Abstract
Emerging results indicate that cancer stem-like cells contribute to chemoresistance and poor clinical outcomes in many cancers, including ovarian cancer (OC). As epigenetic regulators play a major role in the control of normal stem cell differentiation, epigenetics may offer a useful arena to develop strategies to target cancer stem-like cells. Epigenetic aberrations, especially DNA methylation, silence tumor suppressor and differentiation-associated genes that regulate the survival of ovarian cancer stem-like cell (OCSC). In this study, we tested the hypothesis that DNA hypomethylating agents may be able to reset OCSC towards a differentiated phenotype, by evaluating the effects of the new DNA methytransferase inhibitor SGI-110 on OCSC phenotype, as defined by expression of the cancer stem-like marker aldehyde dehydrogenase (ALDH). We demonstrated that ALDH+ OC cells possess multiple stem cell characteristics, were highly chemoresistant, and were enriched in xenografts residual after platinum therapy. Low dose SGI-110 reduced the stem-like properties of ALDH+ cells, including their tumor initiating capacity, resensitized these OCSCs to platinum, and induced re-expression of differentiation-associated genes. Maintenance treatment with SGI-110 after carboplatin inhibited OCSC growth, causing global tumor hypomethylation and decreased tumor progression. Our work offers preclinical evidence that epigenome-targeting strategies have the potential to delay tumor progression by re-programming residual cancer stem-like cells. Further, the results suggest that SGI-110 might be administered in combination with platinum to prevent the development of recurrent and chemoresistant ovarian cancer.
INTRODUCTION
Epithelial ovarian cancer (OC) causes more deaths than any other female reproductive tract cancer (1,2). The majority of women diagnosed with advanced-stage epithelial OC experience tumor recurrence associated with the development of chemoresistance, and platinum-resistant OC is uniformly fatal (3). A new paradigm explaining tumor relapse involves the persistence of “cancer stem cells” which were characterized in several solid tumors, including OC (4–6). While chemotherapy may succeed initially at decreasing the size and number of tumors, it leaves behind residual malignant cells, which we hypothesize are enriched in tumor progenitors or “cancer stem cells”.
Ovarian cancer stem cells (OCSCs) have been isolated from established OC cell lines, ascites, primary and metastatic tumors (4,7,8). They share several characteristics with normal stem cells, including the ability to form anchorage-independent spherical aggregates, express stem cell markers, undergo membrane efflux, form clones in culture and in addition exhibit enhanced tumor-forming ability (9). Although a number of technical approaches have been successfully used to isolate OCSCs (sphere-forming, cell surface markers, stem cell gene reporter assays), the use of an assay measuring aldehyde dehydrogenase isoform 1 (ALDH) enzymatic activity has been recently proposed and is used to define CSCs in multiple other tumor types (10,11).
Ovarian CSCs are hypothesized to be largely (or entirely) responsible for emergence of chemoresistant tumors, because they possess many of the phenotypes associated with drug resistance (e.g., enhanced DNA repair, diminished apoptotic responses, increased efflux mechanisms, quiescent state) (4,12). Moreover similarly to normal embryonic or tissue stem cells, CSC are believed to harbor a significantly altered epigenome (6,13), and it has been hypothesized that DNA hypomethylating agents could “reset” these cells toward differentiation (14). Indeed, several hypomethylating agents were originally characterized as inducers of cancer cell differentiation (6,15). However, it has become clear that hypomethylating agents or other epigenetic modulators alone cannot eradicate relapsed tumors. Pre-clinical studies from our and other groups have established the rationale for combining DNA methylation inhibitors with existing chemotherapeutic agents to overcome acquired drug resistance in OC (16–20). Based on those studies, we recently completed a phase II trial using a DNA methylation inhibitor as a re-sensitizer to traditional chemotherapy in patients with recurrent OC and showed that this combination has clinical and biological activity (21), justifying other rationally designed epigenetic treatment strategies in OC.
Based on the above considerations, we hypothesized that hypomethylating agents, in combination with chemotherapeutics, may target drug-resistant OCSCs, possibly leading to tumor eradication. In the current study, we isolated and characterized ALDH(+) OCSC from OC cell lines and human tumors. ALDH(+) cells were significantly more chemoresistant and tumorigenic compared to ALDH(−) cells in orthotopic tumor initiating assays. Treatment with SGI-110, a second-generation DNA methyltransferase inhibitor (DNMTI), resensitized OCSCs to platinum. A model recapitulating the emergence of recurrent tumors showed an increased percentage of ALDH(+) OCSCs in residual tumors after platinum. Maintenance therapy with SGI-110 during platinum-induced remission inhibited the emergence of platinum resistant tumors. We suggest that epigenomic targeting using SGI-110 may be useful as a “maintenance” clinical strategy after platinum-based therapy in OC.
MATERIALS AND METHODS
Cell lines, patient samples, culture conditions and reagents
OC cell lines (A2780, A2780_CR5, SKOV3) were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) with supplements as described previously (22) and see supplemental methods. Cisplatin-resistant variant A2780_CR5 was established from five-round IC70 survival monoclonal-selection by continuous exposure to increasing concentration of cisplatin (22). A2780 and SKOV3 OC cells were authenticated in 2012 by ATCC (Manassas, VA). Advanced high grade serous ovarian tumors were surgically collected (IRB approved protocol IUCRO-0280), enzymatically disassociated and cultured, as previously described (4). SGI-110 was provided by Astex Pharmaceuticals Inc. (Dublin, CA, USA) and cisplatin (CDDP) was purchased from Calbiochem (Billerica, MA, USA).
Aldefluor Assay and Flow Cytometry
ALDH1 enzymatic activity was measured using the Aldefluor assay (Stemcell Technologies, Vancouver, Canada)(11) (details can be found in supplemental methods).
Cell survival assay
3-(4.5-dimethylthiazol-2-yl)-2.5-diphenyl tetrazolium bromide (MTT) assay was used to evaluate both the chemosensitivity of OC cells (A2780/_CR5, ALDH(+)/(−) derived from A2780_CR5) to CDDP and the platinum resensitization by SGI-110 by determining the 50% growth inhibitory (IC50) dose values (see supplemental methods).
Cell cycle analysis
Cell cycle analysis were conducted as described in supplemental methods.
Sphere and colony formation assays
Sphere formation assays were conducted as described previously (5) and in supplemental methods. Colony formation assay was performed by sorting 500 untreated or drug-treated ALDH(+)/(−) cells into 6-well coated high adhesion plates (Corning). Cells were seeded in 2 ml RPMI (Invitrogen) 1640 medium with 10% FBS (Atlanta Biologicals, Flowery Brance, GA, USA), 1% L-glutamine (Corning) and 1% penicillin/streptomycin (Corning), cultured for 8 days, plates were washed with 2ml PBS, fixed with 3ml 10% formalin (Sigma) for 15 min and stained with crystal violet for 5 min (0.025% w/v, Sigma). The number of colonies was counted in each well, excluding small (< 50 cells) colonies (23).
Differentiation assay
ALDH (+) cells were FACS sorted from control-treated (100nM DMSO) or SGI-110 (100nM per day for 3 days) treated aldefluor-stained A2780_CR5 OC cells, and 50,000 cells then were plated under adherent conditions with either differentiation medium (DMEM/F12 with 10% FBS) or standard RPMI 1640 medium containing 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin, as described (5). The number of ALDH(+) cells on Day 7, 14, 21, 28, and 42 was determined using FACS analysis.
In vivo xenograft experiments
All animal studies were conducted according to a protocol approved by the Institutional Animal Care and Use Committee of Indiana University. Female nude, athymic, BALB/c-nu/nu mice (5–6 weeks old; Harlan, Indianapolis, IN) injected subcutaneously (s.c.) with 20,000 ALDH(+) or ALDH(−) cells sorted either from either SGI-110 (100nM/day for 3 days) or vehicle (DMSO) treated. Aldefluor(+) A2780_CR5 and ALDH(+) or ALDH(−) isolated from three high grade serous human tumors (1,500 cells per mouse). Prior to s.c. injection, cells were resuspended in 100µl 1:1 RPMI 1640 mixed with Matrigel (BD Biosciences), as described (5). Tumor length (l) and width (w) were measured weekly using digital calipers and tumor volume (v) was calculated as v = ½ × l × w2. Mice were euthanized when tumors were > 2cm in diameter or at end of study.
