Abstract
Isocitrate dehydrogenase 1 (IDH1) mutations occur in most lower-grade glioma, and not only drive gliomagenesis but are associated with longer patient survival and improved response to temozolomide (TMZ). To investigate the possible causative relationship between these events, we introduced wild-type (WT) or mutant IDH1 into immortalized, untransformed human astrocytes, then monitored transformation status and TMZ response. TMZ-sensitive parental cells exhibited DNA damage (gamma-H2AX foci) and a prolonged G2 cell cycle arrest beginning 3 days after TMZ (100μM, 3hr) exposure and persisting for greater than 4 days. The same cells transformed by expression of mutant IDH1 exhibited a comparable degree of DNA damage and cell cycle arrest, but both events resolved significantly faster in association with increased, rather than decreased, clonogenic survival. The increases in DNA damage processing, cell cycle progression, and clonogenicity were unique to cells transformed by mutant IDH1, and were not noted in cells transformed by WT IDH1 or an oncogenic form (V12H) of Ras. Similarly these effects were not noted following introduction of mutant IDH1 into Ras-transformed cells or established GBM cells. They were, however, associated with increased homologous recombination and could be reversed by the genetic or pharmacologic suppression of the homologous recombination DNA repair protein RAD51. These results show that mutant IDH1 drives a unique set of transformative events that indirectly enhance homologous recombination and facilitate repair of TMZ-induced DNA damage and TMZ resistance. The results also suggest that inhibitors of HR may be a viable means to enhance TMZ response in IDH1 mutant glioma.
Keywords: IDH1, temozolomide, RAD51, homologous recombination, glioma
Introduction
Isocitrate dehydrogenase 1 (IDH1) mutations are the earliest and most common mutations in lower-grade glioma (1, 2). Mutations at the R132 codon in IDH1, and in particular R132H, are noted in roughly 75% of all lower-grade glioma (2, 3), and more recent studies suggest that IDH mutation precedes other alterations common to these tumors including p53 mutation in astrocytomas and 1p/19q co-deletion in oligodendrogliomas (4,5). The IDH1 gene encodes isocitrate dehydrogenase 1 which is involved in the cytoplasmic oxidative carboxylation of isocitrate to α-ketoglutarate (αKG) (6). The α-KG generated in turn contributes to the production of NADPH in the citric acid cycle as well as to the activity of αKG-dependent enzymes including the 5-methylcytosine hydroxylases and histone methyltransferases that regulate the epigenetic state of the cell (7, 8). Mutations in the IDH1 gene result in the expression of a mutant IDH1 that has the unique ability to convert α-KG to 2-hydroxyglutarate (2HG)(9). The accumulation of 2HG in IDH1 mutant tumors in turn competes with α-KG for binding and activation of α-KG-dependent enzymes, with the result being large-scale alterations in DNA methylation, acetylation, and gene expression that contribute to the tumorigenic process (10-12). IDH1 mutations are noted in lower-grade astrocytomas, oligodendrogliomas, anaplastic oligodendrogliomas, and secondary glioblastoma, but are rare in in pilocytic astrocytomas and primary glioblastoma (1,2). Furthermore, because IDH1-mutant tumors have a very different pattern of genetic alterations than histologically comparable IDH1 WT tumors (13), a consensus has emerged that IDH1 mutation sets cells on a different path to tumorigenesis than cells transformed by other oncogenic insults (14, 15).
IDH1 mutational status also has consequences for therapeutic outcome. In multiple studies, IDH1 mutation correlates with increased overall patient survival and progression-free survival (1, 2, 4, 5, 16), implying that IDH1 mutation alters the underlying biology of the tumor, the chemo/radiosensitivity of the tumor, or both. The DNA methylating agent temozolomide (TMZ) is one of the most frequently used drugs in the therapy of glioma, and indirectly causes DNA double strand breaks, which if not repaired by RAD51-driven homologous recombination (HR) lead to cell cycle arrest and delayed cell death (17-20). A higher rate of response to up-front TMZ has been reported in IDH1-mutant low-grade glioma patients relative to that noted in patients with histologically identical IDH1 WT tumors (21), implying that mutant IDH1 contributes to TMZ sensitivity. Gliomas driven by IDH1 mutations, however, are widely recognized to differ in cell of origin and underlying biology from comparable IDH WT glioma (14, 15), complicating any analysis of the contribution of IDH1 mutation to drug sensitivity. Furthermore, these correlative studies do not address whether mutant IDH1 alters drug sensitivity, or merely sets in motion a series of events that lead to tumorigenicity and altered drug sensitivity. Where examined, exogenous expression of mutant IDH1 had no significant effect on TMZ sensitivity (22). The established glioblastoma cell lines used (U87 and U373MG), however, evolved in the absence of IDH1 mutations (22) and as such may not represent optimal models for the study of the effect of IDH1 status on drug sensitivity.
To more accurately determine if and how IDH1 mutation alters drug sensitivity, we here employed a system that has been widely used not only for the study of cellular transformation and gliomagenesis but also for the study of IDH1 biology (12, 23, 24). This system consists of normal human astrocytes which are immortalized by introduction of virally-encoded E6, E7 and hTERT (23). These immortalized cells can be transformed into cells that grow in an anchorage-independent manner by additional expression of oncogenic H-Ras (23), or alternatively by expression of mutant IDH1, which has previously been shown to change patterns of DNA methylation in a manner similar to that noted in lower-grade glioma (12, 24). We therefore used these cells to determine if mutant IDH1 expression and/or the transformative events it sets in place were sufficient to alter sensitivity to TMZ.
