Abstract
Purpose
Ewing sarcoma is a pediatric bone tumor which absolutely relies on the transcriptional activity of the EWS/ETS family of fusion oncoproteins. While the most common fusion, EWS/FLI, utilizes lysine-specific demethylase 1 (LSD1) to repress critical tumor suppressors, small molecule blockade of LSD1 has not yet been thoroughly explored as a therapeutic approach for Ewing sarcoma. We therefore evaluated the translational potential of potent and specific LSD1 inhibition with HCI2509 on the transcriptional program of both EWS/FLI and EWS/ERG as well as the downstream oncogenic phenotypes driven by EWS/ETS fusions in both in vitro and in vivo models of Ewing sarcoma.
Experimental Design
RNA-seq was used to compare the transcriptional profiles of EWS/FLI, EWS/ERG, and treatment with HCI-2509 in both EWS/FLI and EWS/ERG containing cell lines. We then evaluated morphological phenotypes of treated cells with immunofluorescence. The induction of apoptosis was evaluated using caspase 3/7 activation and TUNEL staining. Colony forming assays were used to test oncogenic transformation and xenograft studies with patient-derived cell lines were used to evaluate the effects of HCI-2509 on tumorigenesis.
Results
HCI2509 caused a dramatic reversal of both the up- and down-regulated transcriptional profiles of EWS/FLI and EWS/ERG accompanied by the induction of apoptosis, and disruption of morphological and oncogenic phenotypes modulated by EWS/FLI. Importantly, HCI2509 displayed single-agent efficacy in multiple xenograft models.
Conclusions
These data support epigenetic modulation with HCI2509 as a therapeutic strategy for Ewing sarcoma, and highlight a critical dual role for LSD1 in the oncogenic transcriptional activity of EWS/ETS proteins.
Keywords: Epigenetics, Ewing sarcoma, EWS/ETS fusion, LSD1, transcriptional reprogramming
Introduction
Dynamic epigenetic regulation, including DNA methylation and posttranslational histone modification, is required for normal development and maintenance of tissue-specific transcriptional programs. Abnormal regulation can lead to altered gene expression and malignant transformation (1,2). Indeed, enzymes which mediate epigenetic modifications are now emerging as therapeutic targets in cancer (1,2). Histone lysine methylation, specifically, can signify both activating and repressive chromatin, depending on the site of methylation (3). For example, methylation at histone H3K4 indicates active chromatin while methylation at H3K9 and H3K27 indicates repressive chromatin (3). While DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) are involved in global epigenetic processes, histone lysine methyltransferases (KMTs) and demethylases (KDMs) regulate histone methylation and gene expression in a manner that is often cell-type specific (4–8). Genetic mutations, chromosomal translocations, and translocation-derived fusion proteins affecting KMTs and KDMs contribute to impaired tumor suppression and altered developmental plasticity in several malignancies (8–14). Strategies targeting individual KMTs and KDMs critical for a particular malignancy may confer increased therapeutic specificity (15).
Lysine-specific demethylase 1 (LSD1) is a KDM implicated in neuroblastoma, acute myeloid leukemia, breast, prostate, bladder, lung, liver, and colorectal tumors (16–21). Recently, high LSD1 expression was reported in certain mesenchymal tumors, including Ewing sarcoma (22,23). Ewing sarcoma is a highly aggressive pediatric malignancy characterized by the presence of a translocation-derived fusion oncoprotein and aberrant transcription factor, EWS/FLI (24). The majority of cases present with the EWS/FLI fusion, while several other related EWS/ETS fusions are occasionally observed as well (25). The most common of these is EWS/ERG, which presents in ~10% of cases (25). While the 5-year overall survival for patients with local disease is 70–80%, for those who present with metastases, or those who have relapsed, this drops to a bleak 10–30% (26,27). Additionally, treatment carries elevated risk for long-term side effects including limb dysfunction, infertility, and secondary malignancies (28). Targeted therapies with increased efficacy and reduced toxicity are imperative. Though the role of LSD1 in Ewing sarcoma pathogenesis was still vague, the LSD1 inhibitor tranylcypromine impaired growth of Ewing sarcoma cell lines in vitro (23). Tranylcypromine is used clinically as a monoamine oxidase inhibitor, but has low efficacy as an LSD1 inhibitor, and has several documented off-target effects precluding widespread clinical use targeting this enzyme.
Further investigation showed LSD1 recruitment by EWS/FLI as a member of the nucleosome remodeling and histone deacetylase (NuRD) complex to repress the critical EWS/FLI targets LOX and TGFBR2 (29). Treatment with the potent LSD1 inhibitor, HCI2509, caused derepression of these genes and impaired tissue culture cell viability in multiple Ewing sarcoma cell lines at physiologically-relevant concentrations (29). The sensitivity of multiple cell lines to LSD1 inhibition suggests a pivotal role for LSD1 in Ewing sarcoma beyond repression of select EWS/FLI targets. The extent of the role that LSD1 plays in the global transcriptional program orchestrated by EWS/FLI, and other EWS/ETS fusions, remains uncharacterized. The experiments herein describe the global transcriptional effects of HCI2509 treatment in Ewing sarcoma and the downstream anti-tumor effects that result.
Materials and Methods
Constructs and Retroviruses
The Luciferase-RNAi (Luc-RNAi), EWS/FLI-RNAi (EF-2-RNAi), 3x-FLAG EWS/FLI, 3x-FLAG Δ22 and 1x-FLAG R2L2 cDNA are previously described (30–32). The 1x-HA HMOX1 cDNA was generated and sub-cloned into the Murine Stem Cell Virus (MSCV) retroviral vector (Clontech). siRNA controls or targeted toward LSD1, CHD4, REST, RCoR1, NCoR and Sin3A are described previously (29).
Antibodies and Reagents
The following antibodies were used for immunodetection: M2-anti-FLAG (HRP; Sigma A8592), anti-FLI-1 (Santa-Cruz sc-356X), anti-α-Tubulin (Calbiochem CP06), anti-HA (Abcam ab9110), anti-H3 total (Abcam ab1791), anti-H3K4 me1 (Abcam ab8895), anti-H3K4 me2 (Millipore, 07-030), anti-H3K4 me3 (Active Motif, 39159), anti-H3K9 me1 (Abcam ab9045), anti-H3K9 me2 (Abcam ab1220), anti-H3K9 me3 (Abcam ab8898), anti-HMOX1 (Sigma SAB1410641), anti-Paxillin (BD Transduction Labs 610619), anti-LSD1 (Cell Signaling Technology 2184) AlexaFluor secondary (Molecular Probes), AlexaFluor Phalloidin (Molecular Probes). HCI2509 is previously described (33).
Cell culture
Ewing sarcoma cell lines harboring the EWS/FLI (A673, TC-71, SK-N-MC, SKES1 and EWS502) or EWS/ERG fusion (TTC-466) were grown in appropriate selection media, as previously described (34,35). NIH 3T3 cells with and without EWS/FLI expression were previously reported (36). Growth assays (3T5) were previously described (35).
Colony formation assays
Soft agar assays were described previously (35). Methylcellulose assays were performed by plating 1×105 cells in a 1:1 mix of 2% methylcellulose and growth media as described previously (29).
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNAeasy kit (Qiagen). Total RNA was then amplified and detected using SYBR green fluorescence for quantitation. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control. The data in each condition was normalized to internal housekeeping control genes, GAPDH and RPL19. Primer sequences are provided in Supplemental Data (Table S3).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (37) using anti-LSD1 antibody (Abcam ab17721). qPCR was performed with HMOX1 gene primers amplifying a region ~29 base pairs upstream of the transcription start site (TSS). BCL2L1 primers were used as normalization controls (38). Primer sequences are provided in the Supplemental Data (Table S3).