For the carboplatin-response studies, mice were injected i.p. with 2 ×106 A2780 cells and subsequently treated with carboplatin (Hospira, INC. Lake Forest, IL) at 50 mg/kg, i.p. or PBS (n= 6–9 animals per group) weekly for 3 weeks beginning 3 days after injection of cells. For the maintenance study, mice were injected with A2780 cells and treated with carboplatin for 3 weeks, as described. At the completion of the carboplatin treatment, mice were randomized to receive SGI-110 (2 mg/kg) or vehicle, s.c. twice-a-week for two weeks (n=12 per group). Mice were euthanized and peritoneal tumors were counted, weighed and volumes determined as described. Tumors were transferred to tubes containing medium RPMI 1640 for immediate isolation of cancer cells, or snap frozen in liquid nitrogen or RLT buffer (Qiagen) and then stored at −80°C until DNA and RNA extraction.
Isolation of tumor cancer cells and growth of spheroids
Xenografts were minced and enzymatically dissociated in Dulbecco’s modified Eagle’s medium/F12 (Invitrogen) supplemented with 5% FBS, collagenase (100 IU/ml, Sigma-Aldrich), and hyaluronidase (100 IU/ml, Sigma-Aldrich), as previously described (5).
qRT-PCR
RNA was isolated from A2780, A2780_CR5, ALDH(+)/(−) cells, normal ovarian epithelial cells (NOSE), and primary tumors using AllPrep DNA/RNA/Protein Mini kit (Qiagen,) following manufacturer’s protocol (see supplemental methods).
DNA extraction, bisulfite conversion and DNA methylation profiling
Genomic DNA was extracted from A2780 xenografts from mice treated with SGI-110 or control, by using QIAamp DNA mini kit (QIAGEN, Valencia, CA). Sodium bisulfite conversion was performed using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA), according to the manufacturer’s instructions. After bisulfite conversion, methylation of CpG sites was determined by Infinium HumanMethylation450 BeadChips (Illumina, San Diego, CA) following a procedure provided by Illumina, at the University of Chicago Genomics Core, Knapp Center for Biomedical Discovery (Chicago, IL). Data quality verification and levels of methylation of the 485,000 CpG sites included in the array were generated by the Illumina GenomeStudio Data Analysis Software. The Illumina Infinium 450k array was used to analyze DNA methylation in promoter site regions. The method measures the methylation levels over 482k CpG probes. The average percentage of methylation levels were expressed as β -values and ranged from 0 (completely unmethylated) to 1 (completely methylated). Data are deposited in GEO (NCBI#1698198).
Western blot
Protein extracts from treated cells were isolated and subjected to western blot analysis as described (24). Antibodies for DNMT1, ALDH1A1 and GAPDH were from Cell Signaling (Danvers, MA, USA). After incubation with horseradish peroxidase labeled secondary antibodies (Cell Signaling) protein bands were visualized using the ECL reaction (Thermo Scientific).
DNA methylation assay by pyrosequencing
Methylation level for CpG islands of selected genes was determined by pyrosequencing assays as described (21) following a procedure provided by EpigenDx (Hopkinton, MA, USA). Average methylation level for each CpG dinucleotide was calculated to indicate the methylation levels of each specific gene.
Statistical analysis
All data are presented as mean values ± SD of triplicate measurements. IC50 dose values were determined by Prism 6 (GraphPad Software, San Diego, CA), using logarithm normalized sigmoidal dose curve fitting. Student’s t-test was used to statistically analyze the significant difference among different groups by using Prism 4.0 (GraphPad Software), P value of 0.05 being considered significant. The genome-wide analysis experiments were conducted using the Partek Genomics Suite (version 6.5). The differences in methylation levels between samples (i.e. the differential methylation levels) were calculated using a mixed-model ANOVA. The resultant P values < 0.05 signified highly significant differential methylation levels at a specific nucleotide site The analysis built a gene integration network, incorporating physical and predicted interactions, co-localization, shared pathways, and shared protein domains. The visualization of the interactions between the genes in the top functional category was realized using Cytoscape (25) (additional informatics analysis can be found in supplemental material).
RESULTS
ALDH(+) cells are enriched in platinum resistant OC
ALDH activity has been demonstrated to be a global and well-established marker for OCSCs(10,11). To determine the baseline level of ALDH(+) subpopulation in OC, the percentages of ALDH(+) cells in OC cell lines A2780, A2780_CR5, SKOV3 and biopsies from chemotherapy naïve high grade serous OC (HGSOC) patients were examined using FACS analysis. The percentage of ALDH(+) in A2780 was 0.3% and enrichment (P<0.05) of ALDH(+) cells was observed in the A2780_CR5 (1.07%, >3-fold increase) and SKOV3 (0.65%, 2-fold increase) (Fig. 1A). In primary tumors, the ALDH(+)% varied (0.4% to 15%), however, the average %ALDH(+) cells was similar to OC cell lines (3.53% vs. 1.07%) (Fig. 1A and Supplemental Table S2). The presence of ALDH(+) cells in OC cell lines and primary tumors and the relative increase in %ALDH(+) cells in platinum resistant cell lines supports the potential contribution of OCSCs to platinum resistance and poor clinical outcome.
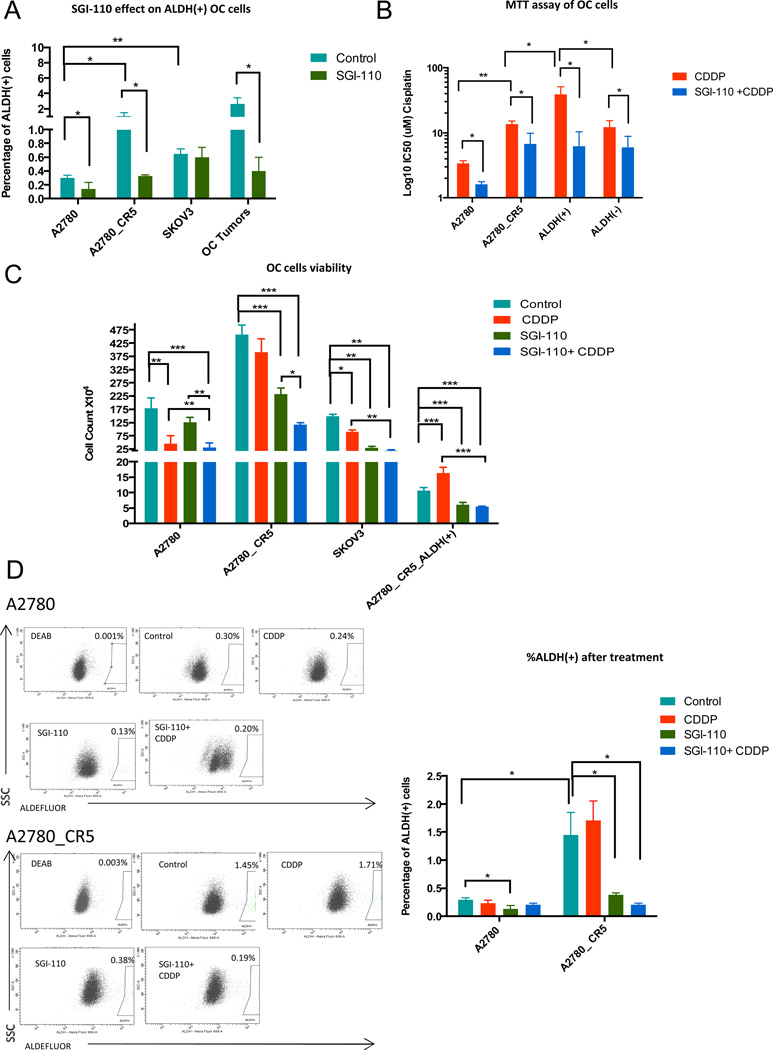
Figure 1. Low dose SGI-110 diminishes tumor-initiating cell populations in cultured OC cells.
(A) Percentage of ALDH(+) cells in untreated/or SGI-11(100nM) treated A2780 ovarian cancer cells (A2780 is the parental/platinum-sensitive and A2780_CR5 is the platinum-resistant subline), SKOV3 and three high grade serous ovarian tumors (patients 10–12). (B) IC50 of A2780 platinum-sensitive and -resistant, and A2780 platinum resistant derived ALDH(+) and ALDH(−) cells after exposure to 24h CDDP (1.67µM) alone or in combination with SGI-110 (100nM) and analyzed by MTT assay. (C) Parental A2780, A2780_CR5, SKOV3 and ALDH(+) cells isolated from A2780_CR5 were treated with SGI-110 (100nM) or CDDP (1.67µM) or the drug combination. Cell viability was measured after drug treatment using trypan blue staining. (D) Percentage of ALDH(+) in platinum-sensitive and-resistant A2780 cells: control (baseline), or after treatment with cisplatin (1.67µM), SGI-110 (100nM) or SGI-110+cisplatin. Mean values ± SD of three independent experiments in triplicate are reported (*: P<0.05, **: P<0.01, ***: P<0.001).
