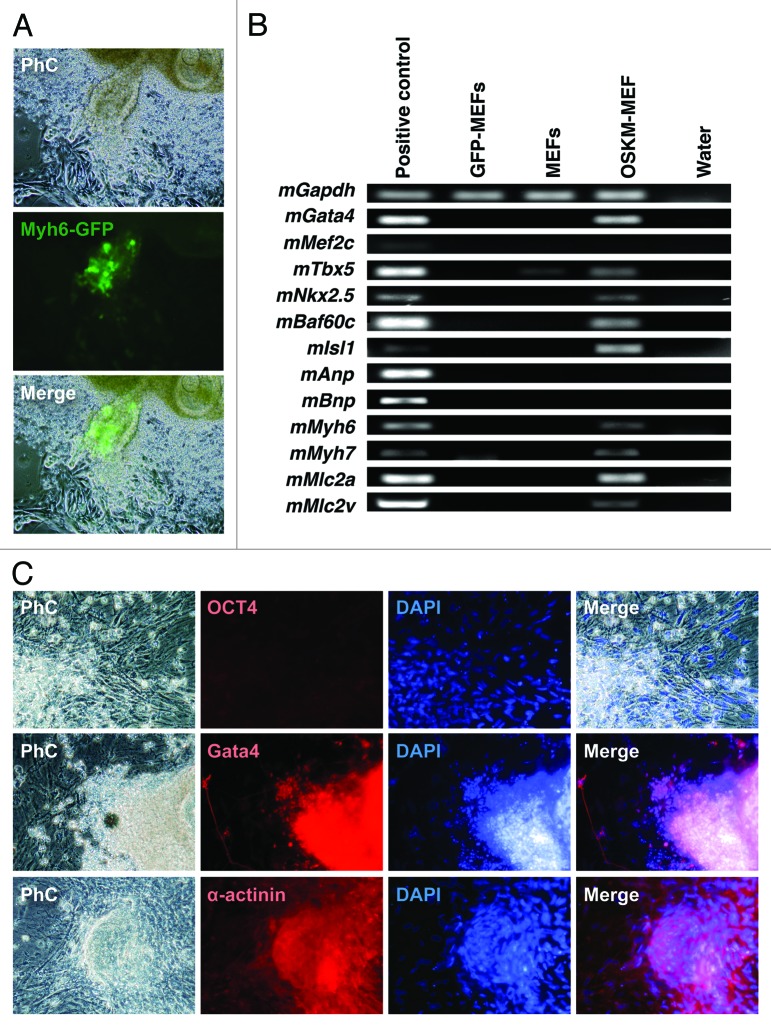
We also examined this primed conversion to cardiomyocytes. Subsequently, mouse embryonic fibroblasts (MEFs) were transduced with a cardiomyocyte-specific myosin heavy chain (Myh6) promoter to drive GFP expression. Fourteen days after infection, OSKM-induced MEFs (OSKM-MEFs) locally formed few cell clusters, which were GFP positive and automatically contractile (Fig. 4A). The genes Gata4, Tbx5, Nkx2.5, Baf60c, and Isl1 were strongly expressed; Myh6, Myh7, Mlc2a, and Mlc2v were also expressed, with the strongest being Mlc2a expression. However, Mef2c, Anp, and Bnp expression was undetectable by reverse transcription polymerase chain reaction (RT-PCR) (Fig. 4B). These results indicated that OSKM-MEFs could be heterogeneous and may have included atrial- or ventricle-like cardiomyocytes. No Oct4-positive cells were detected in the cluster (Fig. 4C). However, OSKM-MEF cell clusters were positive for the cardiac-related proteins Gata4 and α-actinin in immunohistochemical experiments (Fig. 4C). To the best of our knowledge, no primed conversion to human cardiomyocytes has been reported.
Figure 4. Cardiac differentiation of OSKM-mediated mouse embryonic fibroblasts (MEFs) via primed conversion (A) Phase contrast microscopic view of OSKM-mediated MEFs 14 d after OSKM infection. OSKM-mediated MEFs were differentiated into cardiomyocytes. (B) Reverse transcription polymerase chain reaction (RT-PCR) analysis of gene expression. (C) Fluorescent microscopic view with immunofluorescent staining of OSKM-mediated MEFs, which were stained with DAPI, OCT4, Gata4, and α-actinin antibodies.