In vivo Studies
Xenografts
A673, SK-N-MC or SKES1 cells were injected into the right hindflanks of nude mice at 1×106 cells or 1×106 cells or 2.5×106 cells per flank, respectively. For all xenograft studies, ten mice per condition were injected subcutaneously; therefore ten tumors were measured per group. In the SK-N-MC study, one animal perished due to an unrelated rash and was censored from analysis. Tumors were measured using digital calipers and volumes were calculated as follows: (L×W×D)/2. Treatment was initiated on day 7 after bioluminescent imaging confirmed tumor engraftment in the A673 study, while SK-N-MC and SK-ES-1 studies were initiated once tumors reached a volume of >100 mm3. Mice in each group were sacrificed once tumors reached a size limit of 2 cm3. Kaplan-Meier survival curves were plotted using GraphPad Prism. Tumor volume and body weight were recorded for all three models. Harvested tumors harvested were flash frozen, homogenized by mortar and pestle in liquid nitrogen analyzed for RNA or protein. All xenograft experiments were performed in accordance with protocol 11-11003 approved by the University of Utah IACUC. Blood Counts: The facial vein was identified and pierced with a lancet, blood was collected in a heparin capillary tube, and analyzed using a HemaTrue hematology analyzer (Heska).
Immunofluorescence assays
5×104–1.5×105 A673 and TTC-466 cells were seeded onto fibronectin-coated coverslips, allowed ≥24 hours to adhere, treated with vehicle or HCI2509 at 0.5 μM, 1 μM, 2 μM for 3 days in DMEMc+10%FBS, and fixed, stained, and imaged as previously described (39). Briefly, cells were immunostained with paxillin antibody (1:100) or LSD1 antibody (1:400), then with secondary antibody (1:100) and DAPI (0.3 μM). Fluorescent cell images were collected on a Zeiss Axioskop2 mot plus microscope with a 40 X dry objective (NA 0.75 NeoFluor), Axiocam MR camera, and Axiovision v4.8.1 software (Carl Zeiss MicroImaging, Inc.). Cell area analysis was performed with Image J (NIH freeware) and MetaMorph software (Molecular Devices); ≥50 cells were analyzed from ≥10 microscope fields, conversion factor 1 μm = 6.2 pixels. Image data sets were analyzed in GraphPad Prism 5 using unpaired t-tests and graphed as mean and SD.
RNA sequencing analysis, GSEA and Venn overlaps
See Supplementary Methods for a description of RNA-seq data collection. Overlaps between the different gene sets were performed using VennMaster (http://www.informatik.uni-ulm.de/ni/mitarbeiter/HKestler/vennm/doc.html). Statistical significance of the overlaps was determined using Chi square analysis. Gene set enrichment analysis (GSEA) was performed using GSEA v2.0.10 (http://www.broad.mit.edu/gsea/). Functional annotation analysis was performed by DAVID (david.abcc.ncifcrf.gov). Heat maps were generated by converting read counts to fragments per kilobase gene model per million reads (FPKM). Genes were included in the head map if their differential abundance was >3-fold in both experiments and the Benjamini-Hochberg false discovery rate was <0.05. The data were normalized per gene across both experiments and log10 transformed. Heat maps were created using the R gplots package; ranks were based on averages of treatment and control.
Cell viability determination and EC50 shift analysis
A673 cells were stably infected and selected for expression of control Luc-RNAi or EF-2-RNAi. 2×104 cells/well were seeded in a 96-well plate, allowed 24 hours to adhere and treated with either vehicle or HCI2509 for 96 hours. Viability was assayed using CellTiter-Glo (Promega).
In vitro apoptosis assays
Caspase/Viability
2 × 104 cells/well were seeded in a 96-well plate, treated with either vehicle or 2X their respective EC50 for HCI2509. Caspase activation and cell viability were assayed using Caspase-Glo 3/7 (Promega) and CellTiter-Glo (Promega), respectively, at 0, 24, and 48 hours. TUNEL Staining: A673 cells were treated with either vehicle or 2 μM HCI2509 for 48 hours and then assayed using the DeadEnd Colorimetric TUNEL (Promega). Images were collected on an Olympus 1X70 inverted microscope, Olympus EOS Rebel XSi camera, and EOS Utility software (Canon U.S.A., Inc.).
Data availability
Raw sequence reads can be found in the NCBI SRA under numbers SRA096343, SRA096347, SRA096354. Differentially expressed genes from each RNAseq dataset are in Supplementary Table S1.
Results
LSD1 inhibition reverses the EWS/ETS-driven transcriptional program in Ewing sarcoma
HCI2509 is a specific and reversible noncompetitive inhibitor of LSD1 previously shown to derepress the critical EWS/FLI target genes LOX and TGFBR2, and to kill multiple Ewing sarcoma cell lines in vitro (29,30). Interestingly, HCI2509-mediated derepression of LOX and TGFBR2 was dependent on the expression of EWS/FLI. To determine whether the Ewing sarcoma cell death observed with HCI2509 treatment was also dependent on EWS/FLI, we knocked down the fusion protein using retroviral-mediated shRNA, and assessed viability after treatment with HCI2509. Cells expressing EWS/FLI were ~10-fold more susceptible to treatment with HCI2509 compared to cells with EWS/FLI knockdown (Figure 1A), which confirmed the efficacy of HCI2509 is dependent upon the presence of EWS/FLI. Based on previous results showing HCI2509 decreased viability in multiple patient-derived Ewing sarcoma cell lines containing EWS/FLI, we next asked whether the effects were restricted to EWS/FLI-containing lines, or whether they can be generalized to other Ewing sarcoma associated fusions. As was seen for EWS/FLI-containing cell lines, the Ewing sarcoma cell line TTC-466 containing the alternative EWS/ETS fusion, EWS/ERG, was also sensitive to HCI2509 treatment (Figure S1A). Like A673 cells, TTC-466 cells with EWS/ERG knockdown showed decreased sensitivity to HCI2509. We further tested whether introducing EWS/FLI into a heterologous cell line induced sensitization to HCI2509 treatment in NIH 3T3 fibroblasts (Figure S1B). No sensitization was observed, suggesting EWS/ETS-dependent sensitization is unique to Ewing sarcoma cells.
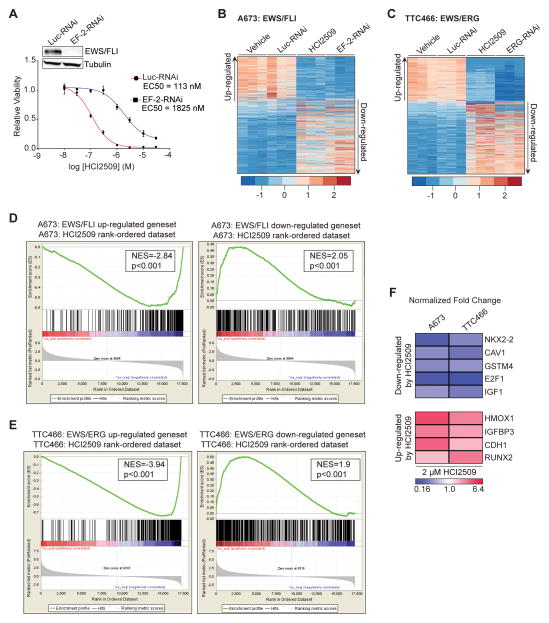
Figure 1. Global EWS/FLI transcriptional activity is disrupted by HCI2509.
A) Cell viability assay showing the difference in HCI2509 sensitivity between A673 cells with control (EC50=113 nM; 95% CI 81.9–158 nM) and EWS/FLI knockdown (EC50=1825 nM; 95%–CI 1111–2999 nM). The dose-response curves were determined after 96 hours of treatment and normalized to the vehicle controls. Mean and SD are shown (n=3). Stable EWS/FLI knockdown was analyzed by Western Blot as shown in the inset.
