Abstract
HLA-DP antigens are beta-alpha heterodimers encoded by polymorphic HLA-DPB1 and -DPA1 alleles, respectively, in strong linkage disequilibrium (LD) with each other. Non-permissive unrelated donor (UD)-recipient HLA-DPB1 mismatches across three different T cell epitope (TCE) groups are associated with increased mortality after hematopoietic cell transplantation (HCT), but the role of HLA-DPA1 is unclear. We studied 1281 onco-hematologic patients after 10/10 HLA-matched UD-HCT facilitated by the National Marrow Donor Program. Non-permissive mismatches defined solely by HLA-DPB1 TCE groups were associated with significantly higher risks of treatment-related mortality compared to permissive mismatches (HR 1.30, CI 1.06–1.53; p=0.009) or allele matches. Moreover, non-permissive HLA-DPB1 TCE group mismatches in the graft versus host (GvH) direction significantly decreased the risk of relapse compared to permissive mismatches (HR 0.55, CI 0.37–0.80; p=0.002) or allele matches. Splitting each group into HLA-DPA1*02:01 positive or negative, in frequent LD with HLA-DPB1 alleles from two of the three TCE groups, or into HLA-DPA1 matched or mismatched, did not significantly alter the observed risk associations. Our findings suggest that the effects of clinically non-permissive HLA-DPB1 TCE group mismatches are independent of HLA-DPA1, and that selection of donors with non-permissive DPB1 TCE mismatches in GvH direction might provide some protection from disease recurrence.
Keywords: Non-permissive mismatch, HLA-DPB1 T cell epitope matching, Unrelated HCT, relapse, transplant related mortality, HLA-DPA1
Introduction
HLA-DP antigens are heterodimers formed by a polymorphic alpha and a polymorphic beta chain encoded by the HLA-DPA1 and -DPB1 loci in the class II region of the major histocompatibility complex (MHC), respectively. With 248 currently described alleles, the HLA-DPB1 locus displays considerably higher polymorphism than the HLA-DPA1 locus, for which only 37 different alleles have been reported so far. Of these, only 12 HLA-DPB1 and 3 -DPA1 alleles are found with a frequency of over 1% in Caucasians.1 The effective HLA-DP antigen polymorphism is further limited by tight linkage disequilibrium (LD) between HLA-DPA1 and -DPB1, with most HLA-DPB1 alleles found in cis with either HLA-DPA1*01:03 or -DPA1*02:01. However, potential heterodimer pairing between trans-encoded alpha and beta chains can increase phenotypic variability, depending on the status of heterozygosity.
In unrelated hematopoietic stem cell transplantation (HCT), donor-recipient allelic HLA-DPB1 disparity occurs in approximately 80% of pairs matched for 10/10 of the non-DP HLA loci (HLA-A, -B, -C, -DRB1 and –DQB1), due to a lack of consideration in donor selection and low LD between HLA-DP and the other HLA class II loci. Donor T cell alloreactivity against allelic HLA-DPB1 mismatches in the recipient is a significant protective factor against leukemia relapse,2–3 however, overall mortality (OM) is not improved due to concomitantly increased acute graft versus host disease (aGvHD).4–5 An innovative approach to defining “non-permissive”, i.e. clinically poorly tolerated HLA-DPB1 mismatches on the basis of alloreactive T cell epitope (TCE) groups, was recently shown to identify subgroups of patients with significant risks of aGvHD and transplant-related mortality.6–8 In this model, HLA-DPB1 alleles are classified into three distinct TCE groups on the basis of T cell crossreactivity patterns. The discrimination of permissive and non-permissive HLA-DPB1 mismatches is made based on whether the donor and recipient alleles are from the same (permissive) or different (non-permissive) TCE groups. The model is based on the rationale that the alloreactive T cell repertoire is shaped by thymic selection, leading to central deletion of clones reactive to self-epitopes. TCE group matching therefore considers not only the mismatch itself, but also the second DPB1 allele carried by patient or donor, which may contribute to the final repertoire of alloreactive T cells. A potential limitation of this model is that there may be differences in the magnitude of the functional effect of mismatches involving TCE groups 1 and 2 compared to 1 and 3, or 2 and 3, all considered as non-permissive. Such subtle differences are difficult to investigate due to statistical power limitations arising from the numerous subgroups involved in such an analysis. A web-based tool for application of the algorithm in unrelated donor searches has recently been made available through the IMGT-HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/dpb.html).9
An unanswered and clinically relevant question is whether or not HLA-DPA1 polymorphism contributes to the immunogenicity of non-permissive HLA-DPB1 mismatches. Interestingly, most HLA-DPB1 alleles belonging to two of the three distinct TCE groups, the highest-risk group containing HLA-DPB1*09:01, *10:01, *14:01, *17:01 and 45:01, that are in LD with HLA-DPA1*02:01, while most frequent alleles from the third, lowest-risk TCE group including HLA-DPB1*02:01, *04:01 and *04:02 that are in LD with HLA-DPA1*01:03. This raises the possibility that the presence of HLA-DPA1*02:01, encoded either in cis or in trans with a non-permissive HLA-DPB1 TCE high-risk group mismatch, might be necessary for the formation of the relevant allo-epitope. Alternatively, donor-recipient allelic HLA-DPA1 disparity recognized by a TCR expressed on donor T-cells might be required to reveal the immunogenicity of non-permissive HLA-DPB1 TCE group mismatches. In the present study, we have tested both hypotheses by retrospective clinical analysis of 1281 10/10 HLA-matched unrelated HCTs facilitated through the National Marrow Donor Program (NMDP).
Methods
Data source
The Center for International Blood and Marrow Transplant Research (CIBMTR) is a research affiliation of the International Bone Marrow Transplant Registry (IBMTR), Autologous Blood and Marrow Transplant Registry (ABMTR) and the NMDP established in 2004 that comprises a voluntary working group of more than 450 transplantation centers worldwide that contribute detailed data on consecutive allogeneic and autologous HCT to a Statistical Center at the Medical College of Wisconsin in Milwaukee and the NMDP Coordinating Center in Minneapolis. Participating centers are required to report all transplants consecutively; compliance is monitored by on-site audits. Patients are followed longitudinally. Computerized checks for discrepancies, physicians’ review of submitted data and on-site audits of participating centers ensure data quality. Observational studies conducted by the CIBMTR are performed in compliance with all applicable federal regulations pertaining to the protection of human research participants. Protected Health Information used in the performance of such research is collected and maintained in CIBMTR’s capacity as a Public Health Authority under the HIPAA Privacy Rule.
Study population
The study included 1281 patients who received a first marrow or peripheral blood stem cell unrelated donor transplantation between 1988 and 2003, for the treatment of hematologic malignancies. The clinical and immunogenetic characteristics of the patients and transplants are detailed in Table 1. The cohort included all those patients and donors for whom complete HLA typing data, including HLA-DPA1 and DPB1, were available through the CIBMTR.
Table 1.
