Abstract
While improved surgical techniques, post-operative care, and immunosuppression regimens have reduced morbidity and mortality associated with orthotopic liver transplantation (OLT), further improvement of outcomes requires personalized treatment and a better understanding of genomic mechanisms involved. Gene expression profiles of ischemia/reperfusion (I/R) injury, regeneration, and rejection, may suggest mechanisms for development of better predictive tools and treatments. The liver is unique in its regenerative potential, recovering lost mass and function after injury from ischemia, resection, and rejection. I/R injury, an inevitable consequence of perfusion cessation, cold storage, and reperfusion, is regulated by the interaction of the immune system, inflammatory cytokines, and reduced microcirculatory blood flow in the liver. Rejection, a common postoperative complication, is mediated by the recipient's immune system through T-cell-dependent responses activating proinflammatory and apoptotic pathways. Characterizing distinctive gene expression signatures for these events can identify therapies to reduce injury, promote regeneration, and improve outcomes. While certain markers of liver injury and regeneration have been observed in animals, many of these are unverified in human studies. Further investigation of these genomic signatures and mechanisms through new technology offers promise, but continues to pose a significant challenge. An overview of the current fund of knowledge in this area is reviewed.
1. Introduction
Orthotopic liver transplantation (OLT) is the accepted treatment for end-stage liver disease (ESLD), and improvements in surgical techniques, immunosuppression, and post-operative care have reduced morbidity and mortality associated with transplantation. Physicians currently rely on laboratory, biopsy and clinical manifestations to assess liver injury and function. Studying gene expression patterns associated with different processes following liver transplantation will provide a genomic basis for evaluating outcomes and identifying patients susceptible to graft dysfunction and may improve long-term survival and outcomes.
I/R injury is an inevitable consequence of organ retrieval, cold ischemic time (CIT), and reperfusion of the graft after implantation. It results from initial injury in the donor and brain death, followed by loss of vascularization of the allograft at procurement, and then reperfusion of the organ in the recipient, leading to further injury. Complications arising from these processes can lead to problems such as early allograft dysfunction and impaired regeneration. I/R injury is regulated by the interaction of immune system cells, the production of inflammatory cytokines, and reduced microcirculatory blood flow in the liver. Activation of intracellular pathways by cytokines, chemokines, and intracellular ionic disturbances contributes to hepatocyte apoptosis and necrosis in I/R injury [1,2]. Cold ischemic injury is the insult to tissue during cold storage and prior to placement in the recipient; warm ischemia occurs during the time the organ is being sewn in, after it is removed from ice and prior to reperfusion. This is followed by early-phase I/R injury immediately after reperfusion of the organ with blood, and is associated with rapid changes in the redox state of liver tissue. Late phase I/R injury is a consequence of inflammation-mediated damage, caused by cytokine and chemokine production, which initiates an immune response [3,4].
Liver regeneration is another process that is critical for recovery after transplantation, as an injured whole liver, or a partial liver graft, seeks to restore lost mass. Liver regeneration mechanisms have been well studied using the partial hepatectomy (PH) model in rodents, where two-thirds PH is carried out by removing two lobes. After two-thirds PH, liver mass is restored within 8–15 days in humans through hypertrophy and hepatocyte proliferation [5,6]. Liver regeneration is a complicated orchestrated event involving a complex network of connected interactions, and is required for hepatocyte recovery following the inevitable I/R injury, as well as to restore lost hepatic mass rapidly while maintaining metabolic functions [7].
While these processes been studied extensively in animal models, it has been difficult to do so in humans. Only a few studies have evaluated human genomic liver expression following liver resection and transplantation [8–10]. Liver transplantation with partial grafts and living donors provide excellent clinical models for the study of human liver regeneration. We will review molecular mechanisms of I/R injury and liver regeneration, explore new findings discovered in animal models, and then describe human hepatic gene expression in recipients of deceased donor and partial liver grafts, and in healthy living liver donors.