B,C) Heat map representation of the HCI2509 expression profile matched to the rank-ordered (B) EWS/FLI- and (C) EWS/ERG-knockdown profiles. Genes were ranked by mean deviation of the log transformed FPKM (Fragments Per Kilobase per Million mapped reads). The columns for each condition represent one independent biological replicate. Each row represents a different gene.
D,E) Gene set enrichment analysis (GSEA) from RNA-seq experiments using (D) the EWS/FLI regulated genes in A673 cells as the rank-ordered dataset and the 281 HCI2509 up-regulated and 376 HCI2509-down-regulated genes as the genesets and (E) EWS/ERG regulated genes in TTC-466 cells as the rank-ordered dataset and the 216 HCI2509 up-regulated and 357 HCI2509-down-regulated genes as the genesets. Normalized enrichment scores (NES) and P values are shown.
F) qRT-PCR validation of NKX2.2, CAV1, GSTM4, E2F1, IGF-1, RUNX2, IGFBP3, HMOX1 and CDH1 as HCI2509 targets in A673 and TTC-466 cells treated for 48 hours with vehicle or HCI2509 at 2xEC50. Normalized fold change is indicated as a heat map. The p-value for each fold-change is < 0.05 (n=3). Individual p-values are reported in Supplementary Table S2.
To assess changes in gene expression caused by LSD1 inhibition in the context of EWS/ETS fusion transcriptional activity, we first generated transcriptional signatures for EWS/FLI and EWS/ERG. A673 and TTC-466 cell lines were subjected to retroviral-mediated shRNA knockdown of EWS/FLI and EWS/ERG, respectively, and the differentially expressed genes were assessed using RNAseq (A673 data is previously reported (40)). We first analyzed the similarity between EWS/FLI and EWS/ERG transcriptional regulation, and found the overlap between these transcriptional profiles was significant by both chi square and Gene Set Enrichment Analysis (GSEA) (Figure S1C–D).
Having established similarity between the EWS/FLI and EWS/ERG transcriptional profiles, we next determined the global transcriptional signature of HCI2509 using RNAseq in both A673 and TTC-466 cells. The comparison of the HCI2509 signature with both fusion proteins showed treatment with HCI2509 comprehensively reversed the transcriptional profiles driven by both EWS/FLI (Figure 1B) and EWS/ERG (Figure 1C). Thus, genes normally upregulated by EWS/FLI and EWS/ERG were downregulated by LSD1 inhibition with HCI2509, and vice versa. Chi square analysis showed a statistically significant overlap between EWS/FLI activated and HCI2509 downregulated genes and vice versa (Figure S1E). This was also true for the TTC-466 cell line (Figure S1F). Importantly, when either EWS/FLI or EWS/ERG upregulated target genes were analyzed with HCI2509 upregulated genes, and vice versa, no significant overlap was observed (data not shown). These results suggest substantial reversal of global transcriptional activity of both EWS/ETS fusions and highlights the importance of LSD1 in EWS/ETS-mediated transcriptional dysregulation, for both EWS/ETS-repressed and EWS/ETS-activated genes.
We alternatively compared HCI2509 and EWS/ETS transcriptional profiles using GSEA with a more stringent cutoff of a 4-fold change and FDR of 1×10−15 for the EWS/FLI geneset and a 3-fold change and FDR of 1×10−10 for the EWS/ERG geneset. The HCI2509 downregulated genes clustered significantly with upregulated EWS/ETS target genes and vice-versa (Figure 1D–E). This confirmed the mechanism of HCI2509 action specifically correlated to the EWS/ETS transcriptional function.
Because EWS/FLI interacts directly with the NuRD/LSD1 complex, we hypothesized the effects of LSD1 inhibition would be observed at genes regulated by direct binding of EWS/FLI. To test this, we compared the HCI2509 regulated gene list to a set of previously identified EWS/FLI direct target genes (38). GSEA showed significant correlation for genes both up- and downregulated directly by EWS/FLI in the HCI2509 regulated gene list (Figure S1G), suggesting that LSD1 is critical to the transcriptional function of EWS/FLI at its direct targets. Since HDAC2 and HDAC3 were previously shown to be directly recruited by EWS/FLI to mediate transcriptional repression (29), we also compared changes in gene expression mediated by the HDAC inhibitor vorinostat (30) to the EWS/FLI direct targets (38). As expected, genes which are directly repressed by EWS/FLI became derepressed with vorinostat treatment (Figure S1H). However, vorinostat had no effect on genes directly activated by EWS/FLI (Figure S1H). Together, these data illustrate that LSD1 inhibition has a unique dual effect in both transcriptional activation and repression mediated by EWS/FLI, while targeted inhibition of HDACs only blocks EWS/FLI mediated transcriptional repression.
We next assessed the functional significance of the HCI2509 and EWS/ETS overlapping gene sets with the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The most significant classes of genes downregulated by HCI2509 are related to DNA replication and cell cycle while the classes of genes upregulated by HCI2509 are related to regulation of cell death and extracellular matrix (Figure S1I, S1J). These are consistent with previously described molecular functions dependent upon EWS/FLI (41). We then used both A673 and TTC-466 cell lines to validate target genes from the HCI2509 RNAseq profiles which have been identified as critical EWS/FLI targets critical for oncogenic transformation, survival, and differentiation (31,42–48). Using qRT-PCR, genes activated by EWS/FLI including NKX2.2, CAV1, GSTM4, E2F1, and IGF1 were all significantly downregulated in both cell lines with HCI2509 treatment. Conversely, genes repressed by EWS/FLI including RUNX2, IGFBP3, CDH1, and HMOX1 were significantly upregulated in both cell lines after treatment (Figure 1F). Using this 9-gene panel we tested other patient-derived Ewing sarcoma cell lines (EWS-502, SK-N-MC, SKES1, TC71) to confirm the transcriptional effects of HCI2509 treatment were not cell-line specific. Each cell line showed results congruent with A673 and TTC-466 cells (Figure S1K) indicating the disruption of EWS/ETS transcriptional activity by HCI2509 occurs in all tested Ewing sarcoma cell lines.