Patient and Transplant characteristics.
| Full matches |
TCE Permissive mismatches |
TCE Non-permissive mismatches |
P-valuea | |
|---|---|---|---|---|
| Variable | N (%) | N (%) | N (%) | |
| Number of Patients | 239 | 585 | 457 | |
| Number of Centers | 54 | 84 | 82 | |
| Median Recipient Age | 36 (1–61) | 36 (<1–65) | 36 (<1–63) | 0.31 |
| Recipient Age | 0.57 | |||
| 0–9 | 17 (7) | 46 (8) | 36 (8) | |
| 10–19 | 17 (7) | 62 (11) | 45 (10) | |
| 20–29 | 39 (16) | 106 (18) | 78 (17) | |
| 30–39 | 61 (26) | 135 (23) | 117 (26) | |
| 40–49 | 62 (26) | 152 (26) | 128 (28) | |
| 50+ | 43 (18) | 84 (14) | 53 (12) | |
| Recipient Race/Ethnicity | 0.25 | |||
| Caucasian | 217 (91) | 551 (94) | 431 (94) | |
| African American | 3 (1) | 9 (2) | 6 (1) | |
| Asian/Pac. Islander | 4 (2) | 5 (1) | 2 (<1) | |
| Hispanic | 12 (5) | 19 (3) | 14 (3) | |
| Native American | 0 | 1 (<1) | 0 | |
| Other/Multiple/Unknown | 3 (1) | 0 | 4 (1) | |
| Sex | 0.79 | |||
| Male | 136 (57) | 331 (57) | 268 (59) | |
| Female | 103 (43) | 254 (43) | 189 (41) | |
| Karnofsky Score | 0.58 | |||
| < 90 | 67 (28) | 134 (23) | 119 (26) | |
| >= 90 | 162 (68) | 426 (73) | 320 (70) | |
| Missing | 10 (4) | 25 (4) | 18 (4) | |
| Disease | 0.03 | |||
| AML | 66 (28) | 153 (26) | 133 (29) | |
| ALL | 39 (16) | 137 (23) | 105 (23) | |
| CML | 102 (43) | 188 (32) | 153 (33) | |
| MDS | 32 (13) | 107 (18) | 66 (14) | |
| Disease Status at Txb | 0.09 | |||
| Early | 167 (70) | 407 (70) | 319 (70) | |
| Intermediate | 11 (5) | 34 (6) | 22 (5) | |
| Advanced | 61 (26) | 132 (23) | 114 (25) | |
| Other | 0 | 12 (2) | 2 (<1) | |
| HLA-DPB1 Matching | < 0.0001 | |||
| Two Mismatches | 0 | 126 (22) | 234 (51) | |
| One Mismatch | 0 | 459 (78) | 223 (49) | |
| Fully Matched | 239 (100) | 0 | 0 | |
| HLA-DPA1*0201 | < 0.0001 | |||
| Absent | 181 (76) | 295 (50) | 183 (40) | |
| Present | 58 (24) | 290 (50) | 274 (60) | |
| HLA-DPA1 allele | < 0.0001 | |||
| Matched | 229 (96) | 321 (55) | 219 (48) | |
| Mismatched | 10 (4) | 264 (45) | 238 (52) | |
| Graft Type | 0.08 | |||
| Bone Marrow | 206 (86) | 513 (88) | 417 (91) | |
| PBSC | 33 (14) | 72 (12) | 40 (9) | |
| GVHD Prophylaxis | 0.004 | |||
| FK506 + (MTX, MMF or Steroids) ± Other | 70 (29) | 147 (25) | 132 (29) | |
| FK506 ± Other | 2 (1) | 2 (<1) | 1 (<1) | |
| CSA + MTX ± Other | 155 (65) | 416 (71) | 294 (64) | |
| CSA ± Other (No MTX) | 8 (3) | 17 (3) | 25 (5) | |
| MTX ± Other (No CSA) | 0 | 1 (<1) | 5 (1) | |
| Other | 4 (2) | 2 (<1) | 0 | |
| In vivo T-cell depletion | 0.49 | |||
| Yes | 31 (13) | 60 (10) | 54 (12) | |
| No | 208 (87) | 525 (90) | 403 (88) | |
| Sex Match | 0.11 | |||
| Male -> Male | 88 (37) | 233 (40) | 200 (44) | |
| Male -> Female | 60 (25) | 127 (22) | 115 (25) | |
| Female -> Male | 48 (20) | 98 (17) | 68 (15) | |
| Female -> Female | 43 (18) | 127 (22) | 74 (16) | |
| CMV Match | 0.92 | |||
| Negative/Negative | 81 (34) | 208 (36) | 167 (37) | |
| Negative/Positive | 64 (27) | 159 (27) | 128 (28) | |
| Positive/Negative | 40 (17) | 98 (17) | 65 (14) | |
| Positive/Positive | 48 (20) | 103 (18) | 88 (19) | |
| Unknown | 6 (3) | 17 (3) | 9 (2) | |
| Median Donor Age | 35 (18–59) | 36 (18–60) | 35 (19–57) | 0.10 |
| Donor Age | 0.45 | |||
| 18–29 | 69 (29) | 154 (26) | 133 (29) | |
| 30–39 | 88 (37) | 228 (39) | 192 (42) | |
| 40–49 | 64 (27) | 167 (29) | 105 (23) | |
| 50+ | 18 (8) | 36 (6) | 27 (6) | |
| Conditioning Regimen | 0.87 | |||
| Cy + TBI | 181 (76) | 421 (72) | 329 (72) | |
| Bu + Cy | 39 (16) | 124 (21) | 96 (21) | |
| TBI 500 single dose or 800 fractionated | 10 (4) | 22 (4) | 19 (4) | |
| Melphalan > 150 mg/m2 | 5 (2) | 9 (2) | 5 (1) | |
| Bu > 9 mg/kg | 4 (2) | 9 (2) | 8 (2) | |
| Year of Transplant | 0.61 | |||
| 1988–1991 | 8 (3) | 23 (4) | 15 (3) | |
| 1992–1994 | 41 (17) | 72 (12) | 63 (14) | |
| 1995–1997 | 49 (21) | 146 (25) | 116 (25) | |
| 1998–2000 | 85 (36) | 189 (32) | 151 (33) | |
| 2001–2003 | 56 (23) | 155 (26) | 112 (25) | |
| Median follow-up of survivors, months | 99 (13 – 200) | 106 (10 – 245) | 102 (22 – 196) | 0.52b |
Abbreviations:
CsA = Cyclosporine; CMV = Cytomegalovirus; FK506 = Tacrolimus; HLA = Human leukocyte antigens; MMF = Mycophenolate mofetil; MTX = Methotrexate; tx = transplant.
Log-rank p-value.
Early stage disease was defined as AML or ALL in first complete remission, CML in first chronic phase, and MDS subtype refractory anemia. Intermediate stage disease was defined as AML or ALL in second or subsequent complete remission, and CML in accelerated phase or second chronic phase. Advanced phase disease was defined as AML in first or higher relapse or primary induction failure, CML in blast phase, MDS subtypes refractory anemia with excess blasts or in transformation.
Surviving patients who did not provide signed, informed consent to allow analysis of their clinical data or HLA typing of stored NMDP Research Repository samples were excluded. All surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program. To adjust for the potential bias introduced by exclusion of non-consenting surviving patients, a modeling process randomly excluded the same percentage of deceased patients using a biased coin randomization with exclusion probabilities based on characteristics associated with not providing consent for use of the data in survivors.10 This procedure is standard for CIBMTR analyses to avoid bias from the retrospective consent process.