Methods and Materials
Cell Culture, Creation of Cell Lines, and Drug Treatment
U87MG human astrocytoma cells and ductal carcinoma T47D cells were obtained from the UCSF Brain Tumor Research Center, and were grown in Dulbecco modified Eagle H-21 medium supplemented with 10% FBS and 1% penicillin/streptomycin (UCSF Cell Culture Facility) at 37°C in a 5% CO2 atmosphere. Normal human astrocytes (NHA) were obtained and maintained in Astrocyte Growth Medium (Clonetics). The generation of U87 cells over-expressing MGMT and NHAs expressing E6/E7/hTERT, and E6/E7/hTERT plus H-RasV12 (Ras astrocytes) has been described previously (23, 25, 26). To generate IDH1-expressing cells, lentiviral constructs encoding GFP and either WT or mut (R132H) IDH1 (27) were co-transfected with VSVG, and ΔVPR plasmids (Open Biosystems) at a 1:0.9:0.1 ratio into 293T packaging cells using Fugene6 (Roche). Lentiviral supernatants were harvested at 48 and 72 hours post-transfection and used to infect U87 cells or NHAs expressing E6/E7/hTERT or E6/E7/hTERT plus H-RasV12. After 72 hours, cells expressing GFP were isolated by fluorescent-activated cell sorting and verified by Western blot for expression of the target protein of interest. Unsynchronized cells were pretreated with DMSO (Sigma) or B02 (10 μM, 1 hr, EMD Millipore), and then treated with TMZ (100 μM, 3 hrs, Sigma) in the presence of drugs. The final DMSO concentration did not exceed 0.1% (vol/vol). After TMZ treatment, cells were washed, incubated in fresh medium containing DMSO or B02 (10μM), then harvested at various time points. The identity of all cells was verified by DNA fingerprinting using the PowerPlex 20 HS kit and comparison to published markers (28). All NHA-derived cultures were similarly verified to be derived from the same parental population.
Genetic suppression of RAD51
E6/E7/hTERT/IDH1 mut cells were plated at 105/mL in 6-well plates in DMEM cell growth media without antibiotics. Twenty four hrs later the cells were transfected with an optimized amount (5 nM) of siRNA targeting human RAD51 (SMARTpool, Dharmacon) or non-targeting siRNAs as a negative control, using DharmaFECT reagent (Dharmacon) according to the manufacturer’s protocol. Twenty-four hours after transfection, the cells were washed, treated with TMZ (100 μM, 3 hours) in complete medium free of siRNA, washed again, and placed in TMZ-free medium until harvested for analysis.
Protein Extraction and Immunoblot Analyses
Cells were washed in cold PBS and lysed in NP40 cell lysis buffer (Life Technologies) supplemented with 1× PhosStop and protease inhibitor cocktail (Roche). The protein was quantitated and used (30 μg) for Western blot analysis using primary antibodies against IDH1 (Dianova), IDH1R132H (Dianova), H-Ras (Santa Cruz Biotechnology), RAD51 (EMD Chemicals, PC130), MGMT, or β-actin (Cell Signaling Technologies) and the appropriate horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology). Antibody binding was detected using ECL reagents (Amersham Pharmacia Biotech).
Cell Cycle and Immunofluorescence Studies
At various time points following TMZ exposure, attached and floating cells were collected and subjected to flow cytometry and analysis using a FACSCalibur (BD Biosciences) and FlowJo data analysis software (TreeStar)(29). For immunofluorescence studies, cells were seeded onto 8-well glass coverslips, incubated with TMZ or TMZ + B02, washed in PBS, fixed with 4% paraformaldehyde in PBS (15 min, room temperature), rinsed with PBS, and blocked in PBS containing 0.1% Triton-X and 10% FBS (1 hr, room temperature). The cells were stained with anti-Ser-139-phosphorylated H2AX antibody (Cell Signaling) at 1:500 dilution followed by Alexa 596 conjugated secondary antibody (Life Technologies)(30 min, room temperature). Cells were washed, counterstained with 4 V,6 V diamidino-2-phenylindole (DAPI), and mounted with Prolong anti-fade reagent (Life Technologies). For RAD51 studies, cells were incubated in blocking solution (PBS containing 20% goat serum, 3% BSA and 0.1% Triton X-100, 1 hr), stained with 1:1000 anti RAD51 antibody in blocking solution (1 hr), then washed in PBS and stained with 1:500 Alexa594 conjugated secondary antibody (Life Technologies)(1hr). For double staining, anti-Ser-139-phosphorylated H2AX antibody and anti-RAD51 antibody (Gene Tex) were used as primary antibodies and Alexia 568 and 647 were used as secondary antibodies. The number of nuclei containing >5 gamma-H2AX or RAD51 foci was quantified by fluorescence microscopy.
Measurement of Cell Viability, Colony Formation Efficiency, and in vitro Transformation
Cell viability, colony formation efficiency, and in vitro transformation of control and TMZ-treated cells were determined by trypan blue-exclusion counting, colony formation assay, and soft agar assay, as previously described (23, 30).