HCI2509 recapitulates morphological phenotypes associated with EWS/FLI knockdown
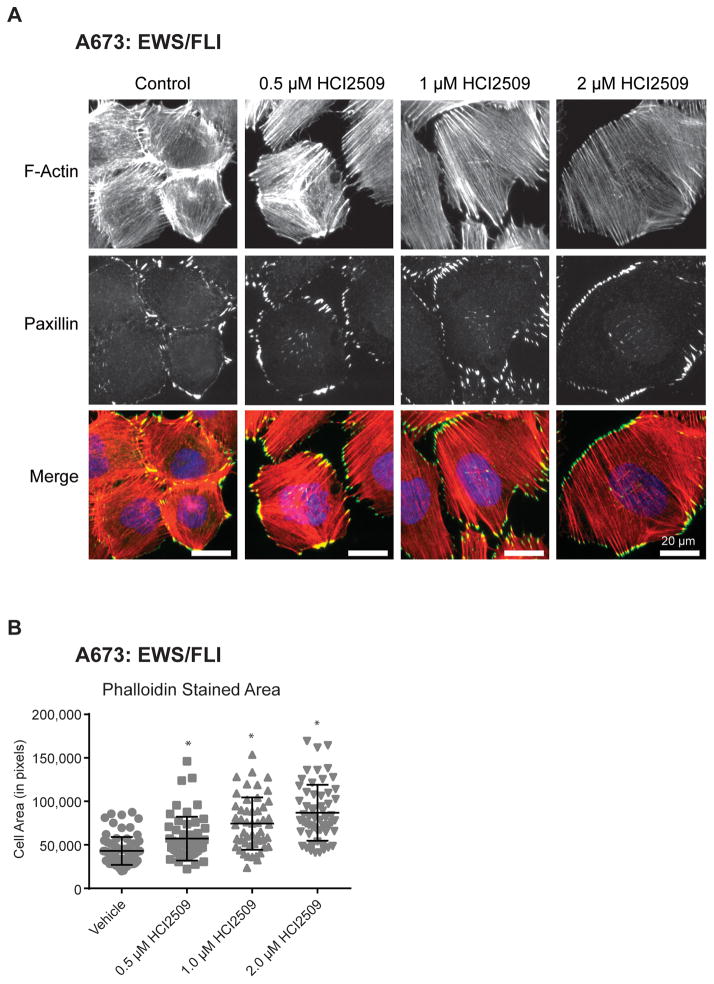
The disruption of EWS/ETS transcriptional activity by HCI2509 should manifest in changes to cellular phenotypes associated with the presence of EWS/ETS. To test this, we first asked whether HCI2509 mimics the morphological phenotypes associated with EWS/ETS knockdown. Ewing sarcoma is characterized histologically by small round blue-staining cells. In the context of EWS/FLI knockdown, cells display morphological phenotypes typical of the putative mesenchymal stem cell cell-of-origin (39). EWS/FLI knockdown induces a robust actin cytoarchitecture with striking actin stress fibers anchored to integrin-based focal adhesions, increased cell adhesion, and spreading, that correlated with increased migration (39). To assess the effect of HCI2509 on cellular architecture we performed immunofluorescence microscopy. Control A673 cells showed the characteristic small round cell phenotype of Ewing sarcoma with short, thin actin fibers (Figure 2A,S2A). Treatment with HCI2509 induced organizing of actin stress fibers throughout a well spread cell, with robust paxillin-containing focal adhesions (Figure 2A,S2A). Cell area of the phalloidin-stained cells was measured to quantify the effect. HCI2509-treated A673 cells showed a dose-dependent increase in cell area (Figure 2B). To show observed morphological changes were on target, we knocked down LSD1 using siRNA and assessed both cytoskeletal architecture and cell area. Consistent with the increased cell spreading phenotypes we observed with HCI2509 treatment, decreased LSD1 protein levels correlated with increased cell spreading (Figure S2B,S2C). TTC-466 likewise showed dose-dependent changes in actin staining, focal adhesions, and cell spreading (Figure S2D–F), suggesting treatment with HCI2509 generated the morphology associated with the loss of EWS/FLI.
Figure 2. Morphological Changes in A673 with HCI2509 Treatment.
A) Immunofluorescence images of A673 cells treated with increasing doses of HCI2509 for 3 days. Staining was performed for F-actin stress fibers (red – phalloidin) and for focal adhesions (green – paxillin), and nuclei (blue). HCI2509 induced a dose-dependent increase in the cell spreading and morphology.
B) Cell area in pixels shows a dose-dependent increase in cell spreading with increasing doses of HCI2509. A673 cells were fixed and stained with phalloidin. Cell area was quantified as previously described (39). Data is plotted as mean with sd and p-values were calculated using a Student’s t-test(* p<0.0001.)
HCI2509 affects oncogenic transformation, histone methylation, and causes apoptosis
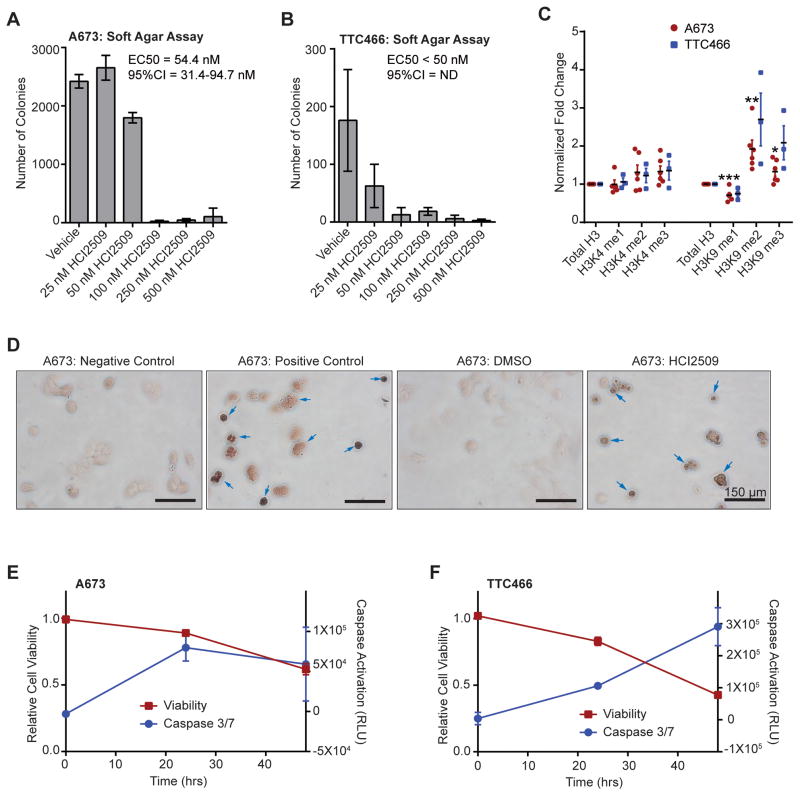
We next tested whether HCI2509 would impair EWS-ETS-driven oncogenic transformation using colony formation assays as a measure of anchorage-independent growth (Figure 3A–B, S3A–C). Interestingly, both the A673 and TTC-466 showed a shift in sensitivity (Figure 3A–B) to the low nanomolar range in 3D cultures. Thus, the EC50 observed was significantly lower than those seen in viability assays (29).
Figure 3. Mechanism of action of HCI2509 in vitro.
A,B) Quantification of colonies formed by (A) A673 cells and (B) TTC-466 cells treated with either vehicle (0.3% DMSO) or increasing doses of HCI2509. Error bars indicate SD of duplicate assays. EC50 values were determined using GraphPad Prism 6.
C) Quantification of global methylation changes at histone H3K4 and H3K9 in A673 (red, n=5) and TTC-466 (blue, n=3) following 48-hr treatment with 2 μM HCI2509. Methyl marks were assayed by western blot and the relative band intensities were quantified using ImageQuant (GE Healthcare Biosciences). Each sample was normalized to total H3 and fold-change was determined by comparison to a vehicle control. Error bars indicate SEM and p values were calculated using a Student’s t-test (*p=4.25E-2, **p=1.07E-2, ***p=6.36E-3).
D) TUNEL staining of A673 cells treated with either vehicle (0.3% DMSO) or 2 μM HCI2509. Negative control indicates lack of labeling and positive control indicates DNase treatment with labeling. Arrows indicated TUNEL-positive cells.
E,F) Cell viability and caspase activation at 0, 24, and 48 hours in (E) A673 and (F) TTC-466 cells treated with 2 μM HCI2509. Measurements were normalized to their respective vehicle (0.3% DMSO) sample at the appropriate time point.
Given the role of LSD1 in H3K4 and H3K9 demethylation, we also characterized changes in global histone methylation at these residues (Figure 3C). Immunodetection of methylation status was quantified using densitometry for statistical analysis. Due to the subtlety of the observed changes, several biological replicates were performed. Interestingly, A673 cells showed no change in H3K4 monomethylation, and while H3K4 dimethylation (H3K4me2) and trimethyation (H3K4me3) trended higher, the effect was not statistically significant (Figure 3C). However, H3K9 monomethylation (H3K9me1) significantly decreased, with a complementary increase in H3K9 dimethylation (H3K9me2) and trimethylation (H3K9me3) (Figure 3C). TTC-466 cells generally recapitulated the effects seen in A673 (Figure 3C), but the results showed more variability.