2. Molecular mechanisms associated with I/R injury
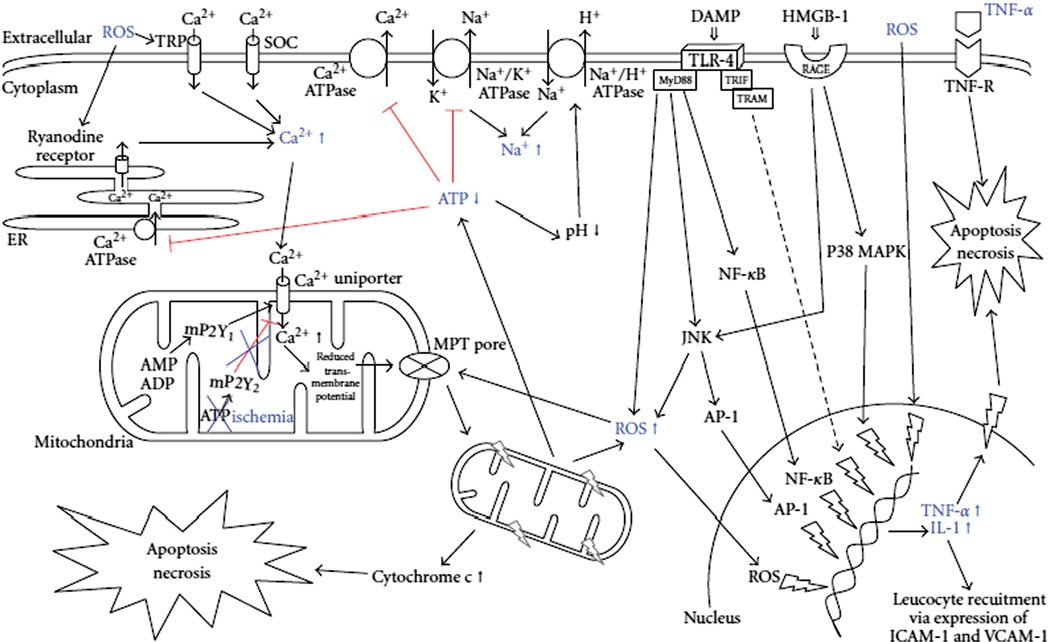
The cascade of inflammatory events following reperfusion is initiated by release of oxidant stress signals from Kupffer cells (KCs), hepatocytes and sinusoidal endothelial cells (SECs), altering the redox state of liver tissue. Damage due to lack of vascularization during cold and warm ischemia depletes liver tissue of ATP, leading to generation of reactive oxygen species (ROS) and disturbances in ionic concentrations of sodium, calcium, and protons, causing hepatocyte swelling and cell death after reperfusion [11,12]. Fig. 1 outlines ionic and mitochondrial disturbances that are involved in I/R injury [4].
Fig. 1.
Intracellular signaling pathways and ionic disturbances engaged during IR injury, resulting in cellular swelling, apoptosis, and necrosis. ADP: adenosine diphosphate; AMP: adenosine monophosphate; AP-1: activator protein-1; ATP: adenosine triphosphate; DAMP: danger-associated molecular pattern; HMGB-1: high mobility group box-1; ICAM-1: intercellular adhesion molecule-1; IL-1: interleukin-1; JNK: c-Jun N-terminal kinase; MAPK: mitogen-activated protein kinase; MPT pore: mitochondrial permeability transition pore; MyD88: myeloid differentiation factor 88; NF-κB: nuclear factor kappa B; RAGE: receptor for advanced gylcation end product; ROS: reactive oxygen species; SOC: store operated calcium channel; TLR: toll-like receptor; TNF: tumor necrosis factor; TRAM: TRIF-related adaptor molecule; TRIF: TIR domain-containing adaptor-inducing interferon; TRP: transient receptor protein; VCAM-1: vascular adhesion molecule-1. Copyright © 2012 Kilian Weigand et al [4]; Open access article distributed under the Creative Commons Attribution License.