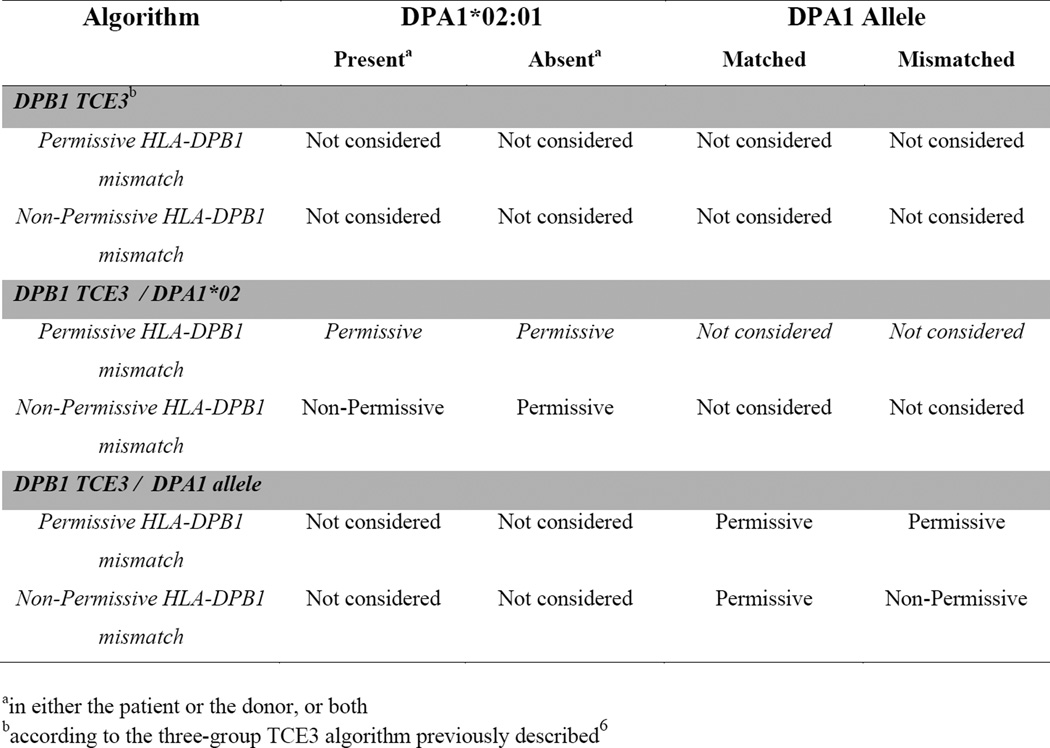
Only patients with high resolution typing of HLA-A, B, C, DRB1, DQB1, DPA1 and DPB1, verified using appropriate DNA-based methods as previously described,11 were included in the study. All cases were high resolution matched for HLA-A, B, C, DRB1 and DQB1 (10/10 allele matched). Cases were divided by HLA-DPB1 type into three TCE categories: HLA-DPB1 matched, permissively HLA-DPB1 mismatched (TCE matched) and non-permissively HLA-DPB1 mismatched (TCE mismatched), according to the three-group model (TCE3) as previously described.6 HLA-DPA1 was classified by the presence or absence of HLA-DPA1*02:01, or as matched and mismatched at the allele level (Figure 1).
Figure 1. Algorithms for permissive or non-permissive HLA-DP TCE group mismatches used in this study.
10/10 HLA allele matched donor-recipient pairs were classified according to three different algorithms: 1) DPB1 TCE3 considers as non-permissive only those pairs in whom the HLA-DPB1 allele mismatch involves a TCE3 group mismatches, regardless of the HLA-DPA1 matching status, as previously described.6 2) DPB1 TCE3/DPA1*02:01 considers as non-permissive only those pairs in whom the HLA-DPB1 allele mismatch involves a TCE3 group mismatch, and in whom either the patient or the donor, or both, carry at least one DPA1*02:01 allele. 3) DPB1 TCE3/DPA1 allele considers as non-permissive only those pairs in whom the HLA-DPB1 allele mismatch involves a TCE3 group mismatch, and in whom a donor-recipient HLA-DPA1 allele mismatch is also present.
Clinical Outcomes
The primary outcomes were overall survival, transplant related mortality, relapse and acute GVHD. Overall survival (OS) considered death from any cause as the event and surviving patients were censored at the date of last contact. Treatment-related mortality (TRM) was defined as death in continuous remission of the primary disease with relapse as a competing risk. Relapse was defined as disease recurrence with death in continuous complete remission as a competing risk. Acute GVHD was assigned using the IBMTR consensus criteria with death as a competing risk.
Statistical methods
To compare pre-transplant characteristics for discrete factors, the number of cases and their respective percentages were calculated and chi-square tests were performed to compare the three histocompatibility groups; HLA-DPB1 matched, permissive mismatched and non-permissive mismatched. For continuous factors, the medians and ranges were calculated and the Kruskal-Wallis test was used to analyze differences between the groups. Probabilities of overall survival were calculated using the Kaplan-Meier estimator. Estimated cumulative incidence was used to describe the probabilities for events with competing risks. These included GVHD, relapse and TRM. Comparisons of survival curves were done with the log-rank test.
Multivariate analyses were performed using the Cox proportional hazards model, evaluating each HLA-DP matching algorithm (Figure 1) independently as the main effect. Models were fit to determine which risk factors were related to a given outcome. All variables were tested for affirmation of the proportional hazards assumption. Factors violating the proportional hazards assumption were adjusted through stratification. A stepwise model building procedure was used to select risk factors for each outcome with a threshold of p≤0.05 for entering into the models. Due to multiple comparisons, p <0.01 was used to determine statistical significance for the main effect. All analyses were performed using SAS version 9.2 (SAS Institute, Inc).
Results
Study Population and Outcomes
A total of 1281 recipients of fully HLA-A, B, C, DRB1, DQB1, DPA1, DPB1 high resolution typed UD-HCTs were included in the analysis. The HLA-DPA1 and –DPB1 alleles found in this study population are listed in supplementary Table 1. All pairs were matched for HLA-A, B, C, DRB1 and DQB1 alleles (10/10), and 239 of them (19%) were also HLA-DPB1 allele matched (12/12), while the remaining 1042 (81%) pairs were HLA-DPB1 allele mismatched. In the latter group, permissive and non-permissive mismatches were determined using the three-group TCE3 algorithm previously described,6 resulting in 585 permissively mismatched and 457 non-permissively mismatched cases (Table 1). Of the latter, 226 and 231 were mismatched in the Graft versus Host (GvH) or Host versus Graft (HvG) direction, respectively. HLA-DPA1 allele mismatches were present in 512 (40%) of the 1281 pairs overall, and in 502 (48.2%) of the 1042 HLA-DPB1 allele mismatched pairs. The HLA-DPB1 allele matched cohort included more CML cases (43%) and fewer ALL cases (16%) than the permissive (32%-CML, 23%-ALL) and non-permissive groups (33%-CML, 23% ALL), and the groups differed slightly for GVHD prophylaxis regimens. All other characteristics were well balanced across the groups (Table 1).
Over 90% of the patients under study were of Caucasian origin and 87% received bone marrow as the stem cell source. 1159 (90%) of the transplants were part of the International Histocompatibility Working Group (IHWG) cohort previously analyzed for the effects of HLA-DPB1 TCE matching in unrelated HCT,6 while 122 (10%) were new cases.
The overall incidence of the four major clinical endpoints of this study were OM 63% at 5 years, TRM 42% at 5 years, severe grade C-D aGvHD 32% at 100 days, and relapse 25% at 5 years with a median follow-up time of 102 months.