Direct Conversion
Cellular dedifferentiation switches a program that drives the specific function of a somatic cell to another program for either proliferation, cell death, or redifferentiation, irrespective of whether the destination is the same as the origin.72 The last avenue without mitosis refers to the conversion. Since the discovery of human iPSCs, regenerative medicine using iPSC-derived differentiated cells has been an enthusiastic target in all areas, particularly cardiology and neurology. The drawbacks of this strategy include the risk of teratoma and time-consuming establishment of bona fide iPSCs that lead to target cells. The direct reprogramming approach provides an additional possibility of differentiation to functional cells such as cardiomyocytes,73-78 hepatocytes,79,80 β-cells,80 brown fat,81 chondrocytes,82,83 chondrosarcoma,84 macrophages,85,86 blood progenitors,87 myocytes,51 and neurons88-97 using lineage-specific combinations of TFs (Table 1). These methods were known as “direct conversion” (Fig. 3).71 Cardiomyocyte-like cells (iCMs) were obtained by introducing Gata4, Mef2c, and Tbx5 (GMT) into murine fibroblasts, despite the rare efficacy.74 Thereafter, a combination of miRNAs 1, 133, 208, and 499 also induced murine iCMs from fibroblasts in vitro.76 Remarkably, in vivo direct gene transfer of GMT98 or GMT and HAND275 resulted in the conversion of cardiac fibroblasts to the cardiomyocyte phenotype and attenuated the cardiac function of the infarcted heart in mice. Recently, human fibroblasts were reprogrammed to iCMs with GMT plus ESRRG and MESP1, and ZFPM2 and myocardin enhanced the conversion efficacy.99 This strategy may offer some advantages such as a reduced cancer risk and shorter time. However, depending on the desired target cells, the factors used to convert them have to be defined for each. Direct conversion has no corresponding physiological process in nature, and it is a purely artificial interventional strategy. Thus, investigations of mechanisms and safety issues, including long-term follow-up prior to any clinical application, are critical.
Table 1. Summary of previously reported cell state conversions.
| Germ layer | Target cell | Species | Parent cell | Inducer | Reference |
|---|---|---|---|---|---|
| Mesoderm | Brown fat | H, M | Myoblast | PRDM16, C/EBPβ | 81 |
| Cardiomyocyte | M | Amniotic mesodermal cell | Tbx5, Gata4, Baf60c | 73 | |
| Cardiomyocyte | M | Cardiac fibroblast | Tbx5, Gata4, Mef2c | 74 | |
| Cardiomyocyte | M | Fibroblast | Tbx5, Gata4, Mef2c, Hand2 | 75 | |
| Cardiomyocyte | M | Fibroblast | miR-1, miR-133, miR-208, miR-499 | 76 | |
| Cardiomyocyte | H | Fibroblast | TBX5, GATA4, HAND2, MYOCARDIN, miR-1, miR-133 | 77 | |
| Cardiomyocyte | H | Fibroblast | TBX5, GATA4, MEF2C, MYOCARDIN, ESRRG, MESP1, ZFPM2 | 78 | |
| Chondrosarcoma | H | Placental cell | T, BCL6, c-MYC, MITF, BAF60C | 84 | |
| Chondrocyte | H, M | Adult dermal fibroblast | SOX9, c-MYC, KLF4 | 82,83 | |
| Macrophage | M | Lymphoid precursor | C/EBPα, PU.1 | 85 | |
| Macrophage | M | β-cell | C/EBPα | 86 | |
| Multilineage blood progenitors | H | Fibroblast | OCT4, Cytokines | 87 | |
| Myocyte | M | Fibroblast | MyoD | 51 | |
| Endoderm | Hepatocyte | M | Fibroblast | Hnf4α, Foxa1, Foxa2, or Foxa3 | 79 |
| Hepatocyte-like cell | M | Fibroblast | Gata4, Hnf1α, Foxa3, and inactivation of p19 (Arf) | 80 | |