We used DAVID analysis to examine the functional relevance of HCI2509 differentially expressed genes apart from the overlap with the EWS/FLI transcriptional profile (Figure S1E). Notably, genes upregulated by HCI2509 related to cell cycle arrest and programmed cell death while downregulated genes associated with S-phase, cell cycle, and proliferation (data not shown). Thus, HCI2509 likely reduces Ewing sarcoma cell viability (29) through induction of apoptosis. We therefore used TUNEL staining and caspase activation to assay whether HCI2509 triggered apoptosis. TUNEL-stained cells treated with HCI2509 for 48 hours showed increased staining as compared to vehicle (Figure 3D). Similarly, treatment with HCI2509 showed increased activation of caspase 3/7 over 48 hours, which corresponded with decreased viability in multiple Ewing sarcoma cell lines (Figure 3E–F, S3D–F). Collectively, these data show HCI2509 not only reversed EWS/FLI-associated oncogenic phenotypes, but also altered H3K9 methylation status and induced apoptosis.
HMOX1 is an on-target response biomarker for HCI2509 treatment in vitro
Screening efforts to identify HCI2509 (33) suggested induction of HMOX1 was an effect proportional to the biochemical potency of the inhibitor (Figure S4A). We evaluated whether HMOX1 constituted biological output to demonstrate target engagement. HMOX1 was significantly upregulated by HCI2509 in both EWS/ETS transcriptional profiles. We assessed the regulation of HMOX1 by alternative EWS/ETS fusions and found HMOX1 was repressed across EWS/ETS fusions (Figure 4A). Moreover, Venn Master analysis revealed HMOX1 was one of 81 genes present in the overlap between EWS/FLI targets upregulated by HCI2509 and genes downregulated by EWS/FLI in primary Ewing sarcoma samples (41) (Figure 4B). Based on the repression of HMOX1 in primary tumors and robust HCI2509-induced derepression across tested cell lines, we asked whether HMOX1 protein also increased. We found HMOX1 protein levels elevated with EWS/FLI knockdown, returned to baseline levels with EWS/FLI reexpression (Figure 4C), and likewise increased in response to HCI2509 (Figure 4D).
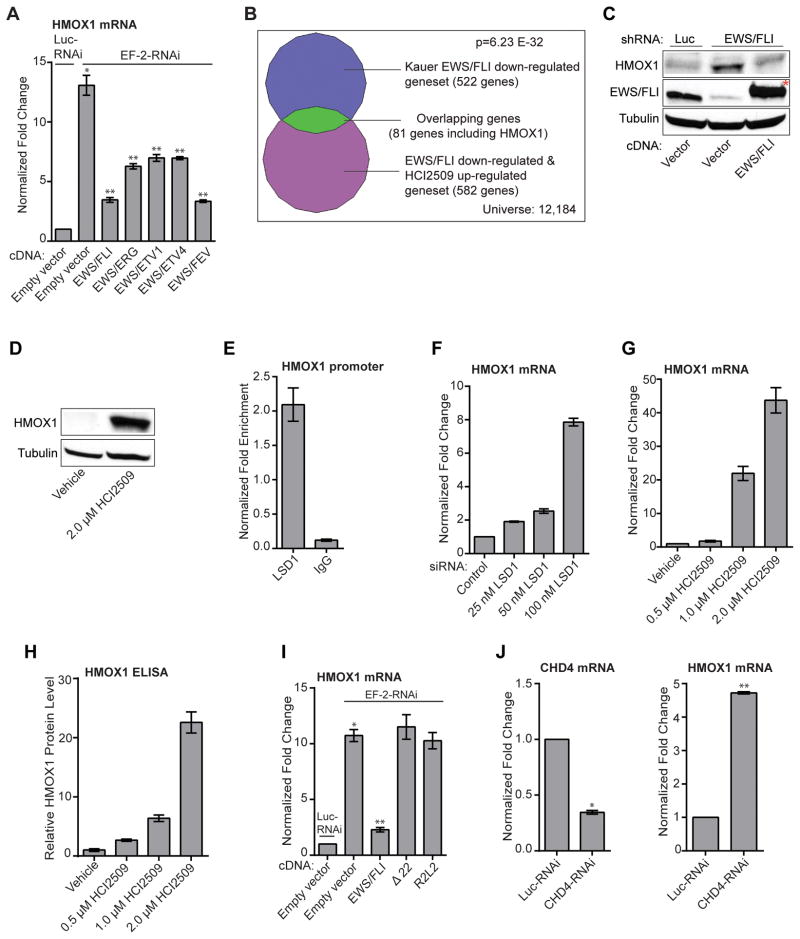
Figure 4. Regulation of HMOX1.
A) Validation of HMOX1 as a target gene of multiple EWS/ETS fusion. qRT-PCR analysis of HMOX1 in A673 cells infected with a control shRNA (Luc) or an EWS/FLI shRNA followed by rescue with an empty vector, an RNAi-resistant EWS/FLI, EWS/ETV1, EWS/ERG, EWS/ETV4, or EWS/FEV cDNA. Error bars indicate SD (n=3). P values were calculated using Student’s t-test. *P value determined against Luc-RNAi/Empty vector (p=1.51E-5). **P values determined against EF-2-RNAi/Empty vector (p≤2.79E-4).
B) Venn diagram representation of the HCI2509-up-regulated and EWS/FLI-down-regulated geneset (from Figure S1E) overlapped with EWS/FLI down-regulated targets in primary tissue samples. The chi square-determined P value is indicated.
C,D) Western blot analysis of HMOX1 protein levels with (B) EWS/FLI knockdown/rescue and (C) HCI2509 treatment. HMOX1 and EWS/FLI levels were assessed in A673 cells using anti-HMOX1 and anti-FLI antibodies. Tubulin was used as a loading control. (*) indicates the 3x-FLAG tagged EWS/FLI cDNA that runs slightly higher than endogenous EWS/FLI.
E) ChIP of LSD1 with the level of enrichment for LSD1 at the HMOX1 transcription start site plotted as normalized fold enrichment compared to the enrichment at BCL2L1 as a negative control. IgG was used as a negative antibody control. The error bars indicate SEM (n=3). The Student’s t-test determined P value is 1.10E-3.
F,G,H,I,J) Validation of HMOX1 repression as on target. P values were calculated using a Student’s t-test. qRT-PCR analysis of HMOX1 in A673 cells treated with: (F) vehicle, 25 nM (p=1.43E-5), 50 nM (p=3.05E-4), and 100 nM LSD1 siRNA (p=7.39E-6) and (G) vehicle, 0.5 uM (p=3.65E-2), 1 uM (5.82E-4), and 2 uM HCI2509 (3.48E-4). (H) Quantification of protein levels by ELISA for A673 cells treated with vehicle, 0.5 uM (p=6E-4), 1 uM (p=1E-4), and 2 uM HCI2509 (p=1E-4). (I) qRT-PCR analysis of HMOX1 in A673 cells treated with (I) knockdown of EWS/FLI and rescue with full-length, Δ22, and R2L2 mutants. *P value determined against Luc-RNAi/Empty vector (P=1.32E-6). **P value determined against EF-2-RNAi/Empty vector (p=3.99E-6). (J) qRT-PCR analysis of both CHD4 and HMOX1 mRNA in the presence of control or CHD4 RNAi. *p=2.95E-7 **p=3.98E-9. Error bars indicate SD (n=3).