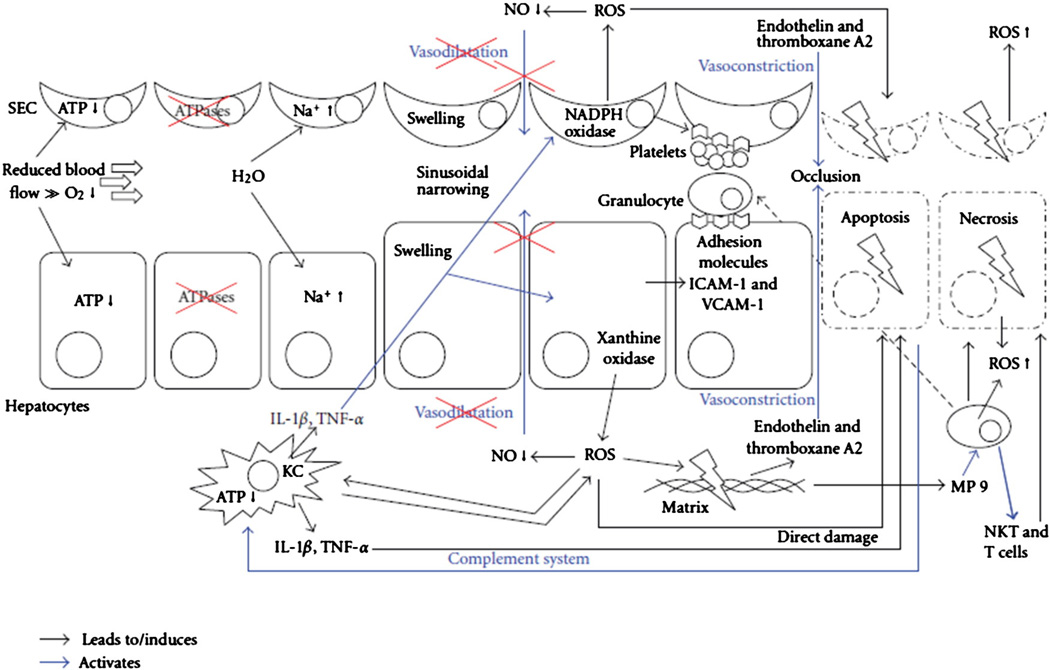
After the immediate I/R injury, an increase in intracellular and extracellular ROS activates the complement system and KCs to release TNF-α, IL-1, IL-6, and platelet-activating factor (PAF). These chemokines further activate ROS generation in KC mitochondria and up-regulate proinflammatory cytokines (IL-1B and TNF-α) and chemokines (IL-8, MCP-1, and regulated on activation, normal T-cell expressed and secreted [RANTES]). This leads to activation and migration of neutrophils and CD4+ lymphocytes from peripheral blood into the liver, generating an inflammatory response. Proinflammatory cytokines up-regulate intercellular adhesionmolecule-1 (ICAM-1), vascular adhesion molecule-1 (VCAM-1), and E-selectin on the surfaces of SECs and hepatocytes, allowing for greater adhesion of leukocytes and platelets to the liver, reducing microcirculatory blood flow and increasing hepatocyte apoptosis [3,4,13,14]. Fig. 2 illustrates cellular interactions which lead to apoptosis, and necrosis in I/R injury [4].
Fig. 2.
Cellular interaction involved in IR injury, resulting in cellular swelling, apoptosis, and necrosis. ATP: adenosine triphosphate; ICAM-1: intercellular adhesion molecule-1; KC: Kupffer cell; IL-1, interleukin-1; NKT: natural killer T cell; NO: nitric oxide; ROS: reactive oxygen species; SEC: sinusoidal endothelial cells; T cell: CD4+ T lymphocyte; TNF: tumor necrosis factor; VCAM-1: vascular adhesion molecule-1. Copyright © 2012 Kilian Weigand et al [4]; Open access article distributed under the Creative Commons Attribution License.
TNF-α, an important component of the I/R injury cascade, binds to specific TNF receptors on the hepatocyte surface, causing up-regulation of cytokine production and activation of the Fas receptor, which initiates apoptosis. Fas receptor activation leads to activation of nuclear factor kappa B (NF-κB), mitogen-activated protein kinase (MAPK), and c-Jun N-terminal kinase (c-JNK). These further activate transcription factors, activator protein-1 (AP-1), heat shock factor, signal transducer and activators of transcription (STATs), antioxidants, inflammation-stimulated inducible enzymes (COX-2), intracellular signaling molecules, and antiapoptotic proteins (Bcl-2, Bcl-x), which travel to the nucleus to up-regulate transcription of IL-1 and TNF-α [4,15]. NF-κB activates other proinflammatory factors (IL-1β and IFN-γ) and chemokines (IL-8,MCP-1, and RANTES), causing a positive feedback loop [16].