Measurement of HR Efficiency
HR efficiency was calculated using a qPCR-based HR assay kit (Norgen Biotek Corp.)(31). Briefly, the system contains two plasmids each with a different mutation in its lacZ coding region. The plasmids were co-transfected, 48 hrs after which the total cellular DNA was isolated and used in a qPCR reaction containing a set of universal primers that amplify all plasmid DNA (and serve as a control for transfection efficiency) or a set of primers that only amplify plasmid DNA generated by HR of the transfected plasmids. The amount of recombinant product for each reaction was calculated by comparing the cycle number at the point of inflection of the amplification curve generated using the HR-specific primers to that using universal primers, and converting the difference in cycle number to a DNA amount by comparison to a standard curve generated using universal primers and different amounts of input DNA. This value was set at 1 in parental/vector cell populations, with the amount of recombinant DNA produced in experimental groups expressed as a HR ratio to this value.
Statistical Analyses
Data are reported as mean ± standard error of at least three experiments. When two groups were compared, the unpaired Student’s t test was applied (P-value). When multiple groups were evaluated, the one-way ANOVA test with post hoc Turkey-Kramer multiple comparisons test was used. P<0.05 was considered statistically significant.
Results
Mutant IDH1 expression changes TMZ response and increases TMZ resistance
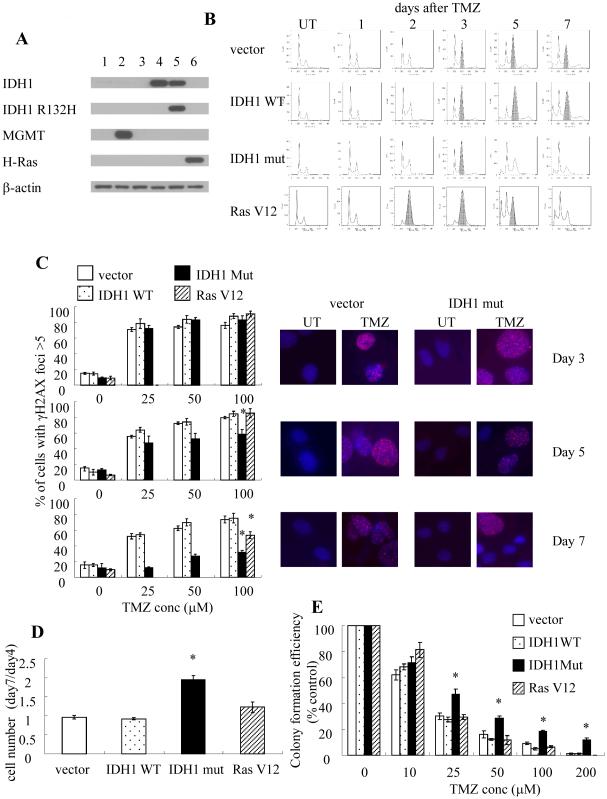
To begin to understand if and how IDH1 mutations influence drug sensitivity, normal human astrocytes immortalized by expression of E6, E7, and hTERT were first lentivirally infected with constructs encoding blank or mutant IDH1, after which the response to TMZ (3hr, 100 μM) was examined. Parental blank vector E6/E7/hTERT cells are MGMT-deficient (Fig 1A, column 3) and as a result exhibited a typical pattern of response to TMZ. Like other MGMT-deficient cells (17, 18), they underwent a G2 arrest (here defined as occurring when the percentage of cells with 4N DNA content was greater than that of cells with 2N DNA content) beginning 3 days after drug exposure, and remained arrested at least 4 days following initiation of the arrest (shaded peaks, Fig 1B). The G2 arrest was also temporally accompanied by the appearance of gamma-H2AX foci indicative of DNA double strand breaks which persisted for the duration of the study following a range of TMZ exposure (Fig 1C). Consistent with this data, the number of viable cells did not significantly change between days 4 and 7 post TMZ exposure (Fig 1D) and the TMZ-treated cells exhibited a dose-dependent loss of clonogenicity (Fig 1E). Introduction of a construct encoding WT IDH1 had no significant effect on any of these parameters (Fig 1A-E). Introduction of a construct encoding mutant IDH1 (Fig 1A) also had no significant effect on the timing of the onset of G2 arrest following TMZ exposure (Fig 1B) or the percentage of gamma-H2AX-positive cells at 3 days post-TMZ (Fig 1C). Cells expressing mutant IDH1, however, exhibited a much shorter TMZ-induced G2 arrest that lasted less than 48 hrs following the initiation of arrest (Fig 1B). Consistent with these findings, the TMZ-induced gamma-H2AX foci disappeared significantly more rapidly (Fig 1C) in mutant IDH1-expressing cells than in empty vector controls. Furthermore, the number of viable cells expressing mutant IDH1 nearly doubled between 4 and 6 days post TMZ exposure(Fig 1D), and the mutant IDH1-expressing cells also exhibited increased clonogenicity relative to uninfected or blank vector controls (Fig 1E). These results show that while the introduction of mutant IDH1 does not affect the extent of TMZ-induced DNA damage, it significantly alters the processing of this damage, and consequently drug action.
Figure 1.