We next asked whether HMOX1 induction depended upon both LSD1 and EWS/FLI function. Targeted LSD1 ChIP showed enrichment at the HMOX1 promoter (Figure 4E). Both siRNA-mediated knockdown of LSD1 and LSD1 inhibition with HCI2509 derepressed HMOX1 in a dose-dependent manner (Figure 4F–G). Moreover, this dose-dependent increase was validated at the protein level using ELISA (Figure 4H). EWS/FLI knockdown resulted in similar HMOX1 induction as 100 nM LSD1 siRNA (Figure 4I). HMOX1 repression was restored with full-length EWS/FLI rescue; however rescue with either the Δ22 mutant (49) lacking most of the EWS domain or the DNA-binding mutant R2L2 (50) failed to repress HMOX1 (Figure 4I, S4B). This is consistent with other EWS/FLI-repressed targets (29), and jointly implicates the repressive function of the EWS domain together with the DNA-binding domain of FLI in suppressing HMOX1 expression.
The necessity of the EWS domain raised the question about which corepressor complex EWS/FLI recruits to repress HMOX1. EWS/FLI is known to recruit the NuRD complex to enact repressive transcriptional activity at some target genes (29) so we first probed whether NuRD was responsible for HMOX1 repression. Knockdown of the NuRD complex with CHD4-RNAi induced a 4.5-fold increase in HMOX1 (Figure 4J). RNAi-mediated knockdown of the REST, Co-REST, NcoR/SMRT, and Sin3A complexes were also evaluated. Only REST knockdown showed a significant, though lower magnitude, increase in HMOX1 mRNA (Figure S4C). These results are consistent with recruitment of LSD1 by REST (51), but highlight the central role of the NuRD complex in repressing HMOX1.
Because HMOX1 protein was strongly induced in cells treated with HCI2509 in addition to those with EWS/FLI knockdown, we investigated whether HMOX1 affects cellular transformation. A673 cells overexpressing HMOX1 (Figure S4D) showed little difference in doubling times compared to control (Figure S4E). Additionally, oncogenic transformation was not affected in cells overexpressing HMOX1 (Figure S4F). These findings support a model in which HMOX1 is directly repressed by EWS/FLI-mediated recruitment of the NuRD complex with its associated LSD1 subunit, but does not play a role in proliferation or the oncogenic phenotype. Thus, in Ewing sarcoma HMOX1 is a useful biological readout of LSD1 inhibition, and associated disruption of EWS/ETS transcriptional activity.
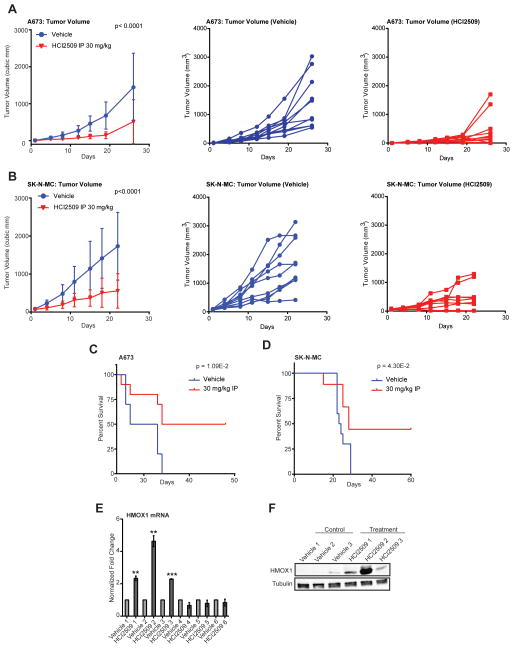
HCI2509 as a single agent significantly reduces tumor growth in vivo
Having observed HCI2509-mediated disruption of global EWS/ETS transcriptional function, reversal of EWS/ETS-associated morphological and oncogenic phenotypes, and induction of apoptosis, we next investigated whether HCI2509 would impair tumorigenesis in vivo. In A673, SK-N-MC, and SK-ES-1 xenograft models daily intraperitoneal treatment with 30 mg/kg HCI2509 delayed tumor growth as a single agent (Figure 5A,5B,S5A). Animal weights were recorded to monitor non-specific toxicity with none observed (Figure S5B–D). Blood counts were also examined due to hematopoietic toxicity associated with LSD1 knockdown (52). During 4 weeks of treatment, no significant difference was observed between vehicle and treated groups (Figure S5E). Importantly, Animals treated with HCI2509 showed improved survival over 60 days (Figure 5C–D) as compared to vehicle in both A673 and SK-N-MC xenograft models.
Figure 5. HCI2509 activity in vivo.
A,B) In vivo subcutaneous hind-flank xenograft studies measuring tumor volume for animals bearing tumors grown from (A) A673 cells and (B) SK-N-MC cells. N=10 for all groups, with the exception of SK-N-MC HCI2509 treated group as noted. For tumor volumes, P-values were determined by 2-way ANOVA comparing the treatment curve to the vehicle curve. Individual tumor growth curves are shown for the vehicle-treated (blue) and HCI2509-treated (red) groups.
C,D) Survival curves for mice bearing subcutaneous hind-flank xenografts of (C) A673 cells or (D) SK-N-MC cells. N=10 for the A673 group. In the SK-N-MC treatment group one mouse died due to a treatment-unrelated rash and was censored from further analysis. Therefore, in the SK-N-MC study 10 and 9 mice were used for the vehicle and HCI2509 treatment groups, respectively. The mice were sacrificed once their tumors reached a size limit of 2 cm3. Percent survival was plotted as Kaplan-Meier survival curves using GraphPad Prism. The log-rank (Mantel-Cox Test) determined P values using GraphPad Prism are indicated.
E,F) Analysis of HMOX1 expression by (E) qRT-PCR and (F) western blot in tumors from both vehicle and treatment A673 groups. Only the three tumors which showed an increase in HMOX1 RNA (E) were used for western blotting with three random vehicle tumors (F). HMOX1 levels were assessed using an anti-HMOX1 antibody. Tubulin was used as a loading control. Error bars indicate SD (*for p<0.01; **for p<0.001).
We also evaluated HMOX1 in tumors from the A673 study using six tumors from the vehicle group and 6 tumors from the treatment group. HMOX1 RNA was elevated in three treatment tumors as compared to the control tumors (Figure 5E). Moreover, this result was validated at the protein level with HMOX protein nearly undetectable in three representative control tumors and expressed in all three HCI2509-treated tumors with elevated HMOX1 mRNA (Figure 5F). This suggested HCI2509 both engages LSD1 and disrupts EWS/FLI transcriptional activity in vivo. When considered with the impaired tumorigenesis and increased survival, these data substantiate LSD1 inhibition with HCI2509 as a potential therapeutic strategy for Ewing sarcoma.
Discussion
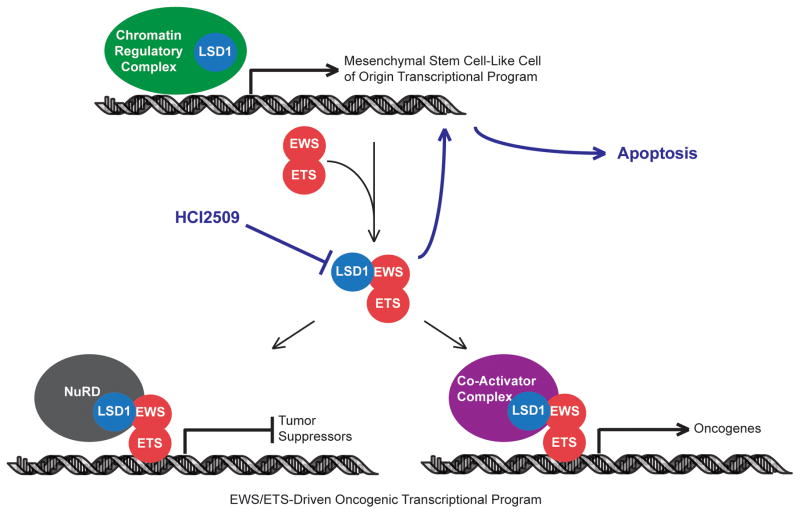
Our findings reveal a novel dual role for LSD1 in mediating both the transcriptional activating and repressive function of EWS/ETS fusions in Ewing sarcoma and modulating the oncogenic phenotypes resulting from the presence of EWS/ETS fusions proteins. Small molecule blockade of LSD1 with the potent and reversible inhibitor, HCI2509, comprehensively disrupted the transcriptional signature of EWS/FLI and EWS/ERG as well as the subsequent downstream malignant characteristics of Ewing sarcoma cells as depicted in Figure 6. Transcription factors are notoriously difficult targets for drug discovery and development programs and identification of small molecules which disrupt EWS/FLI has been an elusive goal (53). Recently, disruption of the c-Myc oncogenic transcription factor was achieved through targeted inhibition of the epigenetic reader BRD4 with the small molecule inhibitor JQ1, which downregulated MYC transcription, decreased c-Myc protein, and disrupted c-Myc transcriptional function (54,55). Our results likewise support an approach undermining oncogenic transcription factors by targeting their associated epigenetic machinery. The observed difference in transcriptional outcomes between LSD1 inhibition and HDAC inhibition with vorinostat (30) highlights the importance of selecting the correct target and demonstrates the potential to tailor epigenetic therapy based on the context of the disease.