Algorithms for the role of HLA-DPA1 in non-permissive mismatches
In order to investigate the role of HLA-DPA1 in associations between clinical risks and non-permissive HLA-DPB1 TCE group mismatches, two different algorithms were analyzed separately. The first, designated DPB1 TCE3/DPA1*02:01, was evaluated due to the observed strong LD between alleles from 2 of the 3 HLA-DPB1 TCE groups and HLA-DPA1*02:01,1 and predicts the presence of HLA-DPA1*02:01 in the patient, the donor or both as a necessary requirement for the assignment of a non-permissive HLA-DPB1 mismatch (Figure 1). The second, designated DPB1 TCE3/DPA1 allele, is based on the hypothesis that allelic mismatches for HLA-DPA1 are a necessary requirement for a HLA-DPB1 TCE3 group mismatch to be classified as non-permissive (Figure 1). In the two algorithms, the number of permissive/non-permissive mismatched cases was 768/274 and 804/238, respectively (Table 1).
Clinical risk associations of non-permissive HLA-DPB1 TCE group mismatches
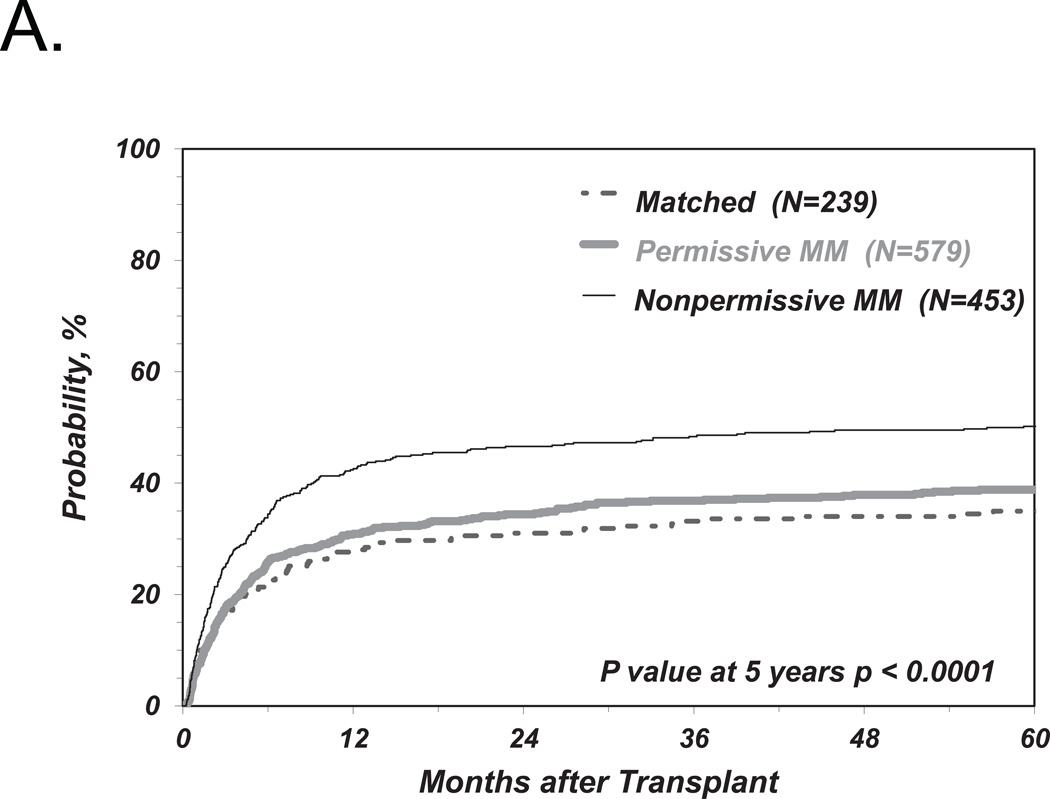
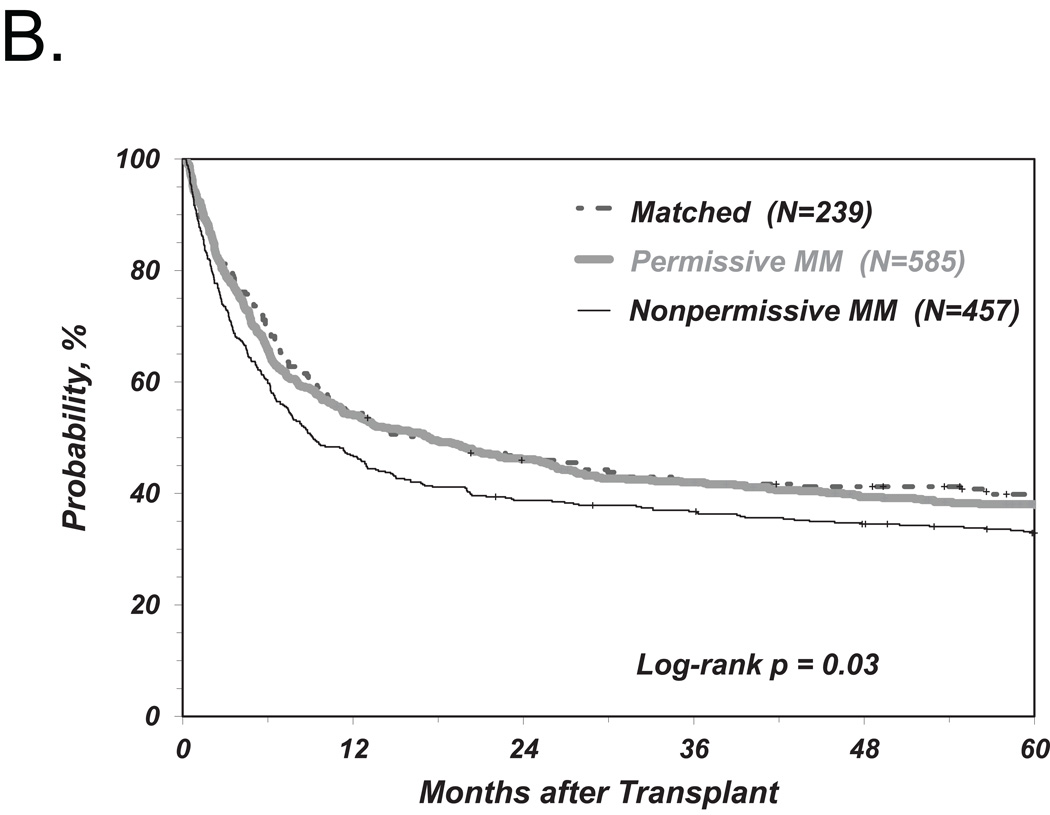