(A) Western blot validation of the identity of the cell lines used in this panel. 1, U87; 2, U87+MGMT; 3, E6/E7/hTERT (vector); 4, E6/E7/hTERT+WT IDH1; 5, E6/E7/hTERT+R132H mutant IDH1; 6, E6/E7/hTERT+RasV12. (B) FACS analysis of cell cycle distribution in various cell lines 1-7 days after TMZ (100μM, 3hr) exposure. Shading, cell populations exhibiting significant G2/M arrest. (C, right panel) Immunofluorescence images of gamma-H2AX foci (pink) in DAPI-stained (blue) control and drug-treated parental E6/E7/hTERT cells at various times after TMZ exposure (100 μM, 3 hrs) and quantitation of data (left panel) in all cells studied. (D) The ratio of cell numbers at 7 days post-TMZ exposure to that 4 days after TMZ exposure (100 μM, 3 hrs) in control and IDH1-modified cells. (E) colony formation efficiency of cells studied following TMZ exposure (0-100 μM, 3 hr). *, p<.05. Values derived are representative of, or derived from, ≥3 independent experiments.
The effects of mutant IDH1 on drug response are not a generalized result of cellular transformation
Tumor cells, as well as normal human cells made tumorigenic by exogenous expression of large T antigen have both been reported to have an increased ability to repair DNA double strand breaks relative to comparable non-transformed cells (32). We therefore questioned whether the effects of mutant IDH1 on TMZ-induced DNA repair and drug action were simply a consequence of mutant IDH1-driven cellular transformation. The parental E6/E7/hTERT cells used were immortalized, but when analyzed for growth in soft agar (a hallmark of in vitro transformation)(33) failed to form colonies (Table 1). The same cells, however, were transformed and readily formed colonies in soft agar after exogenous expression of mutant V12H-Ras (Fig 1A, Table 1). Similarly, exogenous expression of either WT or mutant IDH1 resulted in in vitro transformation of the parental cells, with the mutant IDH1 cells forming fewer colonies than Ras-transformed cells, but more than those expressing WT IDH1 (Table 1). Although the cells expressing mutant H-Ras, WT IDH1, or mutant IDH1 were all transformed, the cells transformed by WT IDH1 were no different than parental cells with regard to the duration of TMZ-induced G2 arrest (Fig 1B), persistence of gamma-H2AX foci (Fig 1C), and TMZ sensitivity (Fig 1D), while the cells transformed by mutant H-Ras exhibited a duration of G2 arrest and gamma-H2AX foci that was intermediate between that of the parental and mutant IDH1 cells (Fig 1B, C) and a TMZ sensitivity no different than parental cells (Fig 1E). The effects of mutant IDH1 on TMZ action were therefore not merely a general result of the transformation process.
Table 1.
Transformation of NHAE6E7hTERT by introduction of IDH1 WT, IDH1 mut, or Ras V12
| cell types | colony numbers |
|---|---|
| vector | 5±1 |
| IDH1 WT | 85±4 |
| IDH1 mut | 189±14 |
| Ras V12 | 1191±78 |
The effects of mutant IDH1 on drug response are not mediated in a direct manner
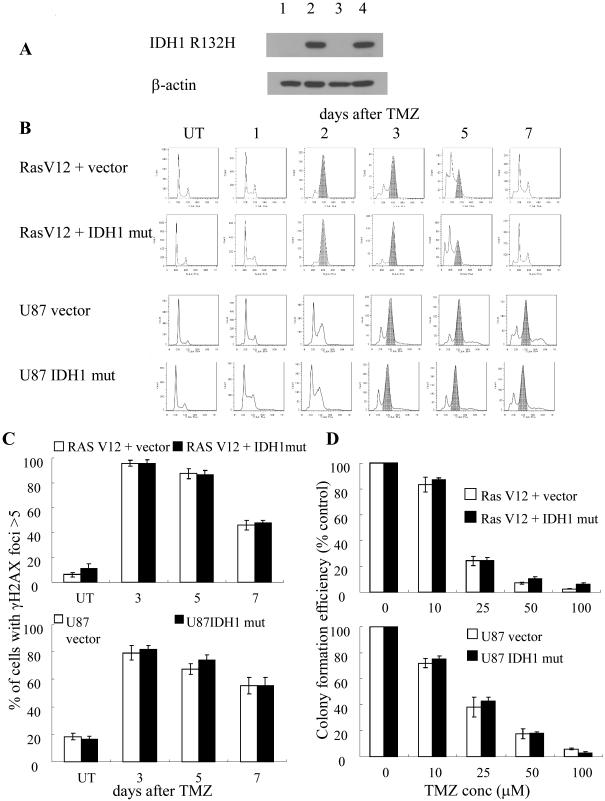
The ability of mutant IDH1 to uniquely alter TMZ response suggests that expression of mutant IDH1 might directly alter cellular mechanisms that contributed to TMZ response. To address this possibility, a blank construct, or a construct encoding mutant IDH1 was lentivirally introduced into the E6/E7/hTERT cells already transformed by expression of mutant Ras, and also into MGMT-deficient tumorigenic U87 glioma cells (Figure 2A), after which effects on TMZ-induced G2 arrest, gamma-H2AX foci formation and persistence, and clonogenicity were examined. Both the parental E6/E7/hTERT+mutant Ras cells and the U87 cells exhibited the expected TMZ-induced G2 arrest beginning 2-3 days after drug exposure (Fig 2B) in association with the formation and persistence of gamma-H2AX foci (Fig 2C) and dose-dependent loss of clonogenicity (Fig 2D). Exogenous expression of mutant IDH1 (Fig 2A) in these already transformed cells had no effect on the extent of TMZ-induced G2 arrest (Fig 2B), gamma-H2AX foci (Fig 2C), or clonogenicity (Fig 2D) in either cell type. These results show that mutant IDH1 does not alter TMZ response in a direct manner, but rather does so only in connection with the mutant IDH1-driven events that lead to cellular transformation.