Endogenously generated carbon monoxide through heme degradation by heme oxygenases (HO) is considered important for the maintenance of cellular processes via the soluble guanylate cyclase (sGC)/cGMP pathway [17]. Activation of HO-1 by I/R injury provides protection against warm and cold I/R injury through the generation of carbon monoxide (CO) [18]. In a rat OLT model, CO inhalation caused a marked down-regulation of early-mRNA expression of TNF-α and IL-6. Cold I/R injury is associated with MAPK phosphorylation; CO inhibited the phosphorylation of ERK1/2,MAPK, upstream MEK1/2, and the downstream transcriptional factor c-Myc [19]. Such studies are difficult to conduct in humans due to the toxicity of CO.
IRF-1 plays a critical role in inducing inflammatory mediators including TNF-α, IL-6, and inducible nitric oxide synthase (iNOS) [20] and may be an important activator of MAPK-mediated damage in cold I/R injury. In a study by Kim et al. [21], I/R injury following OLT was studied in rats using adenovirus-expressing IRF-1 (AdIRF-1) or the control gene vector (Adnull) given pre-harvest. Overexpression of IRF-1 in donor livers pre-transplantation led to more severe I/R injury and post-transplant overexpression of IRF-1 led to increased apotosis and necrosis in rats. IRF-1 overexpression in livers before transplantation is predictive of significant JNK activation 1 hour after transplantation, correlating with a known c-JNK signaling pathway in the I/R injury inflammatory response [21] Analysis of gene expression in the donor liver prior to removal may allow for investigation of possible predictive measures of allograft function in the recipient.
3. Molecular mechanisms of liver regeneration
Liver regeneration has been studied extensively in animal models, and much has been learned in the past 10 years. Following PH, liver regeneration begins with hepatocyte volume increase, as approximately 95% of quiescent hepatic cells re-enter the cell cycle [22]. In a two-thirds PH model, remaining cells undergo rapid proliferation as a round of DNA synthesis is initiated within 24–48 hours [23,24]. Cell-cycle genes linked to cell repair and DNA synthesis, such as c-fos, c-jun, c-myc, and JunB, are highly expressed during regeneration Cytokines (TNF-α and IL-6) and growth factors (hepatocyte growth factor (HGF), epidermal growth factor (EGF), and transforming growth factors (TGFs)) are released by Kupffer cells [22]. After 70% PH, lost hepatic mass is restored through proliferation of mature hepatocytes, which leave the G0 phase and transition into G1 and S phase. Over 100 early genes are activated between G0 and G1, such as TNF-α and IL-6 in KCs. These, and other early genes, activated by apoptosis and inflammation, “prime” hepatocytes to enter the cell cycle (Fig. 3) [22,25]. There is both hepatocyte hypertrophy and cell proliferation, with almost all hepatocytes entering the cell-cycle S phase and half undergoing division to increase cell number [26].
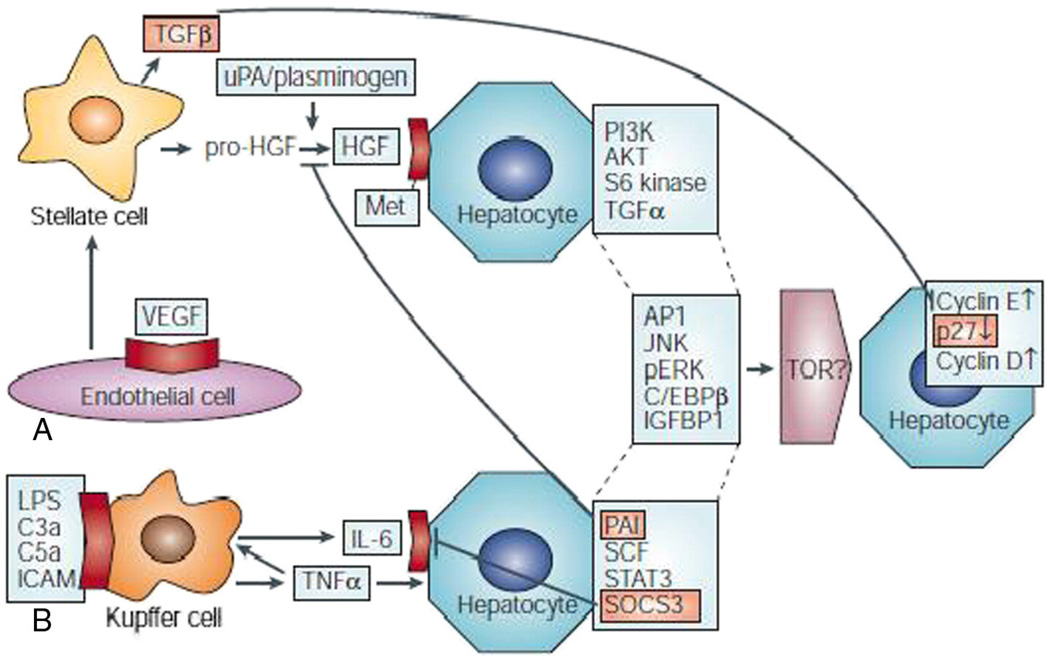
Fig. 3.