| β-cell | M | Adult pancreatic exocrine cell | Ngn3, Pdx1, Mafa | 80 | |
| Ectoderm | Dopaminergic neurons | H, M | Fibroblast | ASCL1, NR4A2, LMX1A | 88 |
| Dopaminergic neurons | H | Fibroblast | ASCL1, BRN2, MYT1L, LMX1A, FOXA2 | 89 | |
| Functional neuron-like cells | H | Fibroblast | ASCL1, NGN2, small molecule | 90 | |
| Functional spinal motor neuron | H, M | Fibroblast | Hb9, ISL1, LHX3, ASCL1, BRN2, MYT1L | 91 | |
| Neuron | M | Embryonic fibroblast | Ascl1, Brn2, Myt1l | 92 | |
| Neuron | H | Fibroblast | ASCL1, BRN2, MYT1L, NEUROD1 | 93 | |
| Neuron | H | Fibroblast | MYT1L, BRN2, miR-124 in differentiation medium | 94 | |
| Neuron | H | Fibroblast | ASCL1, MYT1L, miR-9/9*, miR-124 | 95 | |
| Neuronal cell | H | Fibroblast | ASCL1, BRN2, MYT1L, OLIG2, ZIC1 | 96 | |
| Tripotent neural progenitor | M | Fibroblast | Brn2, Sox2, FoxG1 | 97 |
H, Human; M, Mouse.
Perspective
While dedifferentiation is commonly associated with reentry into the cell cycle, its true nature is to withdraw from a given differentiated state.100 During direct reprogramming to pluripotency, dedifferentiation of donor cells may occur prior to reentry into the cell cycle.101 The fact that supports this notion is that mature B cells need to be dedifferentiated with either C/EBPα or PAX5 before they can be reprogrammed.102 On the other hand, there are few apoptotic cells during blastema formation,103 while many cells that receive OSKM for iPSCs undergo apoptosis.
Because there is no appropriate experimental system to uncouple the stage of dedifferentiation from the cell cycle in animals. One clear example of dedifferentiation is plant protoplasts, which are acquired by the treatment of leaves with cellulase and are a stem cell-like state.72 In tobacco (Nicotiana tabacum), the transition to protoplasts (dedifferentiation) is accompanied with the activation of the transposable element Tnt1.104 In Arabidopsis, the dedifferentiation is associated with large-scale decondensation of the pericentric heterochromatin,105 related to telomerase-independent telomere lengthening, which often involves DNA recombination.106 The increased frequency of somatic recombination in Arabidopsis due to infection and environmental stress107,108 may suggest genetic variation and genome instability during cellular dedifferentiation. The human genome includes transposons and transposon-like repetitive elements, only a small proportion of which remains active. These facts bring attention to the potential risk associated with dedifferentiation processes.109
Unlike dedifferentiation due to artificial intervention such as TF gene transfer and nuclear transplantation, physiological dedifferentiation in amputational regeneration seems to be well organized and regulated to reform tissues. The difference may be an essential cue to develop regenerative medicine based on reprogramming technology. The validation of harmful genetic variation in human iPSCs has been started through whole genome sequencing. Even if a negative impact is detected using the current technology, a thorough evaluation of appendage regeneration should result in progression in the field of medicine using iPSCs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by grants from Ministry of Education, Culture Sports, Science and Technology (MEXT) of Japan.