Figure 2.
(A) Western blot validation of the identity of the cell lines used in this panel. 1, E6/E7/hTERT+RasV12 (RasV12) ; 2, E6/E7/hTERT+RasV12+R132H mutant IDH1; 3, U87; 4, U87+ R132H mutant IDH1. (B) FACS analysis of cell cycle distribution in various cell lines 1-7 days after TMZ (100μM, 3hr) exposure. Shading, cell populations exhibiting significant G2/M arrest. (C) Quantitated data from immunofluorescence analysis of gamma-H2AX foci in control and drug-treated parental and IDH1-modified cells at various times after TMZ exposure (100 μM, 3 hrs). (D) Colony formation efficiency of parental and IDH1-modified cells following TMZ (100 μM, 3 hr) exposure. Values derived are representative of, or derived from, ≥3 independent experiments.
Mutant IDH1-driven transformation increases HR
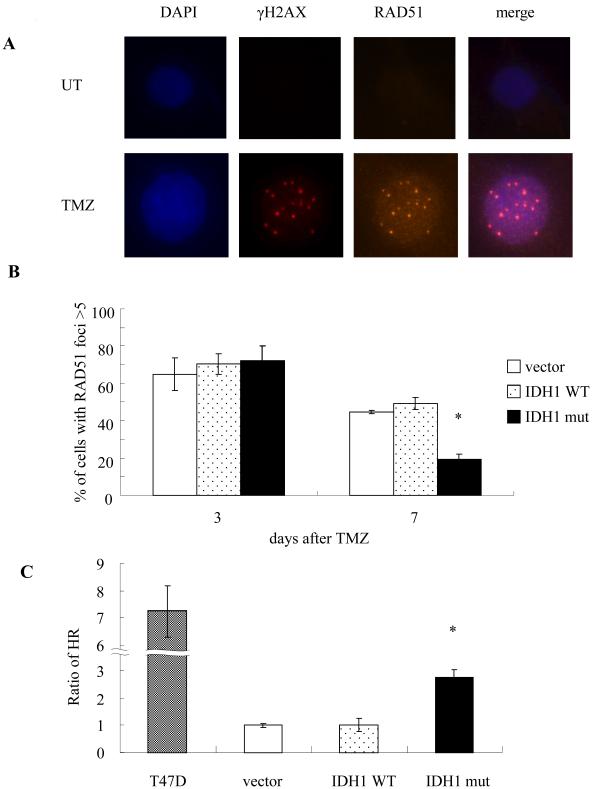
TMZ-induced DNA double strand breaks are repaired in human cells by the process of HR (19, 20). HR in turn requires the collective action of a variety of proteins including RAD51, which accumulates at the gamma-H2AX-marked sites of DNA double strand breaks and contributes to their repair (34, 35). To first determine if alterations in HR contributed to the ability of mutant IDH1-expressing cells to escape from TMZ-induced G2 arrest, we monitored the formation and disappearance of RAD51 foci in TMZ-treated E6/E7/hTERT cells lentivirally infected with a blank vector or a construct encoding WT or mutant IDH1. As shown in Fig 3A, the sites of TMZ-induced RAD51 foci formation in parental cells overlapped those of gamma-H2AX foci formation 3 days following TMZ exposure, supporting the idea that RAD51 accumulates at sites of DNA double strand breaks. All cell groups also exhibited similar percentages of cells with RAD51 foci 3 days after TMZ exposure (Fig 3B) mirroring the results derived from the temporal analysis of gamma-H2AX foci appearance and disappearance in the same cells (Fig 1). Seven days after TMZ exposure, however, the cells expressing mutant IDH1 which had displayed fewer gamma-H2AX foci and had escaped from cell cycle arrest also were less likely to contain RAD51 foci, suggesting a HR-related event was stimulated in the cells expressing mutant IDH1. To directly address this possibility we used an in vivo plasmid recombination-based method (31) to determine if exogenous expression of mutant IDH1 altered the ability of the cells to perform HR. As shown in Fig 3C, control E6/E7/hTERT cells and E6/E7/hTERT cells expressing WT IDH1 had a measurable rate of HR that was significantly less than that of positive control T47D cells, which are known to have a high rate of HR (36). The cells expressing mutant IDH1, however, had a 3-fold higher rate of HR. These results show that mutant IDH1-driven transformation increases HR.
Figure 3.
(A) Immunofluorescence analysis of the physical co-incidence of gamma-H2AX and RAD51 foci in E6/E7/hTERT parental cells three days following TMZ exposure (0 or 100 μM, 3 hrs). (B) Quantitated data from immunofluorescence analysis of gamma-H2AX foci in control and drug-treated parental (E6/E7/hTERT) and IDH1-modified cells at three and seven days after TMZ exposure (100 μM, 3 hrs). (C), Homologous recombination activity as determined by an in vivo plasmid-based recombination reporter assay. Value of the parental cells was set at 1. *, p<.05. Values derived are representative of, or derived from, ≥3 independent experiments.