Growth factor- and cytokine-regulated pathways are activated during liver regeneration. A. The growth factor-mediated pathway. Vascular endothelial growth factor (VEGF) binds to endothelial cells, which triggers the release of the hepatocyte growth factor (HGF) precursor, pro-HGF, from stellate cells. The urokinase-type plasminogen activator (uPA) and plasminogen proteases cleave pro-HGF, which releases HGF. HGF binds to the Met receptor on hepatocytes to activate the phosphatidylinositol 3-kinase (PI3K), AKT and S6 kinase signal-transduction pathways. HGF signaling releases transforming growth factor (TGF) α and other downstream signals that are shared with the cytokine-mediated pathway, such as AP1, Jun amino-terminal kinase (JNK), phosphorylated extracellular signal-regulated kinases (pERKs), CCAAT-enhancer-binding protein (C/EBP) β and insulin-like-growth factor-binding protein (IGFBP) 1. These factors are proposed to activate target of rapamycin (TOR), although this remains to be established, and this leads to cell-cycle transition by increasing the expression of cyclins D and E and reducing p27 levels. B, The cytokine-mediated pathway. Molecules that are crucial for innate immunity, including lipopolysaccharide (LPS), complement factors C3a and C5a and intercellular adhesion molecules (ICAMs), activate Kupffer cells, which produce tumor necrosis factor (TNF)α. This, in turn, up-regulates the expression of interleukin (IL)-6 by Kupffer cells. TNFα and IL-6 activate neighboring hepatocytes, which causes signal transducer and activator of transcription (STAT) 3 activation and the expression of stem-cell factor (SCF) and several proteins that are shared with the growth factor-mediated pathway. Various inhibitory proteins that are important for terminating liver regeneration are also activated (shown in orange), including TGFβ (which is produced by stellate cells), plasminogen activator inhibitor (PAI), suppressor of cytokine signaling-3 (SOCS3) and p27 and other cyclin-dependent kinase inhibitors, and their effects on the two pathways are shown. Cell-surface receptors are shown in red. Reprinted by permission from Macmillan Publishers Limited: Nature Reviews Molecular Cell Biology [22], copyright (2004).
Genes involved in protein synthesis and cell growth are up-regulated throughout the proliferative phase of regeneration [22,27]. Cytokines activate non-mitogenesis-linked transcription factors selectively and are distinct in their action from growth factors. IL-6 knockout mice display liver necrosis and failure, reduced DNA-synthesis response, and G1 abnormalities in hepatocytes. However, recovery of mass is possible, albeit delayed, without IL-6, indicating that other pathways are also involved [22,28]. TNF-α is also crucial for normal proliferation post-PH as it induces IL-6 transcription through up-regulation of NF-κB. When overexpressed, IL-6 functions as a hepatocyte mitogen [29]. C3a andC5a (members of the complement cascade of innate immunity) are also important in regeneration. C3- and C5-deficient animals underexpress IL-6, TNF-α, STAT3, andNF-κB (vital components of liver regeneration). The IGFBP1 gene that encodes a pro-mitogenic and hepatoprotective protein is one of the most highly expressed genes in regeneration. IGFBP1-knockoutmice show impaired liver regeneration similar to IL-6-knockout mice. Since it acts through IGF pathways and is transcriptionally regulated by IL-6, it represents another gene at the intersection of growth factor and cytokine pathways [22,30,31]. Successful liver regeneration depends on an intact TNF-α/NF-κB/IL-6/STAT3 pathway, through interactions between KCs and hepatocytes [22,32]. A20 is an NF-κB-dependent and NF-κB-inhibitory ubiquitin-editing protein and affects genes linked to inflammatory and immune responses, cellular proliferation, energy production, oxidoreductase activity, and lipid and fatty acid metabolism. Overexpression of A20 in the liver provides hepatoprotection in mice against toxic hepatitis, severe I/R injury, and fulminant hepatic failure following extended liver resection [33].