Figure 6. Model for HCI2509 mechanism of action in Ewing sarcoma.
HCI2509 treatment inhibits LSD1, which impairs the ability of EWS/ETS fusions proteins to globally alter gene expression. This leads, in part, to a reversion to the transcriptional program of the putative cell-of-origin and causes apoptosis.
The A673 cell line has been used to investigate the transcriptional effects of EWS/FLI because it is relatively tolerant of EWS/FLI knockdown (31). While A673 cells expressing EWS/FLI shRNA lose transformation (31) and show morphological characteristics similar to the putative cell-of-origin for Ewing sarcoma (39), cell growth and viability are not impaired. HCI2509 treatment largely reproduced the loss of transformation consistent with downregulation of EWS/ETS-activated targets and upregulation of EWS/ETS-repressed targets. However, unlike EWS/FLI knockdown, treatment with HCI2509 triggered caspase-dependent apoptosis in A673 cells. This effect was consistent across cell lines, suggesting additional mechanisms of action for HCI2509. Upon analysis of the genes regulated by HCI2509, but not overlapping with the EWS/FLI transcriptional profile we noted that in this context HCI2509 upregulated genes associated with cell cycle arrest and programmed cell death while genes associated with S-phase, cell cycle, and proliferation were downregulated. This is consistent with both the observed apoptotic phenotype and the reported role for LSD1 in the maintenance of dedifferentiation in cancer (16–19,56) and regulation of the cell cycle in stem-like cells (57–60). Additionally, LSD1 is known to regulate non-histone proteins including p53 (61). LSD1-mediated demethylation of p53 disrupts association of its cofactor 53BP1 and prevents induction of apoptosis (61). Thus, the sensitivity of Ewing sarcoma cell lines to LSD1 inhibition with HCI2509 may be the result of two tiers of LSD1-specific effects and this is reflected in the different EC50s of the phenotypes described with treatment. Sankar et al. previously reported the EC50 for HCI2509 in cell viability assays for multiple Ewing sarcoma cell lines ranging from 0.19 μM–1.4 μM (29). In this report, both A673 and TTC-466 cell lines show a loss of transformation in colony forming assays below 100 nM and 50 nM of HCI2509 treatment, respectively. This suggests a model for HCI2509 mechanism of action in Ewing sarcoma where the first tier of effects disrupt the transforming function of EWS/ETS at concentrations much lower than the second tier of effects, which result in induction of apoptosis. Nonetheless, the proapoptotic effects of HCI2509 may be due to as yet undescribed off-target effects. However, we feel this is unlikely, as the only off-target for HCI2509 known to us is mild inhibition of CYP3A4 with a biochemical IC50 of 2.61 μM (33).
Given its known capability to demethylate both H3K4 and H3K9 mono- and dimethyl marks, and its dual role in modulating activated and repressed genes, we were surprised to observe the only significant changes in global histone methylation occurring at the H3K9 residue. Methylation at H3K9 is typically associated with gene repression so this may be linked to the apparent role of LSD1 in EWS/ETS-mediated gene activation. LSD1-mediated demethylation of MTA1 causes a switch in the associated complex from the repressive NuRD complex to the activated NURF complex (62). In turn, NURF directs the demethylase activity of LSD1 toward H3K9 (62). This presents a plausible mechanism by which EWS/FLI recruits LSD1 as a member of the NuRD complex and is then able to enact both transcriptional activation and repression. Alternatively, LSD1 was recently described to mediate a global reduction in H3K9me2 during the epithelial-to-mesenchymal transition (EMT) (63). Inhibition of LSD1 in this model impaired the cellular migration and chemoresistance resulting from EMT. It is possible that the observed increase in H3K9me2 in Ewing sarcoma cells may be due to a similar phenomenon. Additional studies are required to elucidate the molecular mechanisms choreographing whether LSD1 facilitates EWS/FLI-mediated transcriptional activation or repression and the genome-wide positioning of LSD1 histone substrates in the presence and absence of both EWS/FLI and HCI2509. The absence of significant changes at the H3K4 mark may be due to limitations of our study design to assay global histone marks at the same time point as the transcriptional profiling. In embryonic stem cells, LSD1 was shown to mediate short-term changes at H3K4 important for cell cycle progression and these changes may not have been observed in global methylation analyses at 48 hours (58).
Ewing sarcoma is an aggressive cancer for which few agents show single-agent efficacy in vivo. We assessed HMOX1 induction in the A673 study and while most animals in the treatment group had delayed tumorigenesis, the endpoint study design did not allow us to assay HMOX1 levels during earlier time points of treatment. Importantly, we were able to observe induction of HMOX1 in a subset of tumors from the treated group as compared to the vehicle group. Based on our in vitro results we expected to see a more robust HMOX1 induction in the context of antitumor efficacy. While the presence of an EWS/ETS fusion may predict a favorable response to an LSD1 inhibitor, these results underscore the difficulty in identifying of appropriate treatment response biomarkers in Ewing sarcoma.
Based on the dramatic transcriptional effects of HCI2509 described in vitro, we predict improved dosing will result in enhanced tumor regression in future studies with this class of LSD1 inhibitors. This is a critical hurdle for translation of this strategy to the clinic for Ewing sarcoma. Even with a preliminary preclinical formulation we were able to observe significant delay in tumor growth and improved survival out to 60 days. Optimization of formulation, salt forms, and synthesis of more soluble derivatives will allow fine tuning of drug exposure for each individual. Future studies are required to optimize dosing and determine whether or not LSD1 inhibition with HCI2509 is safe, tolerated, and synergistic with other clinically relevant treatment strategies for Ewing sarcoma, like irinotecan and temozolomide. Taken together, the dramatic effects of HCI2509 on the transcriptional activity of EWS/ETS fusions and its observed single-agent efficacy in vivo validate this class of LSD1 inhibitors as a potential targeted strategy to treat Ewing sarcoma.
Supplementary Material
Statement of Translational Relevance.
Ewing sarcoma is an aggressive cancer, with bleak survival rates (10–30%) for patients with metastatic or relapsed disease. Treatment with the LSD1 inhibitor HCI2509 disrupts the global transcriptional function of EWS/ETS fusions, impairs multiple EWS/ETS-associated oncogenic phenotypes, and shows single agent efficacy in multiple xenograft models of Ewing sarcoma. With several targeted LSD1 inhibitors in pre-clinical development, these results highlight a new therapeutic strategy for this disease.