Low dose SGI-110 treatment depletes ALDH(+) cells in OC
As transient exposure to low doses of the DNMTi decitabine has been shown to target CSCs in leukemia and breast cancer (15), we examined the effect of the second generation DNMTI SGI-110 on OCSCs. OC cell lines and dissociated cells from HGSOC patients’ tumors were treated with SGI-110 (100 nM for 3 days) and the number of (ALDH+) cells was determined by FACS analysis. SGI-110 treatment decreased (P<0.05) the %ALDH(+) cells in A2780 (0.3% to 0.14%), A2780_CR5 (1.07% to 0.33%), and primary ovarian tumors (2.63% to 0.4%), but not in SKOV3 (Fig. 1A), indicating that low dose SGI-110 has the potential to target OCSCs in most OC cells, but not all.
To investigate whether low dose SGI-110 resensitizes OC cells to platinum therapy, MTT assays were conducted after treatment with cisplatin (CDDP) alone or in combination with SGI-110. As expected, A2780_CR5 cells were more (P<0.001) resistant to CDDP than A2780 cells (13.6µM vs. 3.4µM) (Fig. 1B), and low dose SGI-110 treatment decreased (P<0.05) the IC50 for CDDP in both cell lines (A2780, 3.4µM to 1.6µM; A2780_CR5, 13.6µM to 6.7µM) (Fig. 1B). Similarly, ALDH(+) cells derived from A2780_CR5 displayed increased (P<0.05) resistance to CDDP compared to ALDH(−) cells (38.7M vs.12.2M), and SGI treatment increased (P<0.05) CDDP sensitivity of A2780_CR5-derived ALDH(+) cells (38.7µM to 6.2µM), as well as ALDH(−) cells (12.2µM to 6.0µM) but to a lesser extent (Fig. 1B). These results suggest ALDH(+) cells contribute to chemoresistance and can be resensitized to CDDP by epigenetic therap.
To further investigate the impact of SGI-110 on OCSCs, we treated OC cells with CDDP (1.67µM), SGI-110 (100 nM) alone or in combination and examined ALDH(+) cell viability. CDDP treatment alone reduced (P<0.05) the number of viable A2780 (1.8 × 106 vs.4.57 × 105) and SKOV3 (1.5 × 106 vs.8.95 × 105) cells (Fig. 1C), but the number of A2780_CR5 derived ALDH(+) cells increased (1.0 6 × 105 vs.1.64 × 105) after CDDP treatment (Fig. 1C). SGI-110 alone inhibited (P<0.001) the growth of CDDP-resistant A2780_CR5 (4.58 × 106 vs.2.32 × 106), SKOV3 cells (1.50 × 106 vs.2.92 × 105) as well as A2780_CR5-derived ALDH(+) cells (1.06 × 105 vs.6.09 × 104) and reduced (P<0.05) the percentage of ALDH(+) cells in A2780 (0.30% vs.0.14%) and A2780_CR5 (1.5% vs.0.38%) (Fig. 1C, D). As expected, combined treatment with CDDP-SGI-110 effectively inhibited (P<0.001) OC cell viability (A2780, 1.80 × 106 vs.3.14 × 105; A2780_CR5, 4.58×106 vs.1.18 × 106; SKOV3, 1.50 × 106 vs.2.04 × 105) as well as the growth of ALDH(+) cells derived from A2780_CR5 (1.06 × 105 vs.5.43 × 104) and decreased (P<0.05) the ALDH(+) subpopulation in A2780 (0.30% vs. 0.20%) and A2780_CR5 (1.5% vs.0.20%) (Fig. 1C, D).
To determine whether SGI-110 restored CDDP sensitivity, effects of treatment with SGI-110+CDDP were compared to those induced by CDDP only. The number of platinum resistant A2780_CR5, and A2780_CR5 derived ALDH(+) OC cells was reduced (P<0.01) by the combination therapy compared to CDDP alone (Fig. 1C). The total number of viable A2780_CR5 cells was not reduced by treatment with CDDP only, and the number of A2780_CR5 derived ALDH(+) cell increased after CDDP treatment alone (Fig. 1C). The observation that the CDDP response of A2780_CR5 and A2780_CR5 derived ALDH(+) cells to CDDP alone was not significant indicates that the effect of SGI-110+cisplatin on these platinum resistant cells was not simply additive. In addition, modest activity of single agent SGI-110 was evident in platinum resistant cells, inducing more prominent G0/G1 arrest in the platinum-resistant compared to -sensitive OC cells (Supplemental Fig. S1), indicating that low dose SGI-110 exerted a chemo-resensitization effect. As different cellular backgrounds likely contribute to epigenetic therapy response, SKOV3-derived ALDH(+) population appeared to be more resistant to SGI-110 compared to A2780 and primary tumors derived cells (Fig. 1C). However, the overall SKOV3 cell population was responsive to the drug, based on increased G0/G1 arrest in SKOV3 cells treated with SGI-110 (Supplemental Fig. S1). Taken together, these data suggest that low dose SGI-110 exerted anti-tumor and chemo-resensitization effects on OCSC.
Low dose SGI-110 reduces OC self-renewal and clonogenicity
We and others previously demonstrated enhanced sphere forming and self-renewal ability of OCSCs when grown in stem cell-selective culture conditions (26–29). To investigate the effect of SGI-110 on sphere formation, A2780 and A2780_CR5 cells were treated with SGI-110/CDDP alone or in combination, and tumor-sphere formation assays were performed. A2780_CR5 cells demonstrated greater (P<0.001) sphere forming ability than A2780 (224±20 vs. 54±13 spheres; Fig. 2A, B). Moreover, SGI-110 treatment alone markedly inhibited (P<0.001) the spheroid forming ability of A2780_CR5 (224±20 vs.115±11). The combined SGI-110-CDDP treatment inhibited (P<0.05) spheroid formation capability of both the parental line and the resistant subline (A2780: 54±13 vs.27±7; A2780_CR5: 224±20 vs.85±12).
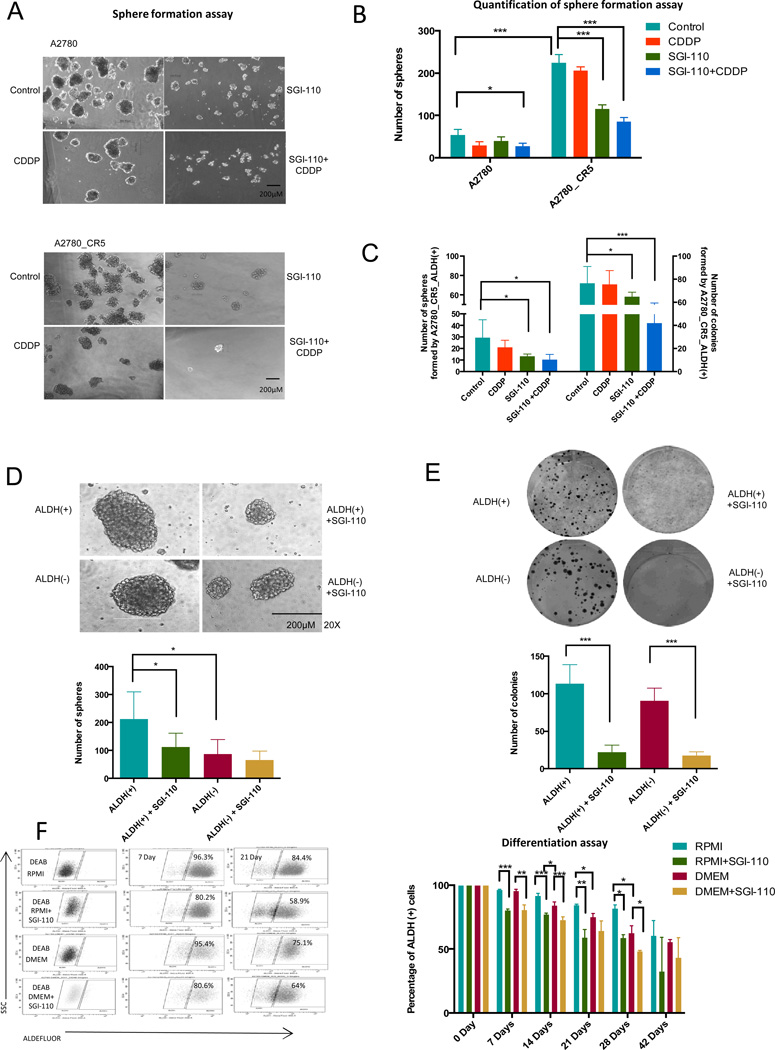
Figure 2. SGI-110 decreases self-renewal and clonogenicity of OC.