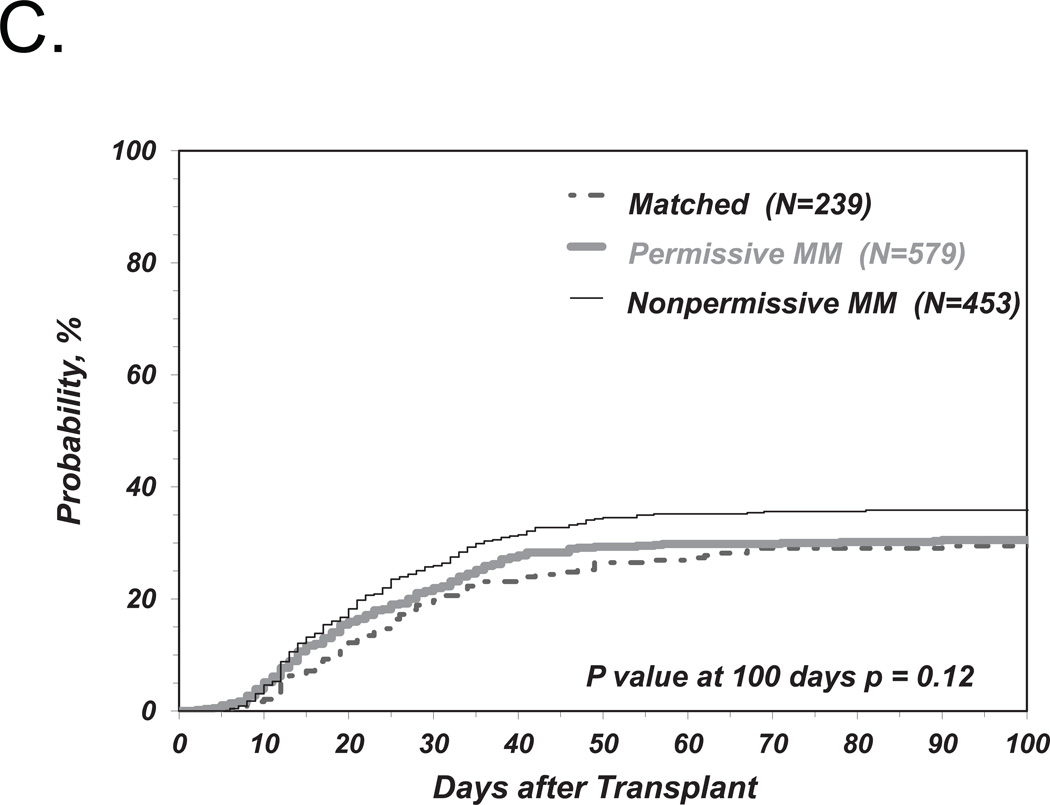
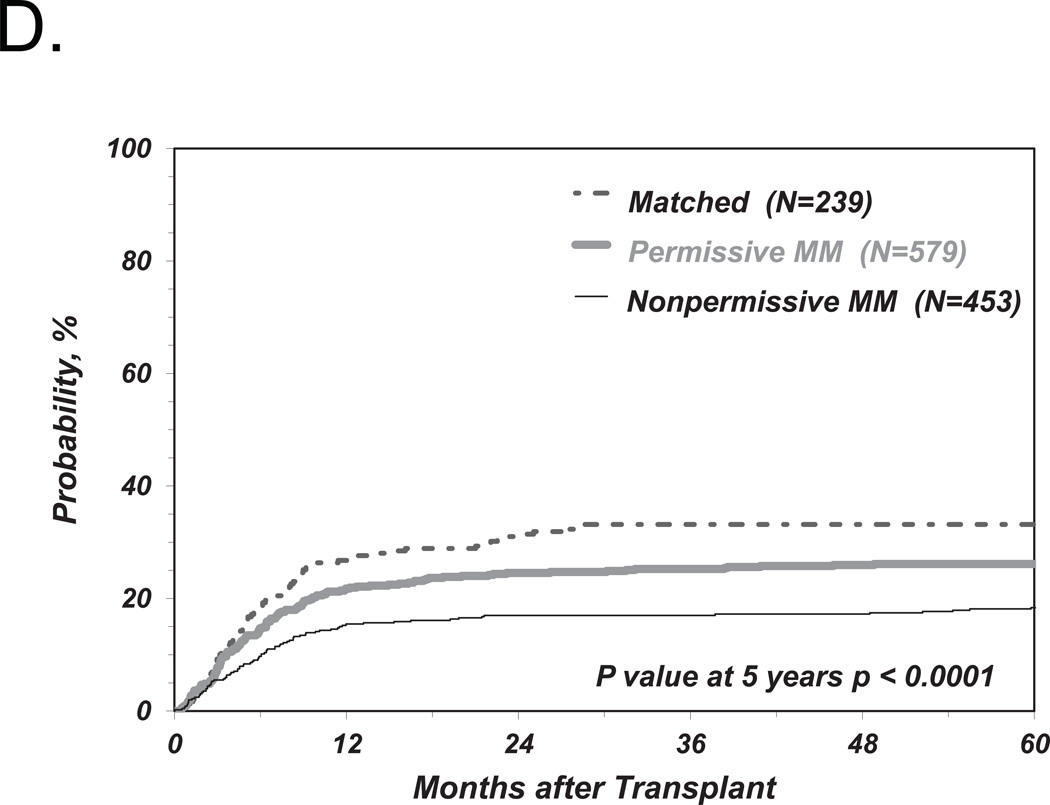
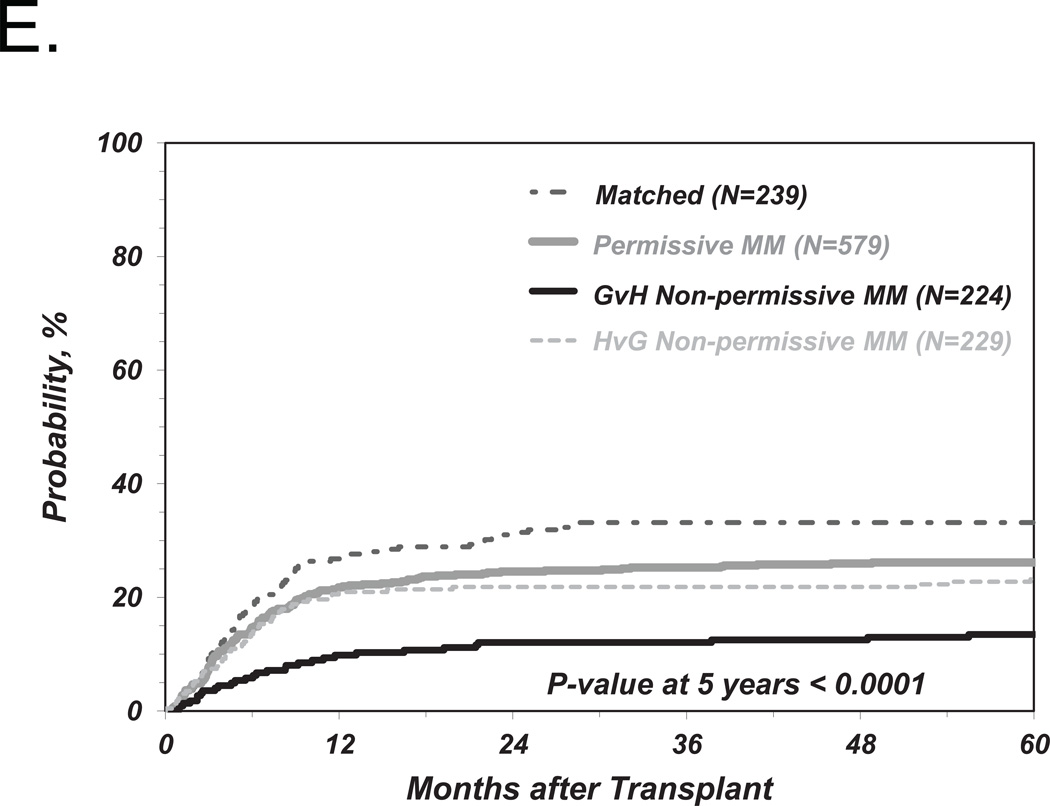
We first investigated the associations between clinical endpoints of UD-HCT and non-permissive HLA-DPB1 TCE3 group mismatches as previously described6, i.e. without consideration of HLA-DPA1 (Figure 1). Consistent with our previous results, we found a significant correlation between non-permissive HLA-DPB1 mismatches in either the GvH or HvG direction and TRM (Table 2 and Figure 2A). Increased risks were also observed for OM and severe aGvHD, with HR similar to those observed in our previous report6, although these were not statistically significant according to the definitions from the present study (Table 2 and Figure 2B, C). Also in line with previous observations, HLA-DPB1 allele matched transplants were associated with an increased risk of relapse compared to permissive or non-permissive transplants in univariate analysis (Figure 2D), but did not reach statistical significance when adjusted for significant co-variates (Table 2).