Inhibition of HR reverses the effects of mutant IDH1 expression on TMZ resistance
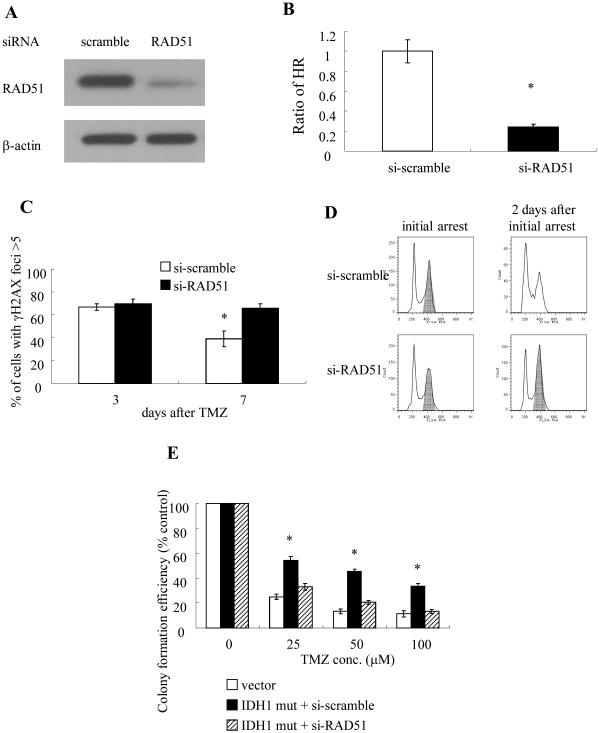
Because the activity of RAD51 is critical for HR, further studies were also performed in which RAD51 activity was genetically or pharmacologically inhibited, after which the levels of HR and effects on TMZ action were measured in the mutant IDH1-expressing cells. Expression of RAD51 siRNA, but not a scrambled control siRNA, in E6/E7/hTERT cells expressing mutant IDH1 suppressed levels of RAD51 by over 85% (Fig 4A), and also reduced HR (Fig 4B) to levels comparable to those seen in parental E6/E7/hTERT cells in Fig 3C. Introduction of the RAD51 siRNA also blocked the ability of mutant IDH1 to enhance the repair of TMZ-induced DNA double strand breaks (Fig 4C), to shorten TMZ-induced G2 arrest (Fig 4D), and to confer resistance to TMZ (Fig 4E).
Figure 4.
(A) Western blot analysis of RAD51 and β-actin levels in E6/E7h/TERT+mutant IDH1 cells three days following exposure to scramble or RAD51-targeting siRNA. (B) Homologous recombination activity as determined by an in vivo plasmid-based recombination reporter assay. Value of the si-scramble cells was set at 1. (C) Quantitated data from immunofluorescence analysis of gamma-H2AX foci in control and drug-treated parental and RAD51-suppressed cells 3 and 7 days after TMZ exposure (100 μM, 3 hrs). (D) FACS analysis of cell cycle distribution in control and RAD51-suppressed TMZ-treated (100 μM, 3 hrs) cells at the time of arrest and two days later. Shading, cell populations exhibiting significant G2/M arrest. (E) Colony formation efficiency of control and RAD51-suppressed cells studied following TMZ exposure (0-100 μM, 3 hr). *, p<.05. Values derived are representative of, or derived from, ≥3 independent experiments.
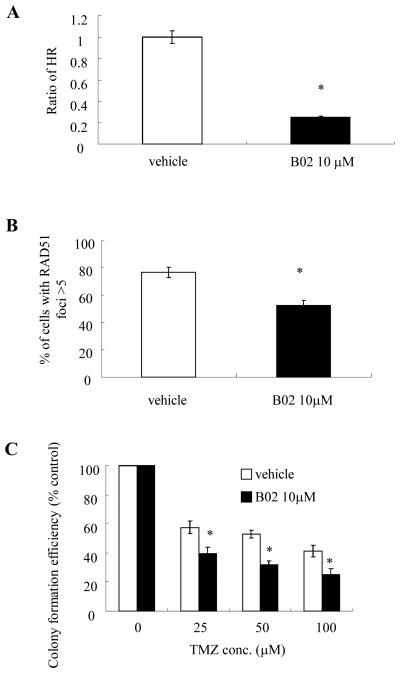
To verify these results, similar experiments were performed using B02, a specific inhibitor of the DNA strand exchange activity of human RAD51 (37, 38). Incubation of E6/E7/hTERT cells expressing mutant IDH1 with non-toxic concentrations of B02 significantly reduced HR in these cells to levels comparable to those seen in control cells (Fig 5A), significantly reduced the percentage of cells with RAD51 foci (Fig 5B), and also significantly reduced the TMZ resistance conferred by mutant IDH1 over-expression (Fig 5C). These results show that the transformation process driven by mutant IDH1 contributes to enhanced HR and TMZ-resistance.
Figure 5.
(A) Homologous recombination activity in vehicle or drug-treated (10 μM B02, 48 hrs) E6/E7/hTERT+mutant IDH1 cells as determined by an in vivo plasmid-based recombination reporter assay. Value of the vehicle cells was set at 1. (B) Quantitated data from immunofluorescence analysis of RAD51 foci in the same vehicle and drug-treated (B02, 10 μM, 48 hrs) cells 7 days after TMZ exposure (100 μM, 3 hrs). (C) Colony formation efficiency of vehicle and drug-treated (B02, 10 μM) cells after TMZ exposure (0-100 uM, 3 hrs). *, p<.05. Values derived are representative of, or derived from, ≥3 independent experiments.