(A) 30,000 dissociated sphere-forming cells derived from A2780 cells (platinum-sensitive and -resistant) were treated with CDDP (1.67µM) or SGI-110 (100nM) alone or in combination. Representative images are shown. Scale bar, 200µm. (B) Quantitative analysis of spheres formation assay. (C) 500 ALDH(+) cells derived from A2780_CR5 were treated with cisplatin (1.67µM), SGI-110 (100nM), alone or in combination, and allowed to recover for 4 days. The number of spheres (left) and colonies (right) was counted in 14 days and 7 days, respectively. (D) Sphere formation assay of 500 untreated and SGI-110 (100nM) treated ALDH(+)/ALDH(−) cells isolated from cultured A2780_CR5 (platinum-resistant). Representative images were shown in the upper panel, Scale bar, 200µm. Quantification of sphere formation assay is shown below the images. Cells were plated in triplicate and spheres were mechanically disassociated every 7 days and counted on the Day 14. (E) Colony formation assay of 500 untreated and SGI-110 (100nM) treated ALDH(+)/ALDH(−) cells isolated from cultured A2780_CR5. Cells were plated in triplicate. Colonies were stained with crystal violet and counted on day 8. (F) ALDH(+) cell differentiation assay. Average number of ALDH(+) population present in untreated or SGI-110 (100nM) treated A2780_CR5_ALDH(+) cells. ALDH(+) cells were cultured in RPMI or DMEM condition for 7, 14, 21, 28, and 42 days. Mean values ± SD of three independent experiments in triplicate are reported (*: P<0.05, **: P<0.01, ***: P<0.001).
To examine the long term impact of SGI-110 on OCSCs, spheroid formation and colony formation assays were performed using ALDH(+) cells derived from A2780_CR5 treated with CDDP and(or) SGI-110 and allowed to recover for 4 days. As shown in Figure 2C and Supplemental Fig. S3, low dose SGI-110, either alone or in combination with CDDP, inhibited (P<0.05) sphere (30±16 vs.13±2 SGI-110 or 30±16 vs.10±5 SGI-110 + CDDP) and colony (72±17 vs.58±5 SGI-110 or 72±17 vs.42±10 SGI-110 + CDDP) formation capability of ALDH(+) cells. To further examine the short term effect of low dose SGI-110 treatment on self-renewal, the same assay was performed on the A2780_CR5 ALDH(+)/(−) cells. Treatment with SGI-110 alone had no effect on ALDH(+) OC cell growth rate (Supplemental Fig. S2), eliminating growth rate as a major contributing factor to the sphere or colony formation capability of ALDH(+) cells. ALDH(+) formed a greater (P<0.05) number of spheroids compared to ALDH(−) cells (87±52 vs.212±97) (Fig. 2D, Supplemental Fig. S4), but no difference in colony formation was observed for ALDH(+) vs. ALDH(−) (Fig. 2E). In addition, low dose SGI-110 treatment inhibited (P<0.05) spheroid-forming capability of ALDH(+) cells (212±97 vs.112±50) (Fig. 2D) and reduced (P<0.001) clonogenicity of ALDH(+) (11±35 vs.23±10) and ALDH(−) (90±17 vs.18±5) cells (Fig. 2E). Moreover, serial passaging indicated that ALDH(+) cells maintained their sphere forming ability over multiple passages, while the number of cell aggregates formed by ALDH(−) cells was reduced from passage 2– (7.7 ± 1.4 vs. 4.0±1.4) (Supplemental Fig. S5). Although ALDH(−) OC cells were incapable of long-term survival in stem cell culture conditions, the cells demonstrated limited survival in anchorage-independent conditions by grouping together to formed loosely adhesive cell clusters (30). Those clusters were unable to undergo serial passage and therefore were not considered true “spheroids”. Thus, the initial 3-day low dose SGI-110 treatment inhibited ALDH(+) sphere-forming ability over 3 passages (P<0.05, Supplemental Fig. S5).
As A2780_CR5 ALDH(+) cells generated a greater (P<0.05) number of ALDH(−) cells in the DMEM medium than in the RPMI medium, and SGI-110 treatment decreased (P<0.01) the ALDH(+) subpopulation under either culture condition (Fig. 2F). Although A2780_CR5 ALDH(+) cells persisted and maintained stemness properties during the initial culture period, the proportion of ALDH(+) cells declined (P<0.05) by 42 days in culture (Fig. 2F), indicating that, A2780_CR5 ALDH(+) cells were able to repopulate ALDH(+) and ALDH(−) and low dose SGI-110 therapy induced ALDH(+) differentiation (Supplemental Fig. S6).
Low dose SGI-110 blunts tumorigenicity of ovarian CSCs by targeting ALDH(+) cells
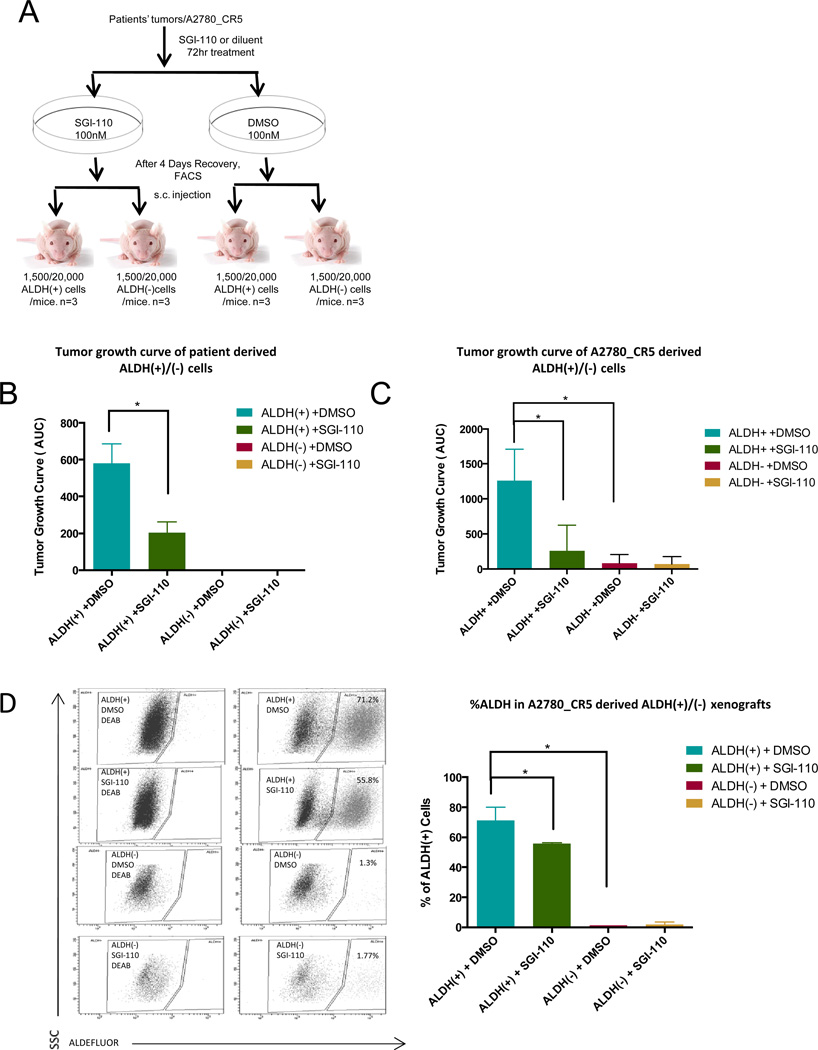
To examine the effect of SGI-110 on OCSCs tumor formation in vivo, untreated and SGI-110 treated ALDH(+)/ALDH(−) cells derived from the A2780_CR5 OC cell line and primary patient tumors were evaluated in a xenograft model (Fig. 3A). ALDH(+) cells from human tumors (1,500 cells/mouse) (Fig. 3B) or A2780_CR5 derived ALDH (+) (20,000 cells/mouse) (Fig. 3C) displayed robust (P<0.05) tumor initiating capacity compared with ALDH(−) cells. Importantly, untreated or SGI-110 pretreated primary tumor-derived ALDH(−) cells were non-tumorigenic under these conditions (Fig. 3B), whereas 20,000 A2780_CR5 derived ALDH(−) cells exhibited reduced (P<0.05) tumor formation ability compared with A2780_CR5 derived ALDH(+) cells (AUC 83±124 vs. 1259±44) (Fig. 3C). Furthermore, increasing the number of ALDH(+)/ALDH(−) cells used in xenograft assays accelerated tumor growth and shortened tumor initiation time for ALDH(+) and ALDH(−) cells (Supplemental Table S3). Treatment with low dose SGI-110 for 3 days in vitro (prior to injection into mice) prolonged (P<0.05) the time to tumor initiation and reduced tumor volume in ALDH(+) xenografts (Supplemental Table S3). To examine the effects of SGI-110 on OCSCs, the percentage of ALDH(+) cells in untreated vs. SGI-110 treated A2780_CR5 derived ALDH(+)/(−) xenograft tumors was assessed by FACS. ALDH(+) cells were highly enriched (P<0.05) in the A2780_CR5 derived ALDH(+) xenografts compared to ALDH(−) tumors (71.2±8.7% vs. 1.3%), supporting that ALDH(−) cells were unable to de-differentiate into ALDH(+) cells. SGI-110 reduced (P<0.05) ALDH(+) subpopulation (71.2±8.6% to 55.8±0.9%) in xenografts (Fig. 3D).