Table 2.
Multivariate regression models for association between non-permissive DPB1 TCE3 mismatches and clinical outcome.
| Permissive HLA-DPB1 mismatch (N = 585) |
HLA-DPB1 match (N = 239) |
Non-Permissive HLA-DPB1 mismatch (N = 457) |
Non-Permissive HLA-DPB1 GvH (N=226) |
Non-Permissive HLA-DPB1 HvG (N=231) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Ref. | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Overall mortalitya | 1 | 0.94 (0.78–1.14) | 0.56 | 1.16 (0.99–1.35) | 0.05 | 1.11 (0.92–1.35) | 0.27 | 1.20 (1.00–1.44) | 0.05 |
| Treatment-related mortalityb | 1 | 0.86 (0.68–1.11) | 0.03 | 1.3 (1.06–1.53) | 0.009 | 1.29 (1.03–1.60) | 0.03 | 1.27 (1.01–1.58) | 0.04 |
| Relapsec | 1 | 1.31 (0.99–1.73) | 0.05 | 0.80 (0.62–1.05) | 0.1 | 0.55 (0.37–0.80) | 0.002 | 1.09 (0.8–1.48) | 0.6 |
| aGVHD C–Dd | 1 | 0.96 (0.73–1.26) | 0.77 | 1.27 (1.02–1.57) | 0.03 | 1.34 (1.03–1.74) | 0.03 | 1.20 (0.92–1.56) | 0.17 |
adjusted for disease, disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen, and recipient age at transplant.
adjusted for disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen and recipient age at transplant.
adjusted for disease and disease status.
adjusted for disease, disease status, graft type and year of transplant.
Figure 2. Risk-associations between HLA-DPB1 TCE group mismatches and outcome of UD-HCT.
Shown are the cumulative incidence curves of TRM (A), KM curves of OS (B), cumulative incidence curves of grade 3–4 GvHD (C) and cumulative incidence curves of Relapse (D, E) in patients matched for 10/10 of the non-HLA-DP alleles and matched for both HLA-DPB1 alleles (“Matched”), HLA-DPB1 allele mismatched but TCE group matched (“Permissive”), HLA-DPB1 allele mismatched and TCE group mismatched (“Non-Permissive”) (A-D), or HLA-DPB1 allele mismatched and TCE group mismatched in the GvH (“Non-Permissive GvH”) or HvG (“Non-Permissive HvG”) direction (E).
The risk stratifications for the endpoints TRM and aGvHD were evenly distributed between non-permissive mismatches in the GvH or HvG direction (Table 2) which, again, is in line with previous results. Interestingly however, the risks of relapse were significantly lower after transplantation from a donor non-permissively mismatched in the GvH but not in the HvG direction, compared to transplants from a permissively mismatched donor (Table 2 and Figure 2E). However, we did not find a significantly lower risk of OM associated with non-permissive GvH compared to HvG mismatches (Table 2).
Similar results were obtained for the four-group model of HLA-DPB1 TCE4 group matching previously described (data not shown).8
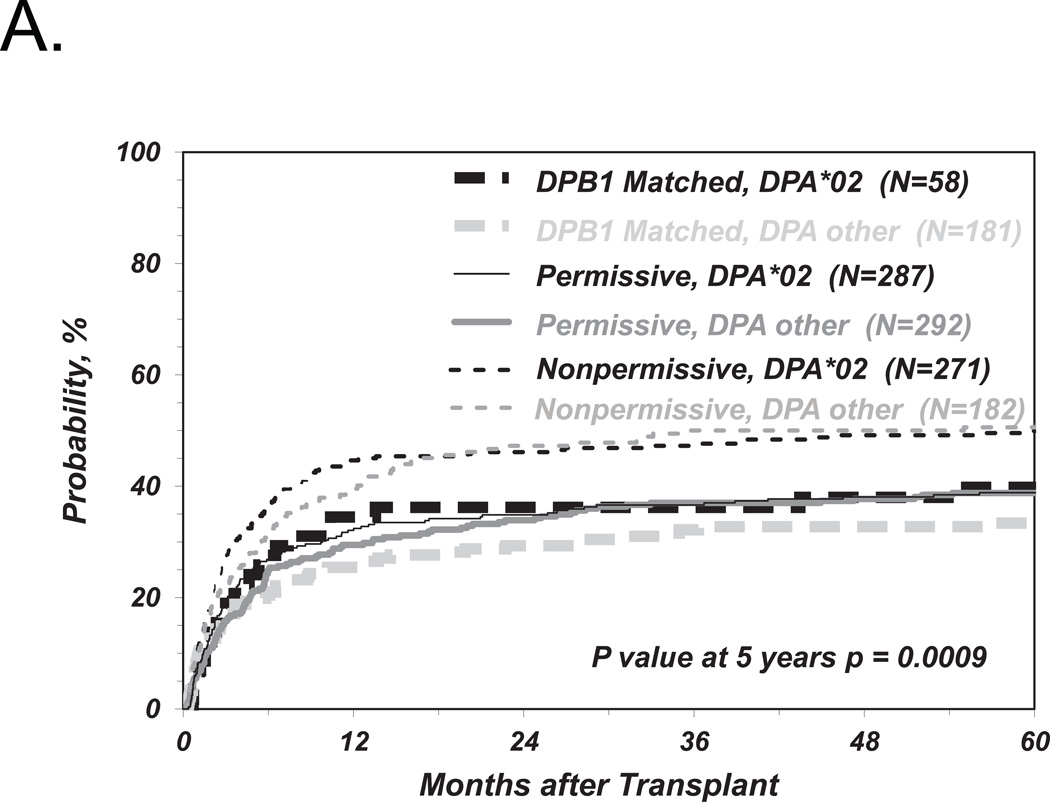
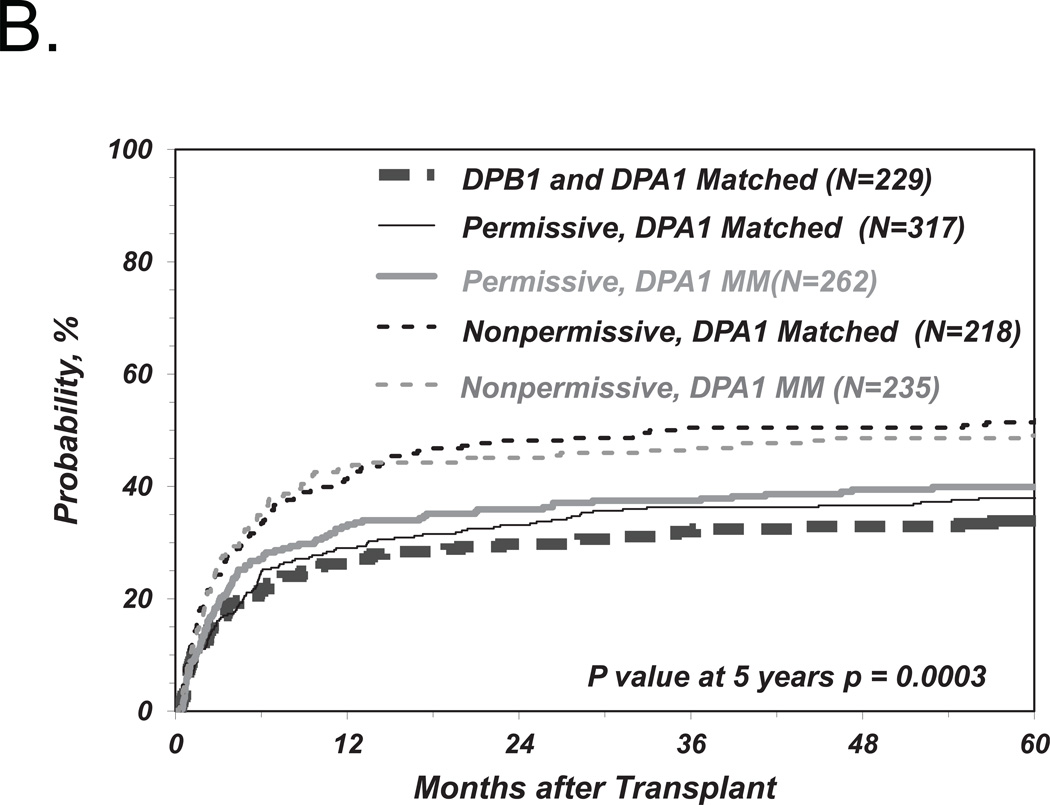
Role of HLA-DPA1 in non-permissive HLA-DP TCE group mismatches
In order to investigate the impact of HLA-DPA1 polymorphism on non-permissive HLA-DP mismatches, we further stratified HLA-DPB1 allele matched and permissively or non-permissively mismatched groups according to two algorithms of HLA-DPA1 matching status as described above (Figure 1). Neither the presence or absence of HLA-DPA1*02 in the patient or the donor, nor the presence or absence of HLA-DPA1 allele mismatches between patient and donor, when added as a requirement in addition to HLA-DPB1 TCE3 group disparity in the definition of non-permissive HLA-DP mismatches, had a significant impact on the risks of TRM (Figure 3A, B) or any of the other clinical endpoints (Tables 3 and 4). Similar results were obtained for the four-group model of HLA-DPB1 TCE4 group matching previously described (data not shown).8 Interestingly, when the HLA-DPB1 allele mismatched group was considered as a whole (i.e. HLA-DPB1 permissive and non-permissive combined), the presence of an allelic HLA-DPA1 mismatch was associated with a reduced relapse risk which was significant in univariate analysis (p= 0.02) but not after adjustment for disease type and status at transplantation (HR 0.73; 95%CI 5.56–0.94, p=0.016). This trend was similar in the HLA-DPB1 permissive (HR 0.74; 95%CI 0.54–1.04; p=0.08; Table 4) and in the non-permissive group (HR 0.74; 95%CI 0.49–1.13; p=0.16).