Glossary
Abbreviations:
- iPSCs
induced pluripotent stem cells
- dpa
day post-amputation
- FGF
fibroblast growth factor
- c-Myc
myelocytomatosis oncogene
- Klf4
Krüppel-like factors 4
- Oct4 (POU domain
class 5, transcription factor 1
- Sox2
sex-determining region Y-box 2
- GFP
green fluorescent protein
- TF
transcription factor
- EMT
epithelial–mesenchymal transition
- plk1
polo-like kinase 1
- SCNT
somatic cell nuclear transfer
- PSC
pluripotent stem cell
- OSKM
Oct4, Sox2, Klf4, and c-Myc
- MK
c-Myc, Klf4
- OSK
Oct4, Sox2, Klf4
- H3K4me2
histone H3 that is dimethylated at lysine 4
- Fbxo15
F-box only protein 15
- pfkl
phosphofructokinase liver B-type
- Gpi
glucose phosphate isomerase
- Thy-1
thymus cell antigen 1
- Col5a2
collagen type V alpha 2
- MEFs
mouse embryonic fibroblasts
- Myh6
myosin heavy chain
- RT-PCR
reverse transcription polymerase chain reaction
- iCMs
Cardiomyocyte-like cells
- GMT
GATA4, MEF2C, and TBX5
- NT-ESCs
nuclear transfer-embryonic stem cells
References
- 1.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–22. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stocum DL, Cameron JA. Looking proximally and distally: 100 years of limb regeneration and beyond. Dev Dyn. 2011;240:943–68. doi: 10.1002/dvdy.22553. [DOI] [PubMed] [Google Scholar]
- 3.Song F, Li B, Stocum DL. Amphibians as research models for regenerative medicine. Organogenesis. 2010;6:141–50. doi: 10.4161/org.6.3.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–7. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–5. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 6.Bader D, Oberpriller J. Autoradiographic and electron microscopic studies of minced cardiac muscle regeneration in the adult newt, notophthalmus viridescens. J Exp Zool. 1979;208:177–93. doi: 10.1002/jez.1402080206. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agata K, Inoue T. Survey of the differences between regenerative and non-regenerative animals. Dev Growth Differ. 2012;54:143–52. doi: 10.1111/j.1440-169X.2011.01323.x. [DOI] [PubMed] [Google Scholar]
- 11.Tal TL, Franzosa JA, Tanguay RL. Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish - a mini-review. Gerontology. 2010;56:231–40. doi: 10.1159/000259327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao N, Jhamb D, Milner DJ, Li B, Song F, Wang M, Voss SR, Palakal M, King MW, Saranjami B, et al. Proteomic analysis of blastema formation in regenerating axolotl limbs. BMC Biol. 2009;7:83. doi: 10.1186/1741-7007-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly DJ, Tassava RA. Cell division and ribonucleic acid synthesis during the initiation of limb regeneration in larval axolotls (Ambystoma mexicanum) J Exp Zool. 1973;185:45–54. doi: 10.1002/jez.1401850105. [DOI] [PubMed] [Google Scholar]
- 14.Loyd RM, Tassava RA. DNA synthesis and mitosis in adult newt limbs following amputation and insertion into the body cavity. J Exp Zool. 1980;214:61–9. doi: 10.1002/jez.1402140109. [DOI] [PubMed] [Google Scholar]
- 15.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–89. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 16.Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–58. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 17.Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–60. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 18.Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–9. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008;18:381–6. doi: 10.1016/j.gde.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knapp D, Schulz H, Rascon CA, Volkmer M, Scholz J, Nacu E, Le M, Novozhilov S, Tazaki A, Protze S, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart R, Rascón CA, Tian S, Nie J, Barry C, Chu LF, Ardalani H, Wagner RJ, Probasco MD, Bolin JM, et al. Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol. 2013;9:e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–24. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison JI, Borg P, Simon A. Plasticity and recovery of skeletal muscle satellite cells during limb regeneration. FASEB J. 2010;24:750–6. doi: 10.1096/fj.09-134825. [DOI] [PubMed] [Google Scholar]
- 25.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–86. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisúa Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 28.Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139:1921–30. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- 29.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A. 2010;107:17206–10. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spemann H. Experimentelle Beitrage zu einer Theorie der Entwicklung. Berlin: Springer, 1936. [Google Scholar]
- 32.Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proc Natl Acad Sci U S A. 1952;38:455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–5. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 34.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–34. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–20. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waddington CH. The strategy of the genes: a discussion of some aspects of theoretical biology. London: Allen & Unwin, 1957. [Google Scholar]
- 40.Gurdon JB, Byrne JA. The first half-century of nuclear transplantation. Proc Natl Acad Sci U S A. 2003;100:8048–52. doi: 10.1073/pnas.1337135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–6. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 42.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–74. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 43.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–38. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Vodyanik MA, He P, Slukvin II, Thomson JA. Human embryonic stem cells reprogram myeloid precursors following cell-cell fusion. Stem Cells. 2006;24:168–76. doi: 10.1634/stemcells.2005-0292. [DOI] [PubMed] [Google Scholar]