Acknowledgments
Funding: S.L.L. was supported by NIH/NCI grant R01 CA140394. S.L.L and S.Sh. were supported by a Developmental Research Program subaward from the SARC Sarcoma SPORE (U54 CA168512), The Sunshine Project from the Pediatric Cancer Foundation, the Make Some Noise: Cure Kids Cancer Foundation, and the Alan B. Slifka Foundation. S.Sa. was supported by the HHMI Med into Grad program at the University of Utah (U2M2G), E.R.T was supported by the American Foundation for Pharmaceutical Education 2013 Pre-Doctoral Fellowship (AFPE), and T.M. was supported by the University of Utah MD/PhD Program. L.M.H. and M.C.B. were supported by NIH grant R01 GM50877. We also acknowledge support of funds in conjunction with NCI grant P30 CA042014 awarded to the Huntsman Cancer Institute, as well as funds from the Experimental Therapeutics Program at Huntsman Cancer Institute.
Footnotes
Competing interests: S.L.L is on the scientific advisory board for and S.Sh. is a founder and Chief Medical Officer of Salarius Pharmaceuticals, a company focused on epigenetic therapies for cancer.
References
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–24. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 4.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Kubicek S, Gilbert JC, Fomina-Yadlin D, Gitlin AD, Yuan Y, Wagner FF, et al. Chromatin-targeting small molecules cause class-specific transcriptional changes in pancreatic endocrine cells. Proc Natl Acad Sci USA. 2012;109:5364–69. doi: 10.1073/pnas.1201079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10:413–24. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–13. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 10.Popovic R, Zeleznik-Le NJ. MLL: How complex does it get? J Cell Biochem. 2005;95:234–42. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 11.Hormaeche I, Licht JD. Chromatin modulation by oncogenic transcription factors: new complexity, new therapeutic targets. Cancer Cell. 2007;11:475–78. doi: 10.1016/j.ccr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimi A, Kurokawa M. Key roles of histone methyltransferase and demethylase in leukemogenesis. J Cell Biochem. 2011;112:415–24. doi: 10.1002/jcb.22972. [DOI] [PubMed] [Google Scholar]
- 13.Hudlebusch HR, Santoni-Rugiu S, Simon R, Ralfkiær E, Rossing HH, Johansen JV, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–33. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 14.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian X, Zhang S, Liu H-M, Zhang Y-B, Blair CA, Mercola D, et al. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013;13:558–79. doi: 10.2174/1568009611313050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–71. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 17.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–87. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 19.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–7. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 20.Hayami S, Kelly JD, Cho H-S, Yoshimatsu M, Unoki M, Tsunoda T, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–86. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z-K, Yu H-F, Wang DR, Dong P, Chen L, Wu W-G, et al. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol. 2012;18:6651–6. doi: 10.3748/wjg.v18.i45.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schildhaus H-U, Riegel R, Hartmann W, Steiner S, Wardelmann E, Merkelbach-Bruse S, et al. Lysine-specific demethylase 1 is highly expressed in solitary fibrous tumors, synovial sarcomas, rhabdomyosarcomas, desmoplastic small round cell tumors, and malignant peripheral nerve sheath tumors. Hum Pathol. 2011;42:1667–75. doi: 10.1016/j.humpath.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Bennani-Baiti I-M, Machado I, Llombart-Bosch A, Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcoma. Hum Pathol. 2012;43:1300–7. doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–5. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 25.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351–65. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linabery AM, Ross JA. Childhood and adolescent cancer survival in the U.S. by race and ethnicity (diagnostic period 1975–1999) Cancer. 2008;113:2575–96. doi: 10.1002/cncr.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall RL, Lessnick SL, Jones KB, Gouw LG, Cummings JE, Cannon-Albright L, et al. Is there a predisposition gene for Ewing’s sarcoma? J Oncol. 2010 doi: 10.1155/2010/397632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Adamo DR. Appraising the current role of chemotherapy for the treatment of sarcoma. Semin Oncol. 2011;38 (Suppl 3):S19–29. doi: 10.1053/j.seminoncol.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2012;32:5089–100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2. 2 during oncogenic transformation in Ewing’s sarcoma. PLoS ONE. 2008;3:e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, et al. Expression profiling of EWS/FLI identifies NKX2. 2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5:2753–9. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 33.Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, et al. High-throughput virtual screening identifies novel N′-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible inhibitors of LSD1. J Med Chem. 2013;56:9496–508. doi: 10.1021/jm400870h. [DOI] [PubMed] [Google Scholar]
- 34.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol Cancer Res. 2006;4:851–9. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 35.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 36.May WA, Lessnick SL, Braun BS, Kiemsz M, Lewis BC, Lunsford LB, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–8. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–94. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gangwal K, Sankar, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci USA. 2008;105:10149–54. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC. The EWS/FLI oncogene drives changes in cellular morphology, adhesion, and migration in Ewing sarcoma. Genes Cancer. 2012;3:102–16. doi: 10.1177/1947601912457024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankar S, Gomez NC, Bell R, Patel M, Davis IJ, Lessnick SL, et al. EWS and RE1-silencing transcription factor inhibit neuronal phenotype development and oncogenic transformation in Ewing sarcoma. Genes Cancer. 2013;4:213–23. doi: 10.1177/1947601913489569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kauer M, Ban J, Kofler R, Walker B, Davis S, Meltzer P, et al. A molecular function map of Ewing’s sarcoma. PLoS ONE. 2009;4:e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirado OM, Mateo-Lozano S, Villar J, Dettin LE, Llort A, Gallego S, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006;66:9937–47. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 43.Luo W, Gangwal K, Sankar S, Boucher KM, Thomas D, Lessnick SL. GSTM4 is a microsatellite-containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene. 2009;28:4126–32. doi: 10.1038/onc.2009.262. [DOI] [PubMed] [Google Scholar]
- 44.Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman BE, et al. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene. 2001;20:3258–65. doi: 10.1038/sj.onc.1204437. [DOI] [PubMed] [Google Scholar]
- 45.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–83. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cironi L, Riggi N, Provero P, Wolf N, Suvà ML, Suvà D, et al. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS ONE. 2008;3:e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, McGee-Lawrence ME, Decker M, Westendorf JJ. The Ewing’s sarcoma fusion protein, EWS-FLI, binds Runx2 and blocks osteoblast differentiation. J Cell Biochem. 2010;111:933–43. doi: 10.1002/jcb.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang H-G, Jenabi JM, Zhang J, Keshelava N, Shimada H, May WA, et al. E-Cadherin cell-cell adhesion in Ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007;67:3094–105. doi: 10.1158/0008-5472.CAN-06-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci USA. 1993;90:5752–6. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailly RA, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, et al. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–41. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10:R9. doi: 10.1186/gb-2009-10-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprüssel A, Schulte JH, Weber S, Necke M, Händschke K, Thor T, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–51. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 53.Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, et al. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4:e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238–49. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chau CM, Deng Z, Kang H, Lieberman PM. Cell cycle association of the retinoblastoma protein Rb and the histone demethylase LSD1 with the Epstein-Barr virus latency promoter Cp. J Virol. 2008;82:3428–37. doi: 10.1128/JVI.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nair VD, Ge Y, Balasubramaniyan N, Kim J, Okawa Y, Chikina M, et al. Involvement of histone demethylase LSD1 in short-time-scale gene expression changes during cell cycle progression in embryonic stem cells. Mol Cell Biol. 2012;32:4861–76. doi: 10.1128/MCB.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem. 2007;282:15471–5. doi: 10.1074/jbc.M701023200. [DOI] [PubMed] [Google Scholar]
- 60.Cho H-S, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–60. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 61.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–8. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 62.Nair SS, Li D-Q, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013;49:704–18. doi: 10.1016/j.molcel.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial to mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads can be found in the NCBI SRA under numbers SRA096343, SRA096347, SRA096354. Differentially expressed genes from each RNAseq dataset are in Supplementary Table S1.