Figure 3. Role of HLA-DPA1 matching status in risk-associations between HLA-DPB1 TCE group mismatches and TRM after UD-HCT.
Shown are the cumulative incidence curves of TRM in HLA-DPB1 allele matched, permissively or non-permissively mismatched transplants in the presence or absence of HLA-DPA1*02 in either the patient or the donor, or both (A), or in the presence or absence of HLA-DPA1 allele mismatching (B).
Table 3.
Multivariate regression models for association between non-permissive DPB1 TCE3 / DPA1*02:01 mismatches and clinical outcome.
| Permissive HLA-DPB1 mismatch + HLA-DPA1*02:01 (N = 290) |
HLA-DPB1 match + HLA-DPA1*02:01 (N = 58) |
Non-Permissive HLA-DPB1 mismatch + HLA-DPA1*02:01 (N = 274) | |||
|---|---|---|---|---|---|
| Ref. | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Overall mortalitya | 1 | 0.88 (0.74–1.06) | 0.18 | 0.98 (0.81–1.19) | 0.83 |
| Treatment-related mortalityb | 1 | 0.78 (0.61–0.98) | 0.03 | 1.02 (0.81–1.27) | 0.89 |
| Relapsec | 1 | 1.40 (1.05–1.78) | 0.02 | 0.75 (0.52–1.08) | 0.12 |
| aGVHD C–Dd | 1 | 0.91 (0.69–1.18) | 0.48 | 1.29 (0.99–1.66) | 0.05 |
adjusted for disease, disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen, and recipient age at transplant.
adjusted for disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen and recipient age at transplant.
adjusted for disease and disease status.
adjusted for disease, disease status, graft type and year of transplant.
Table 4.
Multivariate regression models for association between non-permissive DPB1 TCE 3/ DPA1 allele mismatches and clinical outcome.
| Permissive HLA-DPB1 mismatch + DPA1 match (N = 321) |
Permissive HLA-DPB1 mismatch + DPA1 mismatch (N = 264) |
HLA-DPB1 match + HLA- DPA1 match (N = 229) |
HLA-DPB1 match + HLA- DPA1 mismatch (N=10) |
Non-Permissive HLA-DPB1 mismatch + HLA-DPA1 match (N = 219) |
Non-Permissive HLA-DPB1 mismatch + HLA-DPA1 mismatch (N = 238) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | HR (95% CI) | p- value |
HR (95% CI) | p- value |
HR (95% CI) | p- value |
HR (95% CI) | p- value |
HR (95% CI) | p- value |
|
| Overall mortalitya | 1 | 0.92 (0.75–1.13) | 0.42 | 0.87 (0.71–1.08) | 0.22 | 2.31 (1.22–4.41) | 0.011 | 1.22 (0.99–1.50) | 0.06 | 1.03 (0.84–1.27) | 0.76 |
| Treatment-related mortalityb | 1 | 1.01 (0.78–1.31) | 0.93 | 0.84 (0.63–1.10) | 0.2 | 1.97 (0.86–4.50) | 0.11 | 1.39 (1.08–1.78) | 0.01 | 1.19 (0.93–1.53) | 0.16 |
| Relapsec | 1 | 0.74 (0.54–1.04) | 0.08 | 1.13 (0.83–1.54) | 0.44 | 2.70 (0.981–7.39) | 0.05 | 0.84 (0.59–1.19) | 0.33 | 0.62 (0.43–0.89) | 0.01 |
| aGVHD C–Dd | 1 | 1.11 (0.83–1.49) | 0.49 | 1.00 (0.74–1.37) | 0.98 | 1.02 (0.32–3.21) | 0.98 | 1.22 (0.90–1.66) | 0.19 | 1.42 (1.06–1.89) | 0.02 |
adjusted for disease, disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen, and recipient age at transplant.
adjusted for disease status, GVHD prophylaxis, use of in vivo T cell depletion, preparative regimen and recipient age at transplant.
adjusted for disease and disease status.
adjusted for disease, disease status, graft type and year of transplant.