Discussion
The overwhelming majority of low grade glioma and secondary glioblastoma express a mutant form of IDH1, and mutant IDH1 expression is thought to play a driving role in the genesis of these tumors (1,2). Because IDH1 mutant glioma appear to be more chemosensitive than histologically identical but genetically distinct IDH1 WT tumors, it was suggested that IDH1 mutation conferred a degree of sensitivity to the drugs used in the treatment of these tumors, including TMZ. The present results show that in an extensively used model of gliomagenesis, expression of mutant IDH1 confers TMZ resistance rather than TMZ sensitivity. This drug resistance is not a direct effect of mutant IDH1 expression but rather is an indirect effect of mutant IDH1-driven cellular transformation associated with changes in RAD51-driven HR, and can be reversed by exposure to inhibitors of the HR process.
The present results differ in important ways from those of previous studies of the effect of mutant IDH1 expression on drug sensitivity. In previous studies, introduction of mutant IDH1 into U87 and 373MG cells had no effect on TMZ resistance (22). In the present studies, introduction of mutant IDH1 into cells transformed independently of mutant IDH1, including U87, also did not increase TMZ resistance. Because expression of mutant IDH1 drives the tumorigenesis process, however, cells transformed in an IDH1-independent manner are not an appropriate model in which to determine how mutant IDH1 function contributes to tumor behavior. Although cells derived from IDH1 mutant tumors could be used, IDH1 mutant tumor cells have proven to be extremely difficult to culture or grow as xenografts, frequently undergo loss of mutant IDH1, and do not allow for a direct isogenic comparison between WT and IDH1 mutant populations (39, 40). The immortalized human astrocyte cell system used in the present studies therefore represents perhaps the only viable approach to study the effects of mutant IDH1 expression on drug sensitivity, particularly as in these cells the exogenous expression of mutant IDH1 generates epigenetic alterations similar to those noted in IDH1 mutant lower grade gliomas (12, 24), contributes to the transformation of the cells (12, 24), and generates cells which when injected intracranially in mice generate tumors which histologically resemble grade III astrocytomas (unpublished observation). In this proper context, mutant IDH1 expression was shown to increase, rather than decrease, TMZ resistance, suggesting that the context in which IDH1 mutations occur are critical in determining the effect of the alteration on drug sensitivity.
The present results also show that the effects of exogenous mutant IDH1 on TMZ sensitivity are not direct, but rather are part of a larger indirect cellular reprogramming. Exogenous expression of mutant IDH1 has been shown to alter the concentrations of a variety of metabolites and also to change the levels of various enzymes involved in energy regulation (11, 12, 27, 41). Furthermore direct mutant IDH1-driven decreases in NADPH levels have been suggested to result in increased susceptibility to oxidative stress and drug-induced apoptosis (42). In the present study, however, such changes were not sufficient in and of themselves to alter TMZ sensitivity. Rather, expression of mutant IDH1 only had effects on TMZ sensitivity in cells that were also transformed by mutant IDH1. As such, the effects of mutant IDH1 on TMZ sensitivity appear to be a result of the action of a larger indirect mechanism that contributes to transformation.
The results from the present studies show that expression of exogenous mutant IDH1 increases RAD51-driven HR, which in turn contributes to increased TMZ resistance as well as perhaps the transformation process itself. While the mutant IDH1-driven changes in HR and TMZ sensitivity occur in the absence of changes in the protein level of RAD51 (relative to WT IDH1 groups, not shown), a number of proteins contribute to HR, and modulation of any or all of these could lead to changes in the rate of HR. HR has been suggested to contribute to genomic stability and suppression of tumorigenesis (43), and as such it’s not immediately apparent why the expression of transforming IDH1 mutants increases HR. It’s worth considering, however, that mutant IDH1 tumor cells are typically also defective in telomere maintenance, and rely on an alternative (ALT) mechanism rather than telomerase up-regulation to elongate their telomeres (13, 44). Because HR has been suggested to play a role in the ALT process (45), it may be possible that the increased HR driven by mutant IDH1 contributes to the early events required for gliomagenesis, while the epigenetic alterations driven by mutant IDH1 and 2HG substitute for the frank mutations and genomic instability that drive tumorigenesis in HR-deficient cells. These possibilities remain to be examined.
From a clinical perspective, the relative chemosensitivity of IDH1 mutant tumors has been a therapeutic bright spot, and there has been little consideration of means to increase drug sensitivity in IDH1 mutant tumors. The present study suggests that the chemosensitivity of mutant IDH1 tumors may be somewhat of an illusion, and that this sensitivity only exists in comparison to genetically unrelated but histologically identical IDH1 WT tumors. The present studies further suggest that targeting components of HR, including RAD51, may be as effective as targeting mutant IDH1 itself in increasing the effectiveness of standard therapy. A careful examination of the use of TMZ, or perhaps other double strand break-producing agents, in combination with inhibitors of HR might be an effective way to further improve response in IDH1 mutant tumors. The better understanding of the underlying mechanism of mutant IDH1 provided here may therefore pave the way for better combinatorial approaches in otherwise difficult to treat lower grade gliomas.