In transplant, the loss of hepatic mass due to injury, alloimmune responses, and volume deficiencies in segmental grafts, necessitates a balance between required metabolic function and replication to ensure survival [7]. Failure to maintain this balance may result in graft dysfunction as seen in small-for-size syndrome [34]. In the early post-PH period, the liver must prioritize protein synthesis to recover lost mass and bear heavy metabolic demands of the body. Plasma proteins and intermediate metabolism are up-regulated later once the liver has initiated restoration of necessary hepatic mass to meet the metabolic load [22,27,35]. As expected, gene expression associated with protein biosynthesis and cytoskeletal assembly remains high through the first round of proliferation [36,37]. Many liver-specific immediate early genes are responsible for encoding enzymes and proteins of gluconeogenesis. Gluconeogenesis allows the liver to meet homeostatic glucose demands of the body even with reduced mass, while deprioritizing fatty acid metabolism. Two types of transcription factors, growth-induced (STAT-3 and AP-1) or tissue-specific (hepatic nuclear factor-1 (HNF-1)), consort to amplify gene expression that maintains homeostasis during repair, overcoming reduced liver mass [22]. In contrast, lipid, fatty acid, and hormone metabolism are down-regulated early after PH and remain in this state through the first round of cell division.
4. Mechanism overlap between I/R injury and regeneration
The pathways associated with the regenerative cascade show significant overlap with immune and inflammatory response pathways in I/R injury [38]. Growth factors (IL-6, TNF-α, EGF, and HGF), transcription factors (STAT-3, AP-1, and NF-κB), and immediate early genes (c-fos, c-myc, and cyclins) are markers of I/R injury as well as cellular repair and regeneration. More extensive I/R injury results in enhanced up-regulation of cytokines, transcription factors, and early genes, leading to a more significant hepatocellular replication response [38,39]. While these proinflammatory factors are necessary for triggering regeneration, the liver is only capable of withstanding I/R injury to the point where enough critical mass is available to maintain the balance of homeostatic and regenerative activities [7]. Animal models of the interplay between the regenerative response and I/R injury demonstrated that grafts with a weight-to-liver-volume ratio below 40% showed functional impairment and poor survival [40].
To maintain graft integrity, the host response must be inhibited through immunosuppression. However, this may also impact natural regenerative pathways. Glucocorticoids inhibit cell-cycle progression in PH models as well as in I/R injury models, and sirolimus interferes with replication through its inhibition of the mTOR pathway [41–46]. Thus, immunosuppression should be carefully chosen and monitored in the early post-transplant period to ensure that the regenerative response is not inhibited along with the immune response.
5. Gene expression patterns of I/R injury and regeneration in human liver transplantation
One of the first studies describing gene expression following human liver transplantation was by Berberat et al. [47]. In this study, the authors investigated the expression of 67 genes from 59 post-perfusion biopsies (52 primary transplants; 7 secondary; median MELD = 15.2) using real-time polymerase chain reaction (PCR). They proposed that early gene expression can predict early allograft dysfunction (EAD). The genes selected were believed to play a role in the acute inflammatory process and the authors tried to correlate expression with graft dysfunction. Assessment of the correlation between a limited set of inflammatory genes in the reperfusion biopsy and graft-related clinical outcomes within 30 days of transplantation yielded six genes associated with graft-related complications in the first month. These included increased expression of C-reactive protein (CRP) (a marker of ongoing proinflammatory response), and reduced expression of CTGF, WWP2, CD274, VEGF and its receptor FLT1 (genes related to vascular endothelial physiology). High TNF-α expression was strongly correlated with complications requiring early retransplantation [47]. This suggests that certain gene expression patterns detected as early as 1 hour post-reperfusion could predict liver EAD in the early post-operative period.