Discussion
The present study is the first to address the influence of HLA-DPA1 matching in the context of the HLA-DPB1 TCE group model. The results show that the presence of HLA-DPA1*02:01 does not appear to be necessary for the formation of immunogenic, clinically non-permissive HLA-DPB1 TCE group mismatches. Likewise, allelic donor-recipient HLA-DPA1 disparity does not seem to be required for that purpose. The HLA-DP alpha chains encoded by HLA-DPA1*01:03 and DPA1*02:01 differ by three amino acids at positions 31, 50 and 83 in the peptide antigen binding groove which potentially could have an effect on peptide presentation and/or T cell receptor recognition. Accordingly, we found that a number of T cells alloreactive to HLA-DPB1 encoded antigens display a preference for HLA-DPB1 heterodimers with either HLA-DPA1*01:03 or DPA1*02:01, as shown by transfection experiments of the relevant DPB1 coding sequence into HLA-DPA1 homozygous B lymphoblastoid cell lines12. In line with this experimental evidence, we found that allelic HLA-DPA1 mismatches appeared to increase the well-known protective effect of HLA-DPB1 disparity on relapse. This was shown by a suggestive, though not statistically significant trend in multivariate analysis, towards reduced hazards of relapse in the presence of combined HLA-DPB1-DPA1 allelic mismatches, compared to HLA-DPB1 mismatches but –DPA1 matches. This effect was similar in the HLA-DPB1 permissive and non-permissive transplants after separate analysis, suggesting that the relevant mismatch is indeed at the HLA-DPA1 level. This association may have gone undetected in previous studies which compared risk-associations between HLA-DPA1 matched or mismatched transplants regardless of the HLA-DPB1 matching status13–18, due to the confounding effect of concomitant HLA-DPB1 mismatches in these analyses. Based on these findings, HLA-DPA1 typing in unrelated stem cell donor searches might be considered for patients at high risk of relapse, a notion that is in accordance also with recommendations from a previous report.18
The results from this study confirm our previous findings regarding the associations of permissive or non-permissive HLA-DPB1 TCE mismatches with clinical outcome of HCT.6 In fact, the HR for all clinical endpoints – OM, TRM, aGvHD and Relapse – observed here are similar to those observed in the IHWG transplant cohort, although not all were statistically significant according to the statistical definitions of the present study, likely reflecting lower statistical power. It should be noted that 90.5% of the patients analyzed here overlap with the IHWG cohort that included a large number of transplants coordinated by the Japanese Marrow Donor Program. Interestingly, the reduced risk of leukemia relapse associated with non-permissive HLA-DPB1 TCE group mismatches was more marked in the present study (HR 0.55; p=0.002) than in the IHWG report (HR 0.80; p=0.02)6 and highly significant, possibly reflecting greater disease type homogeneity for AML and MDS patients. This observation could be relevant for the selection of stem cell donors for high risk AML patients who might benefit from GvL mediated by T cells alloreactive to non-permissive DPB1 TCE mismatches, in particular if in correlation with allelic HLA-DPA1 disparity. Additional prospective analyses of independent transplant cohorts are warranted to better investigate this important point.
Supplementary Material
Acknowledgements
This work was supported by a grant (no. IG12042) from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to K.F. The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-12-1-0142 and N00014-13-1-0039 from the Office of Naval Research; and grants from Allos, Inc.; Amgen, Inc.; Angioblast; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Children’s Leukemia Research Association; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Genzyme Corporation; GlaxoSmithKline; HistoGenetics, Inc.; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Merck & Co, Inc.; Millennium: The Takeda Oncology Co.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Optum Healthcare Solutions, Inc.; Osiris Therapeutics, Inc.; Otsuka America Pharmaceutical, Inc.; RemedyMD; Sanofi; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; StemCyte, A Global Cord Blood Therapeutics Co.; Stemsoft Software, Inc.; Swedish Orphan Biovitrum; Tarix Pharmaceuticals; Teva Neuroscience, Inc.; THERAKOS, Inc.; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. Government.
Funding: This work was supported by a grant (no. IG12042) from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to Katharina Fleischhauer.
Footnotes
The online version of the article contains a data supplement.
Conflict of Interest Disclosures
None of the authors has any conflict of interest to disclose.
Supplementary Information
Supplementary information is available at BMT's website
Authorship Contributions
KF, MFV and SS drafted the research plan; MH and TW performed statistical analyses; KF, MFV, SL, SS, MB, LBL, FC, JD, JG, GH, MH, SRM, PM, DM, MO, MP, VR, DS, BES and EKW reviewed and interpreted the analysis; KF and SS wrote the manuscript; KF, MFV, SL, SS, MB, LBL, FC, JD, JG, GH, MH, SRM, PM, DM, MO, MP, VR, DS, BES and EKW critically revised the manuscript.
References
- 1.Hollenbach JA, Madbouly A, Gragert L, et al. A combined DPA1-DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics. 2012;64(8):559–569. doi: 10.1007/s00251-012-0615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutten CE, van Luxemburg-Heijs SA, Griffioen M, et al. HLA-DP as specific target for cellular immunotherapy in HLA class II-expressing B-cell leukemia. Leukemia. 2008;22(7):1387–1394. doi: 10.1038/leu.2008.90. [DOI] [PubMed] [Google Scholar]
- 3.Rutten CE, van Luxemburg-Heijs SA, Halkes CJ, et al. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19(1):40–48. doi: 10.1016/j.bbmt.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Shaw BE, Gooley TA, Malkki M, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110(13):4560–4566. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 5.Stevanovic S, van Bergen CA, van Luxemburg-Heijs SA, et al. HLA-class II upregulation during viral infection leads to HLA-DP directed Graft-versus-Host Disease after CD4+ donor lymphocyte infusion. Blood. 2013;122(11):1963–1973. doi: 10.1182/blood-2012-12-470872. [DOI] [PubMed] [Google Scholar]
- 6.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zino E, Frumento G, Marktel S, et al. A T cell epitope encoded by a subset of HLA-DPB1 alleles determines non-permissive mismatches for hematological stem cell transplantation. Blood. 2004;103(4):1417–1424. doi: 10.1182/blood-2003-04-1279. [DOI] [PubMed] [Google Scholar]
- 8.Crocchiolo R, Zino E, Vago L, et al. Non-Permissive HLA-DPB1 Disparity is a Significant Independent Risk Factor for Mortality after Unrelated Hematopoietic Stem Cell Transplantation. Blood. 2009;114(7):1437–1444. doi: 10.1182/blood-2009-01-200378. [DOI] [PubMed] [Google Scholar]
- 9.Shaw BE, Robinson J, Fleischhauer K, Madrigal JA, Marsh SG. Translating the HLA-DPB1 T-cell epitope-matching algorithm into clinical practice. Bone Marrow Transplant. 2013;48(12):1510–1522. doi: 10.1038/bmt.2013.91. [DOI] [PubMed] [Google Scholar]
- 10.Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006;12(8):876–884. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Spellman S, Setterholm M, Maiers M, et al. Advances in the selection of HLA-compatible donors: refinements in HLA typing and matching over the first 20 years of the National Marrow Donor Program Registry. Biol Blood Marrow Transplant. 2008;14(9Suppl):37–44. doi: 10.1016/j.bbmt.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Lauterbach N, Crivello P, Wieten L, et al. Allorecognition of HLA-DP by CD4+ T cells is affected by polymorphism in its alpha chain. Mol Immunol. 2014;59(1):19–29. doi: 10.1016/j.molimm.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Varney MD, Lester S, McCluskey J, Gao X, Tait BD. Matching for HLA DPA1 and DPB1 alleles in unrelated bone marrow transplantation. Hum Immunol. 1999;60(6):532–538. doi: 10.1016/s0198-8859(99)00004-x. [DOI] [PubMed] [Google Scholar]
- 14.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339:1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 15.Petersdorf EW, Kollman C, Hurley CK. Effect of HLA class II gene disparity on clinical outcome in unrelated donor hematopoietic cell transplantation for chronic myeloid leukemia: the US National Marrow Donor Program Experience. Blood. 2001;98(10):2922–2929. doi: 10.1182/blood.v98.10.2922. [DOI] [PubMed] [Google Scholar]
- 16.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 17.Schaffer M, Aldener-Cannavá A, Remberger M, Ringdén O, Olerup O. Roles of HLA-B, HLA-C and HLA-DPA1 incompatibilities in the outcome of unrelated stem-cell transplantation. Tissue Antigens. 2003;62(3):243–250. doi: 10.1034/j.1399-0039.2003.00089.x. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Viña MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121(22):4603–4610. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.