- 45.Foshay KM, Looney TJ, Chari S, Mao FF, Lee JH, Zhang L, Fernandes CJ, Baker SW, Clift KL, Gaetz J, et al. Embryonic stem cells induce pluripotency in somatic cell fusion through biphasic reprogramming. Mol Cell. 2012;46:159–70. doi: 10.1016/j.molcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–73. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 48.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 49.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–8. doi: 10.1016/S0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 50.Surani MA. Nuclear reprogramming by human embryonic stem cells. Cell. 2005;122:653–4. doi: 10.1016/j.cell.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 51.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–32. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–92. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Rana TM. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nat Commun. 2012;3:1085. doi: 10.1038/ncomms2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma T, Xie M, Laurent T, Ding S. Progress in the reprogramming of somatic cells. Circ Res. 2013;112:562–74. doi: 10.1161/CIRCRESAHA.111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–8. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–6. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okita K, Hong H, Takahashi K, Yamanaka S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc. 2010;5:418–28. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- 64.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 65.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 66.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 67.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–39. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–22. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 71.Morris SA, Daley GQ. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell Res. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grafi G. How cells dedifferentiate: a lesson from plants. Dev Biol. 2004;268:1–6. doi: 10.1016/j.ydbio.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 73.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–11. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–93. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu J-D, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Reports. 2013;1:235–47. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–3. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 80.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 81.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–8. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Outani H, Okada M, Yamashita A, Nakagawa K, Yoshikawa H, Tsumaki N. Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS One. 2013;8:e77365. doi: 10.1371/journal.pone.0077365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hiramatsu K, Sasagawa S, Outani H, Nakagawa K, Yoshikawa H, Tsumaki N. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J Clin Invest. 2011;121:640–57. doi: 10.1172/JCI44605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishii R, Kami D, Toyoda M, Makino H, Gojo S, Ishii T, Umezawa A. Placenta to cartilage: direct conversion of human placenta to chondrocytes with transformation by defined factors. Mol Biol Cell. 2012;23:3511–21. doi: 10.1091/mbc.E11-10-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–62. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/S0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 87.Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 88.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 89.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Björklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kögler G, Müller FJ, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–8. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 91.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–8. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–71. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Lujan E, Chanda S, Ahlenius H, Südhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–32. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian L, Berry EC, Fu JD, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–15. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- 99.Fu J-D, Stone Nicole R, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau Benoit G, Srivastava D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Reports 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Echeverri K, Tanaka EM. Mechanisms of muscle dedifferentiation during regeneration. Semin Cell Dev Biol. 2002;13:353–60. doi: 10.1016/S1084952102000915. [DOI] [PubMed] [Google Scholar]
- 101.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 102.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–64. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mescher AL, White GW, Brokaw JJ. Apoptosis in regenerating and denervated, nonregenerating urodele forelimbs. Wound Repair Regen. 2000;8:110–6. doi: 10.1046/j.1524-475x.2000.00110.x. [DOI] [PubMed] [Google Scholar]
- 104.Pouteau S, Huttner E, Grandbastien MA, Caboche M. Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 1991;10:1911–8. doi: 10.1002/j.1460-2075.1991.tb07717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci. 2007;120:1200–8. doi: 10.1242/jcs.000026. [DOI] [PubMed] [Google Scholar]
- 106.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 107.Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet. 2002;30:311–4. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- 108.Lebel EG, Masson J, Bogucki A, Paszkowski J. Stress-induced intrachromosomal recombination in plant somatic cells. Proc Natl Acad Sci U S A. 1993;90:422–6. doi: 10.1073/pnas.90.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grafi G. The complexity of cellular dedifferentiation: implications for regenerative medicine. Trends Biotechnol. 2009;27:329–32. doi: 10.1016/j.tibtech.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 110.Yechoor V, Liu V, Espiritu C, Paul A, Oka K, Kojima H, Chan L. Neurogenin3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev Cell. 2009;16:358–73. doi: 10.1016/j.devcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manohar R, Lagasse E. Transdetermination: a new trend in cellular reprogramming. Mol Ther. 2009;17:936–8. doi: 10.1038/mt.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]