Figure 3. SGI-110 decreases tumorigenesis by targeting ALDH(+) cells.
(A) Schematic diagram of the approach used to study in vivo tumorigenesis of low dose SGI-110 untreated and treated ALDH(+)/(−) cells. (B) Primary xenograft tumor growth curve of 1,500 patient-derived ALDH(+) or ALDH(−) cells pretreated with SGI-110 (100nM) for 72h or DMSO in mice (n=3 for each group) during 8 weeks. Average of area under the curve (AUC) was calculated and shown in the histogram. ALDH(+)/(−) cells isolated from three high grade serous human tumors (1,500 cells per mouse). (C) Xenograft tumor growth curve of 20,000 A2780_CR5-derived ALDH(+) or ALDH(−) cells pretreated with SGI-110 (100nM) for 72h or DMSO in mice in 7 weeks (n=3 for each group). AUC was calculated and shown in the histogram. (D) Average number of ALDH(+) population present in untreated or SGI-110 (100nM) pre-treated A2780_CR5-derived ALDH(+) or ALDH(−) xenograft tumors. Mean values ± SD of three independent experiments (*: P<0.05, **: P<0.01, ***: P<0.001).
ALDH(+) cells overexpress stemness-associated genes
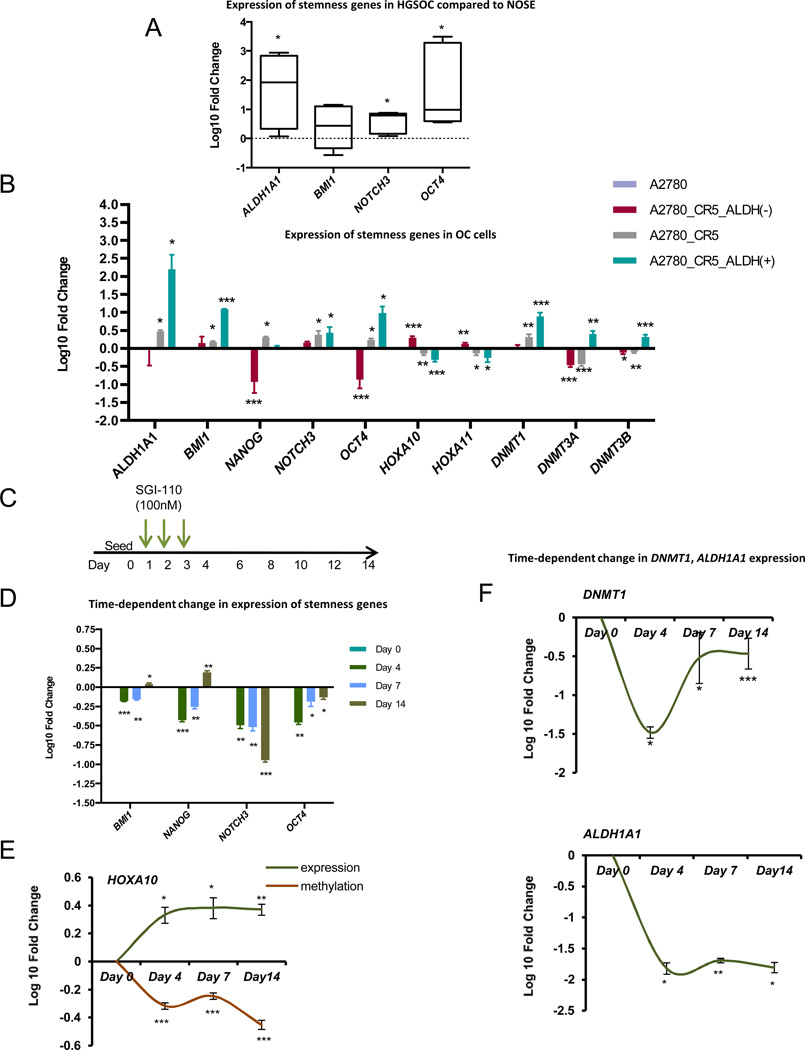
To confirm the stem-like properties of the ALDH(+) OC cells, we examined the mRNA levels of several known stem-cell associated genes (4,11,30). Compared to normal ovarian surface epithelial cells (nOSE), HGSOCs (n=5) displayed increased (P<0.05) expression of stem-cell associated genes, NOTCH, OCT4 and ALDH1A1 (Fig. 4A). Importantly increased (P<0.05) expression levels of ALDH1A1 (162.42±8.8-fold), BMI1 (8.8±0.2-fold), NANOG (9.5±0.8-fold), NOTCH3 (1.9±0.6-fold) and OCT4 (71.4±1.7-fold) and decreased (P<0.05) expression levels of the differentiation-related gene HOXA10 (4.1±0.7-fold) and HOXA11 (2.4±0.8-fold) were observed in A2780_CR5- derived ALDH(+) compared to ALDH(−) cells, (Fig. 4B, Supplemental Fig. S7).
Figure 4. SGI-110 decreases expression of pluripotency genes and induces differentiation-associated genes in OC.
(A) Average expression of stemness genes (ALDH1A1, BMI, NOTCH3, and OCT4) was measured in 5 primary high-grade serous ovarian epithelial cancers from patients compared with normal ovarian epithelial cells. (B) Average expression of stemness genes (ALDH1A1, BMI, NANOG, NOTCH3, and OCT4), DNMT isoforms (DNMT1, DNMT3A and DNMT3B) and differentiation associated genes (HOXA10 and HOXA11) were measured by qRT-PCR in A2780_CR5 (platinum-resistant), ALDH (+) and ALDH(−) cells derived from A2780_CR5 compared with parental A2780 cells. (C) Scheme of low dose SGI-110 time-course treatment. (D) Average expression of stemness genes (BMI, NANOG, NOTCH3, and OCT4) were measured by qRT-PCR in A2780_CR5-derived ALDH(+) cells on the Day 4, 7 and 14 after the initial 3 days-SGI-110 (100nM) treatment compared with untreated ALDH(+) cells. (E) Average expression of differentiation-associated gene HOXA10 mRNA measured by qRT-PCR in A2780_CR5-derived ALDH(+) cells over SGI-110 (100nM) treatment compared with untreated ALDH(+) cells. Average of DNA methylation level of HOXA10 was measured using pyrosequencing. Average expression of mRNA DNMT1 (F, upper) and ALDH1A1 (F, lower) was measured by qRT-PCR in A2780_CR5-derived ALDH(+) cells at indicated time points over SGI-110 (100nM) treatment compared with untreated ALDH(+) cells. Three independent experiments were performed and mean values ± SD are calculated (*: P<0.05, **: P<0.01, ***: P<0.001).