Acknowledgments
Grant Support: This work was supported in part by National Institutes of Health Grants CA136774 and CA097257 (SPORE) to ROP
Footnotes
There are no relationships that could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to this manuscript.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in glioma. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 4.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–54. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 5.Van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1598–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of the human cytosolic NADP-dependent isocitrate dehydrogenase from pig heart. J Biol Chem. 2004;279:33946–57. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reitman ZJ, Parsons DW, Yan H. IDH1 and IDH2: not your typical oncogenes. Cancer Cell. 2010;17:215–6. doi: 10.1016/j.ccr.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1 alpha. Science. 2009;324:261–5. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH1 mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y, Killela PJ, Reitman ZJ, Rasheed BA, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–22. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh KM, Rice T, Decker PA, Kosel ML, Kollmeyer T, Hansen HM, et al. Genetic variants in telomerase-related genes are associated with an old age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. NeuroOnc. 2013;15:1041–7. doi: 10.1093/neuonc/not051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan H, Bigner DD, Velculescu V, Parsons DW. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69:9157–9. doi: 10.1158/0008-5472.CAN-09-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–50. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 17.Hirose Y, Berger MS, Pieper RO. p53 Effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61:1957–63. [PubMed] [Google Scholar]
- 18.Hirose Y, Berger MS, Pieper RO. Abrogation of the Chk1-mediated G2 checkpoint pathway potentiates temozolomide-induced toxicity in a p53-independent manner in human glioblastoma cells. Cancer Res. 2001;61:5843–49. [PubMed] [Google Scholar]
- 19.Liu X, Han EK, Anderson M, Shi Y, Semizarov D, Wang G, et al. Acquired Resistance to Combination Treatment with Temozolomide and ABT-888 Is Mediated by Both Base Excision Repair and Homologous Recombination DNA Repair Pathways. Mol Cancer Res. 2009;7:1686–92. doi: 10.1158/1541-7786.MCR-09-0299. [DOI] [PubMed] [Google Scholar]
- 20.Roos WP, Nikolova T, Quiros S, Naumann SC, Kiedron O, Zdzienicka MZ, et al. Brca2/Xrcc2 dependent HR, but not NHEJ, is required for protection against O-6-methylguanine triggered apoptosis, DSBs and chromosomal aberrations by a process leading to SCEs. DNA Repair. 2009;8:72–86. doi: 10.1016/j.dnarep.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Houillier C, Wang X, Kaloshi G, Mokhtari K, Giullevin R, Laffaire J, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–6. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Chou AP, Chen W, Chen R, Deng Y, Phillips HS, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro-Oncol. 2012;15:57–68. doi: 10.1093/neuonc/nos261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61:4956–60. [PubMed] [Google Scholar]
- 24.Rohle R, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos c, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito M, Ohba S, Gaensler K, Ronen SM, Mukherjee J, Pieper RO. Early Chk1 phosphorylation is driven by temozolomide-induced, DNA double strand break- and mismatch repair-independent DNA damage. PLoS One. 2013;8(5):e62351. doi: 10.1371/journal.pone.0062351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonoda Y, Ozawa T, Aldape KD, Berger MS, Deen DF, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674–78. [PubMed] [Google Scholar]
- 27.Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429–33. doi: 10.1038/ncomms3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bady P, Diserens A-C, Castella V, Kalt S, Heinimann K, Hamou M-F. DNA fingerprinting of glioma cell lines and considerations on similarity measurements. Neuro Oncol. 2012;14:701–11. doi: 10.1093/neuonc/nos072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirzoeva OK, Kawaguchi T, Pieper RO. The Mre11/Rad50/Nbs1 complex interacts with the mismatch repair system and contributes to temozolomide-induced G2 arrest and cytotoxicity. Mol Cancer Ther. 2006;5:2757–66. doi: 10.1158/1535-7163.MCT-06-0183. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura JL, Garcia E, Pieper RO. S6K1 plays a role in glial transformation. Cancer Res. 2008;68:6516–23. doi: 10.1158/0008-5472.CAN-07-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasileva A, Linden RM, Jessberger R. Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Research. 2006;34:3345–60. doi: 10.1093/nar/gkl455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia SJ, Shammas MA, Reis RJS. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol Cell Biol. 1997;17:7151–58. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacPherson I, Montagnier L. Agar suspension culture for the selective assay of cells transformed by polyoma virus. Virology. 1964;23:291–4. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- 34.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 35.Rossi MJ, Mazina O, Burgreev DV, Mazin AV. The RecA/RAD51 protein drives migration of Holliday junctions via polymerization on DNA. Proc Natl Acad Sci. 2011;108:6432–37. doi: 10.1073/pnas.1016072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009;11:683–91. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, et al. An Optimized RAD51 Inhibitor That Disrupts Homologous Recombination without Requiring Michael Acceptor Reactivity. J Med. Chem. 2013;56:254–63. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang F, Mazini OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J Med Chem. 2012;55:3011–20. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 39.Borodovsky A, Salmasi V, Turcan S, Fabius AWM, Baia GS, Eberhart CG. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–47. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol. 2013 doi: 10.1093/neuonc/not243. doi: 10.1093/neuonc/not243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitmana ZJ, Jina G, Karolyd ED, Spasojevice I, Yangh J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci. 2011;108:3270–75. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert MR, Liu Y, Neltner J, Pu H, Morris A, Sunkara M, et al. Autophagy and oxidative stress in gliomas with IDH1 mutations. Acta Neuropathologica. 2014;127:221–33. doi: 10.1007/s00401-013-1194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genetic. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]