Low dose SGI-110 induces differentiation of ALDH(+)
Methyltransferases DNMT1, 3A and 3B are the main effectors of DNA methylation. Deregulated levels of DNMTs have been reported in cancer (31) and in OC (32,33), and in association with platinum resistance. Therefore, DNMT1, 3A and 3B expression levels were measured in platinum sensitive (A2780) and resistant (A2780_CR5) sublines and in A2780_CR5 derived ALDH(+) and ALDH(−) cells. DNMT1 was (P<0.05) significantly upregulated in cisplatin resistant A2780_CR5 and ALDH(+) cells, and DNMT3A and 3B were overexpressed in ALDH(+) cells, suggesting that aberrant methylation patterns may be associated with increased or altered DNMT activity in OCSCs (Fig. 4B). We then assessed stem-cell, differentiation-related genes and DNMTs expression- levels in response to treatment with low dose SGI-110 in a time-course experiment (Fig. 4C). In ALDH(+) cells derived from A2780_CR5, SGI-110 suppressed (P<0.05) expression of stemness genes BMI, NANOG, NOTCH3 and OCT4 (Fig. 4D), and induced upregulation (P<0.05) of the differentiation gene HOXA10 (Fig. 4E). The latter was accompanied by HOXA10 promoter CpG island demethylation (Fig. 4E), consistent with SGI-110 hypomethylating effect. SGI-110 also reduced (P<0.05) the mRNA expression levels of DNMT1 (Fig 4F) DNMT3A and 3B (Supplemental Fig. S8A, B) and ALDH1A1 (Fig. 4F) in ALDH(+) cells up to 14 days (P<0.05). Three-day low dose SGI-110 treatment also resulted in decreased DNMT1 and ALDH1A1 protein levels in A2780, A2780_CR5 and SKOV3 cells (Supplemental Fig. S8C–E). These data support that low doses of SGI-110 promote differentiation of ALDH(+) OC cells and suppress their stem like properties.
Enrichment in ALDH(+) cells after platinum in OC xenografts
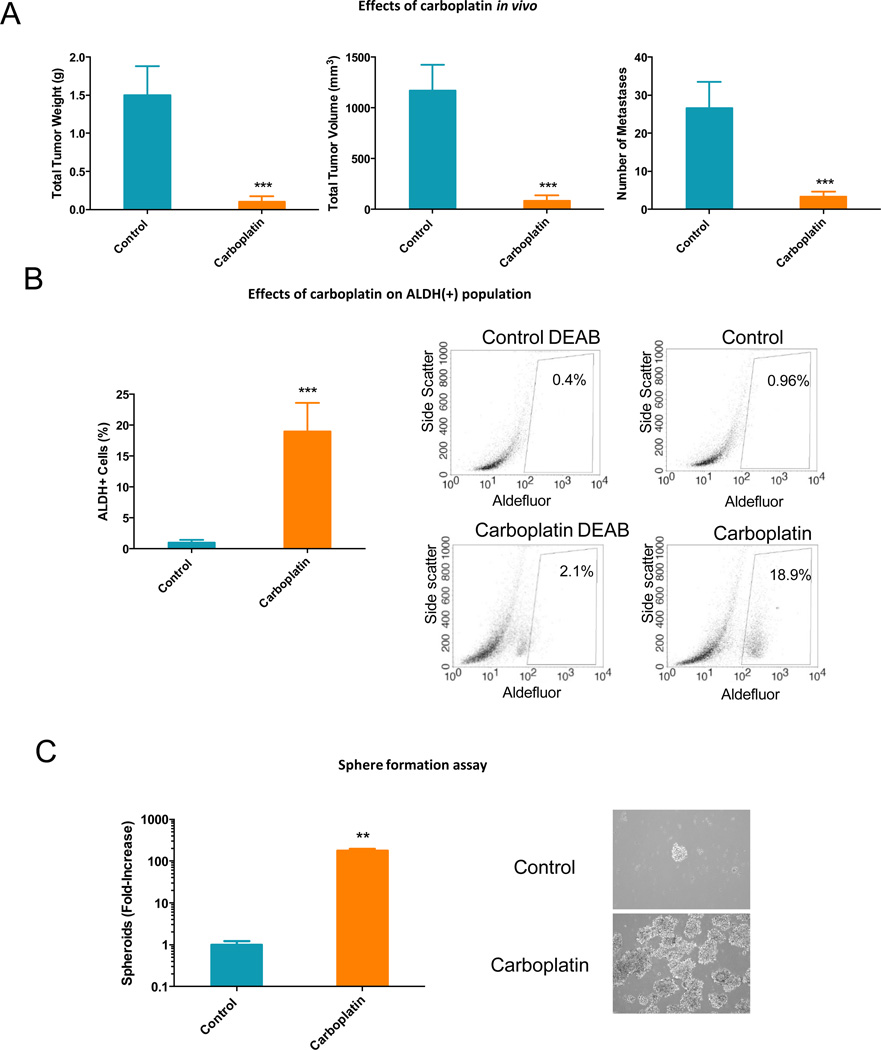
To test the hypothesis that CSCs persist in ovarian tumors after platinum-based chemotherapy, we used an ip xenograft model derived from parental (platinum-sensitive) A2780 OC cells treated with carboplatin or vehicle (control). Tumor volume, weight and number of metastases were significantly decreased (P < 0.001) by weekly treatment with carboplatin at 50mg/kg (Fig. 5A). Vehicle and carboplatin-treated tumors were dissociated to single cell suspension at the end of treatment and cells were analyzed for aldefluor positivity, and for ability to form spheres in anchorage-independent conditions. The percentage of ALDH(+) cells was increased (P < 0.001) ~20-fold in tumors residual after carboplatin compared to vehicle treated tumors (Fig. 5B). Cells dissociated from carboplatin-treated tumors formed increased (P<0.001) numbers and size of spheres compared to cells dissociated from control tumors (Fig. 5C), consistent with an OCSC phenotype. In all, carboplatin significantly decreased tumor growth in vivo but also contributed to enriching the OCSC population in residual tumors.
Figure 5. In vivo effects of carboplatin on xenograft growth and ovarian CSCs.
(A) Effects of carboplatin on weights, volumes and metastases sites of xenograft tumors derived from A2780 cells. Bars represent average measurements +/−SD; *** P < 0.001 (n=6 per group). (B) Percentage of ALDH(+) cells in control or carboplatin-treated xenografts. Cells were isolated by mechanical and enzymatic digestion and ALDH (+) cells were detected by FACS. Bars represent average of 4 measurements +/−SD; *** P < 0.001 (left panel). Representative FACS histograms are shown in the right panels. (C) Spheroid formation by cells dissociated from control or carboplatin-treated xenografts; Bars represent average of 3 measurements +/−SD; ** P < 0.01 (left panel). Phase microscopy shows morphology of spheres formed by cells dissociated from control or carboplatin-treated xenografts (100× magnification, right panels).
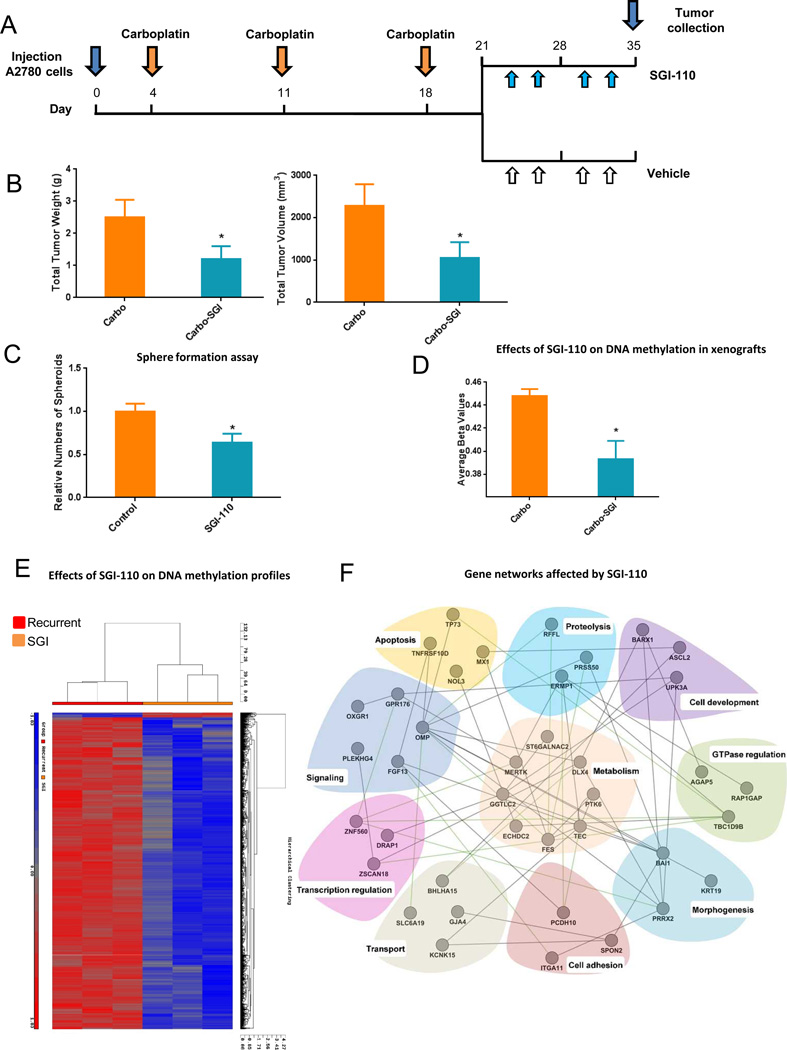
DNA demethylation induced by SGI-110 delayed recurrence of OC xenografts
To determine whether DNA hypomethylation induced by SGI-110 prevents tumor recurrence after maximal response to platinum therapy, mice bearing ip A2780 derived xenografts and treated with carboplatin were randomized to 2 week treatment with SGI-110 or vehicle (Fig. 6A, n=12 mice per group). Treatment with SGI-110 decreased (P < 0.05) total tumor weight and volume compared to control treated mice (Fig. 6B). Cells dissociated from SGI-110 treated tumors significantly reduced spheroid formation capability in vitro (Fig. 6C, P<0.05), consistent with inhibition of stem cell properties. To demonstrate that SGI-110 induced global DNA hypomethylation consistent with its DNMT inhibitory properties, Illumina Infinium HumanMethylation450 arrays were used to quantify DNA methylation in control and SGI-110 treated ovarian xenografts. More than 62,000 methylation sites and 10,000 CpG islands were found to be significantly hypomethylated in SGI-100 treated tumors compared to controls (Table 1). The substantial global DNA hypomethylation induced by SGI-110 in vivo was also demonstrated by a decrease of 6% in β-values across all CpG islands (P<0.0001, Fig. 6D) and through unsupervised hierarchical clustering analysis of methylation sites in control and SGI-100 treated xenografts (Fig. 6E). To understand biological processes represented by the genes whose promoter CpG islands were significantly hypomethylated in response to SGI-110, we grouped those genes into well-defined functional GO categories, using DAVID (A Database for Annotation, Visualization, and Integrated Discovery)(34). Of the 84 genes meeting the criteria (described in Materials and Methods, Supplementary Table S4), 65 had well-defined GO categories, and 40 could be grouped into 10 functional categories containing at least three assigned genes. These categories represent important biological processes, including metabolism, apoptosis, proteolysis, cell development, morphogenesis, cell adhesion, transport, signaling, transcriptional regulation and GTPase regulation (Fig. 6F), suggesting that hypomethylation induced by SGI-110 alters critical pathways in cancer. Genes included in these networks include PCDH10, a gene known to be downregulated in cancer through DNA methylation (35), miR-203 that is epigenetically silenced in myeloma and involved in apoptosis control (36), PTK6, involved in epithelial to mesenchymal transition (37) and others.
Figure 6. In vivo effects of SGI-110 as maintenance therapy following carboplatin treatment.
(A) Diagram illustrating the experimental design including the carboplatin treatment phase followed by randomization to either SGI-110 (2 mg/kg twice weekly) or diluent. (B) Effects of SGI-110 on tumor weight and volume. Bars represent average measurements +/−SD; * P < 0.05 (n=12 per group). (C) Spheroid formation by cells dissociated from control or SGI-110-treated xenografts. Bars represent average of 3 measurements +/−SD; * P < 0.05. (D) Mean β-value calculated across all CpG sited measured using Infinium 450 human methylation arrays in control and SGI-110 treated xenografts (P<0.001). (E) Hierarchical clustering displays differential DNA methylation profiles of SGI-110 or control treated xenografts (n=3 replicates) measured using Infinium 450 human methylation arrays. Columns represent individual samples and rows represent methylation sites. Each cell corresponds to the level of methylation at a specific site in a given sample. A visual dual color code is utilized with red and blue indicating high and low expression levels, respectively. The scale of color saturation, which reflects the methylation levels, is included. (F) Functional relationships between genes significantly hypomethylated by SGI-110 treatment were determined by using GeneMANIA and visualized by Cytoscape, as described.
Table 1.
Number of DNA methylation sites and regions showing significant changes in methylation in xenografts treated with SGI-110 or vehicle.
| Control vs. | Decreased Methylation | Increased Methylation | ||
|---|---|---|---|---|
| SGI-110 | Total | Δ Beta ≥.2b | Total | Δ Beta ≥.2b |
| CpG Sites | 62,964 | 9,971 | 781 | 165 |
| CpG Regionsc | 10,570 | 54 | 15 | 0 |
Methylation of CpG sites was determined using Infinium HumanMethylation450 arrays and was expressed as beta values ranging from 0 (unmethylated) to 1 (totally methylated). Significant changes: P<.01 (ANOVA) and FDR<.05.
ΔBeta = Difference in beta values between Control and SGI-110 groups.
CpG islands plus shores and shelves.
DISCUSSION
Our results demonstrate that ALDH(+) OC cells possessing stem cell characteristics are enriched in platinum-resistant OC cell lines, human tumors and xenografts residual after platinum therapy. The novel DNMTI SGI-110 inhibits ALDH(+) cell viability, sphere formation, and tumor initiating capacity, represses stem-cell associated gene transcription, and resensitizes platinum-resistant OC cells to platinum. In vivo, maintenance treatment with SGI-110 after carboplatin induces profound global hypomethylation and delays tumor progression. Collectively, our data suggest that a strategy targeting DNA methylation in OC exerts potent anti-tumor activity by allowing elimination of ALDH(+) cells enriched in residual, platinum resistant tumors. Our data have several implications.
First, we assert that ALDH1A1 expression and activity characterizes OC cells with stem cell properties, in agreement with reports from other groups (6,30,38). Aldefluor positivity detectable by FACS identifies the enzymatic activity of ALDH1, a member of the ALDH family that metabolizes reactive aldehydes (39). While ALDH positivity has been recognized as a stem cell marker in various tissues, the role of the enzyme in the functions of CSC remains elusive. A potential function relates to its regulatory role in the synthesis of retinoids, which play a critical role in cellular differentiation. Whether the enzyme has other functions important to the maintenance of cancer stem cells remains not known. Here we show that ALDH(+) cells derived from OC cell lines and from primary ovarian tumors are more resistant to platinum, express stem cell restricted transcription factors, and are able to generate spheres and tumors in vivo.
Second, we demonstrate that ovarian xenografts residual after treatment with platinum are enriched in ALDH(+) cells suggesting that cells with stem cell characteristics escape traditional cytotoxic treatments. Our model is consistent with the proposed concept that stem cells elude the effects of traditional anticancer strategies and can reconstitute recurrent tumors which become recalcitrant to chemotherapy (12). We use A2780, a tumorigenic and one of the few available platinum sensitive OC cell lines, to recapitulate the clinical evolution of OC, with massive initial response to chemotherapy, followed by inevitable resurgence of resistant tumors While it has been suggested that the genomic signature of A2780 does not fully match that of HGSOC (40,41), previous studies demonstrated aggressive in vivo growth of A2780 cells (26,42) and response to platinum, resembling the human disease. Our model supports that tumor recurrence could be attributed to the persistence of platinum refractory stem cells at the end of initial treatment. We propose a novel strategy to target these resistant cells through epigenomic reprogramming by using a novel DNMTI. We show that SGI-110 suppressed the viability of ALDH(+) cells, their ability to form spheres in vitro, and their tumorigenic potential in vivo. Importantly, treatment with SGI-110 resensitized platinum resistant ALDH(+) cells to platinum, providing proof of concept for further investigating hypomethylating strategies as means to resensitize tumors to chemotherapy.
Third, it has been recognized that embryonic and cancer stem cells harbor distinct DNA methylation profiles (43,44) that enable tight control of cell differentiation and self-renewal capacity. Therefore, treatment of a stem cell enriched cell population with DNA hypomethylating agents would remove the repressive epigenetic brakes, allowing stem cells to undergo differentiation and leave the pluripotent undifferentiated state. While, this concept has been tested in leukemia models (45), it remains unexplored in solid tumors. Here we show for the first time that the expression levels of all 3 DNMT isoforms is significantly increased in ALDH(+) cells derived from platinum resistant OC cells and that SGI-110 is able to re-set these cells towards differentiation. As histone modifications have been associated with DNA methylation and regulation of stemness-associated genes, and an association between EZH2 and ALDH1A1 expression has been reported (46), it seems plausible that additional epigenetic regulatory mechanisms contribute to maintaining stem cell characteristics. Thus our studies provides the first proof of principal that epigenomic strategies efficiently target OC stem cells
Our results also demonstrate that SGI-110 induces profound hypomethylation in vivo, with tens of thousands of CpG sites becoming demethylated in response to treatment. A distinctly hypomethylated DNA profile emerges, providing reassurance that the novel DNMTI hits its biologic targets in solid tumors in vivo. Future studies will strive to identify the critical genes or pathways responsible for tumor growth inhibition and chemotherapy resensitization in response to this DNMT inhibitor. It is likely that not a single gene, or pathway, but a complex program is re-engaged by targeting the epigenome, as we show here by pathway analyses.
Lastly, we demonstrate that treatment with SGI-110 after platinum decreases recurrent tumor burden in a platinum-sensitive OC ip xenograft model which recapitulates firstly the response to therapy, and secondly the recurrence of disease after chemical debulking using carboplatin. This data support exploring maintenance treatment with a hypomethylating agent after maximal response induced by traditional treatment. Maintenance strategies after chemotherapy have been investigated with variable level of success in OC (47–49) and remain an area of active exploration. Our study provides the first evidence that epigenome targeting strategies decrease tumor progression by targeting and reprogramming residual cancer stem cells, supporting further refinement of this intervention and translating these findings to the clinic.
Supplementary Material
Acknowledgements
We thank Dr. Mohammad Azab and Gavin Choy (Astex Pharmaceuticals, Inc.) for providing SGI-110 and helpful scientific comments, and Mrs. Andrea Caperell-Grant and Christiane Hassel for technical assistance, and recognize the use of the IUB Flow Cytometry Core Facility. This work was made possible by funding from the Ovarian Cancer Research Fund [PPDIU01.2011; KN and DM], National Cancer Institute Awards CA133877 and CA113001, US Department of Veterans Affairs (to DM) and the Walther Cancer Foundation (Indianapolis, IN).
Footnotes
Conflict of interest:
KN and DM received research funding from Astex Pharmaceuticals Inc. and PT is employed by Astex Pharmaceuticals Inc.
REFERENCES
- 1.Bukowski RM, Ozols RF, Markman M. The management of recurrent ovarian cancer. Seminars in oncology. 2007;34:S1–S15. doi: 10.1053/j.seminoncol.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S, Coward JI, Bast RC, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu CM. Cancer of the ovary. The New England journal of medicine. 2005;352:1268–1269. author reply 68–9. [PubMed] [Google Scholar]
- 4.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan CT, Guzman ML, Noble M. Cancer stem cells. The New England journal of medicine. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 6.Shah MM, Landen CN. Ovarian cancer stem cells: are they real and why are they important? Gynecologic oncology. 2014;132:483–489. doi: 10.1016/j.ygyno.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 8.Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 9.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annual review of cell and developmental biology. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 10.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PloS one. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landen CN, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting Aldehyde Dehydrogenase Cancer Stem Cells in Ovarian Cancer. Mol Cancer Ther. 2010;9:3186–3199. doi: 10.1158/1535-7163.MCT-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balch C, Fang F, Matei DE, Huang THM, Nephew KP. Epigenetic Changes in Ovarian Cancer. Endocrinology. 2009;150:4003–4011. doi: 10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2'-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60:6039–6044. [PubMed] [Google Scholar]
- 17.Frost P, Abbruzzese JL, Hunt B, Lee D, Ellis M. Synergistic cytotoxicity using 2'-deoxy-5-azacytidine and cisplatin or 4-hydroperoxycyclophosphamide with human tumor cells. Cancer Res. 1990;50:4572–4577. [PubMed] [Google Scholar]
- 18.Fang F, Balch C, Schilder J, Breen T, Zhang S, Shen C, et al. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116:4043–40453. doi: 10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecologic oncology. 2010;116:195–1201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balch C, Yan P, Craft T, Young S, Skalnik DG, Huang TH, et al. Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol Cancer Ther. 2005;4:1505–1514. doi: 10.1158/1535-7163.MCT-05-0216. [DOI] [PubMed] [Google Scholar]
- 21.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, et al. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC medical genomics. 2009;2:34. doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafehi H, Orlowski C, Georgiadis GT, Ververis K, El-Osta A, Karagiannis TC. Clonogenic assay: adherent cells. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan MY, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–11966. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao YP, Chen LY, Huang RL, Su PH, Chan MW, Chang CC, et al. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Human molecular genetics. 2013 doi: 10.1093/hmg/ddt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shank JJ, Yang K, Ghannam J, Cabrera L, Johnston CJ, Reynolds RK, et al. Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecologic oncology. 2012;127:390–397. doi: 10.1016/j.ygyno.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda T, Hirohashi Y, Torigoe T, Yasuda K, Takahashi A, Asanuma H, et al. ALDH1-High Ovarian Cancer Stem-Like Cells Can Be Isolated from Serous and Clear Cell Adenocarcinoma Cells, and ALDH1 High Expression Is Associated with Poor Prognosis. PloS one. 2013;8:e65158. doi: 10.1371/journal.pone.0065158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saw YT, Yang J, Ng SK, Liu S, Singh S, Singh M, et al. Characterization of aldehyde dehydrogenase isozymes in ovarian cancer tissues and sphere cultures. BMC cancer. 2012;12:329. doi: 10.1186/1471-2407-12-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva IA, Bai SM, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde Dehydrogenase in Combination with CD133 Defines Angiogenic Ovarian Cancer Stem Cells That Portend Poor Patient Survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 32.Bai X, Song Z, Fu Y, Yu Z, Zhao L, Zhao H, et al. Clinicopathological significance and prognostic value of DNA methyltransferase 1, 3a, and 3b expressions in sporadic epithelial ovarian cancer. PloS one. 2012;7:e40024. doi: 10.1371/journal.pone.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbosh PH, Montgomery JS, Starkey JA, Novotny M, Zuhowski EG, Egorin MJ, et al. Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res. 2006;66:5582–5591. doi: 10.1158/0008-5472.CAN-05-3575. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Li W, Xie J, Wang Y, Tang A, Li X, et al. Epigenetic inactivation of PCDH10 in human prostate cancer cell lines. Cell biology international. 2011;35:671–676. doi: 10.1042/CBI20100568. [DOI] [PubMed] [Google Scholar]
- 36.Wong KY, Liang R, So CC, Jin DY, Costello JF, Chim CS. Epigenetic silencing of MIR203 in multiple myeloma. British journal of haematology. 2011;154:569–578. doi: 10.1111/j.1365-2141.2011.08782.x. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y, Wang Z, Bie W, Brauer PM, Perez White BE, Li J, et al. PTK6 activation at the membrane regulates epithelial-mesenchymal transition in prostate cancer. Cancer Res. 2013;73:5426–5437. doi: 10.1158/0008-5472.CAN-13-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasuda K, Torigoe T, Morita R, Kuroda T, Takahashi A, Matsuzaki J, et al. Ovarian Cancer Stem Cells Are Enriched in Side Population and Aldehyde Dehydrogenase Bright Overlapping Population. PloS one. 2013;8:e68187. doi: 10.1371/journal.pone.0068187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Human genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4 doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-specific cell line models for type-specific ovarian cancer research. PloS one. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rattan R, Graham RP, Maguire JL, Giri S, Shridhar V. Metformin suppresses ovarian cancer growth and metastasis with enhancement of cisplatin cytotoxicity in vivo. Neoplasia. 2011;13:483–491. doi: 10.1593/neo.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altun G, Loring JF, Laurent LC. DNA methylation in embryonic stem cells. Journal of cellular biochemistry. 2010;109:1–6. doi: 10.1002/jcb.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trowbridge JJ, Orkin SH. DNA methylation in adult stem cells: New insights into self-renewal. Epigenetics: official journal of the DNA Methylation Society. 2010;5:pii11374. doi: 10.4161/epi.5.3.11374. [DOI] [PubMed] [Google Scholar]
- 45.Gattei V, Aldinucci D, Petti MC, Da Ponte A, Zagonel V, Pinto A. In vitro and in vivo effects of 5-aza-2'-deoxycytidine (Decitabine) on clonogenic cells from acute myeloid leukemia patients. Leukemia. 1993;7(Suppl 1):42–48. [PubMed] [Google Scholar]
- 46.Li H, Bitler BG, Vathipadiekal V, Maradeo ME, Slifker M, Creasy CL, et al. ALDH1A1 is a novel EZH2 target gene in epithelial ovarian cancer identified by genome-wide approaches. Cancer prevention research. 2012;5:484–491. doi: 10.1158/1940-6207.CAPR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. The New England journal of medicine. 2011;365:2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 48.Hess LM, Rong N, Monahan PO, Gupta P, Thomaskutty C, Matei D. Continued chemotherapy after complete response to primary therapy among women with advanced ovarian cancer: a meta-analysis. Cancer. 2010;116:5251–5260. doi: 10.1002/cncr.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. The New England journal of medicine. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.