Another early study by Conti et al. [48] examined gene expression using Affymetrix HG-U133A Plus 2.0 oligonucleotide arrays (Affymetrix, Santa Clara, CA) in 10 biopsies from 5 cadaveric donor livers used in human OLT. They identified 795 genes with significantly modified expression between reperfused livers (RL) biopsied within 3 hours after reperfusion and basal liver (BL) expression prior to retrieval from the donor. Twelve percent of these were involved in apoptotic pathways, 12.5% in inflammatory pathways, and 22.5% encoded for heat shock proteins [48]. Considering the role of inflammation in I/R injury, the cytokine–cytokine receptor interaction pathway showed significant up-regulation in RL samples, with the most important factors involved being IL-6, IL-1, IL-8, and chemokine ligands 3 and 4 [48]. The pathway associated with these inflammatory response genes regulates WNT signaling, apoptosis, and MAPK signaling, which act together to regulate the cell-cycle pathway, suggesting a role in liver regeneration following OLT [48]. Furthermore, an analysis of enriched Gene Ontology (GO) categories for biological processes demonstrated significant up-regulation in signal transduction, intracellular signaling cascade, JAK-STAT and MAPK activity, as well as apoptosis, cell death and cell growth pathways. A more recent study by Conti et al. [49] compared the expression of 33,000 genes across three groups (deceased donor livers, transplanted livers 2 hours post-reperfusion, and control livers) to differentiate between changes caused by I/R injury and variations due to brain death and other donor factors. They demonstrated that approximately 900 genes show dysregulation in deceased donors versus control livers. The up-regulated genes are involved in apoptosis, immune response, and inflammation; the down-regulated genes are involved in metabolism and electron transport. An additional 317 genes, previously undetectable due to similar dysregulation in the donor and in the transplanted liver, were identified.
Evidence of EAD may also be reflected by alterations in expression of serum proteins associated with an inflammatory response in the peri-operative period. In a study by Friedman et al. [50], serum levels of 25 cytokines, chemokines, and immunoreceptors were measured by Luminex multiplex assays pre- and post-liver transplantation. Levels of each cytokine biomarker were compared in adult recipients with or without EAD at serial time points using samples collected pre-operatively and at specific intervals post-transplant. EAD was defined according to standard criteria with elevated transaminases, bilirubin, and/or INR [51]. Multivariable analyses showed that patients experiencing EAD had lower pre-operative IL-6 and higher IL-2R levels. Patients with EAD also showed higher MCP-1 (CCL2), IL-8 (CXCL8), and RANTES (CCL5) chemokine levels in the early post-operative period, suggesting up-regulation of the NF-kB pathway, in addition to higher levels of chemokines and cytokines associated with T-cell immunity, including MIG (CXCL9), IP-10 (CXCL10) and IL-2R. These findings identified several possible biomarkers and pathways associated with EAD, that can guide future validation studies and investigation of specific cellular and molecular mechanisms of graft dysfunction, and contribute to peri-operative prediction of the occurrence of EAD leading to identification of potential interventional therapies.
In another study examining early gene expression in living donor liver transplantation (LDLT), Borozan et al. [8] utilized a 19K-human cDNA microarray in 24 consecutive LDLT. They showed increased expression of 129 genes and decreased expression of 106 genes post-reperfusion compared to expression prior to organ removal. Differential gene expression patterns were observed in acute versus chronic stress in biopsies of grafts compared to unstressed donor livers prior to operation. A 25-gene subset was identified as a molecular signature for two forms of acute liver stress (brain death and reperfusion following LDLT), which were not seen in chronic liver stress such as hepatitis or PBC. Although the two studies differed in length of ischemia time as well as the use of living (Borozan et al.) versus deceased donors (Conti et al.), these studies showed strong agreement for the up-regulated genes in the signature, and a similar, but less statistically significant agreement with the dowregulated genes. The poorer agreement with down-regulated genes may be due to impairment of these pathways in cadaveric livers prior to retrieval [8,48].
A study from our group described early gene expression in livers from deceased donor (DD), as well as those from LDs. In a comparison of 8 LDs to 13 DDs examining gene expression profiles using Affymetrix microarrays and quantitave PCR and immunopathology, we demonstrated 579 genes differentially expressed after reperfusion in DD and 1324 genes differentially expressed after reperfusion in LDs. There was significant up-regulation of inflammatory, immune, and cell-cycle genes associated with I/R injury in DD grafts, including inflammatory chemokine IL-8, CCL2 (MCP-1), and TNFAIP3/A20 and ICAM-1, correlating well with the work from Conti et al. (Table 1) [10,48].
Table 1.
Comparison of immune and inflammatory genes differentially expressed in LD and DD grafts in POST biopsies compared to PRE.
| Genes differentially expressed in LD POST vs. LD PRE | Genes differentially expressed in DD POST vs. DD PRE | ||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Description | Fold change LD post vs LD pre |
GO category | Gene symbol | Description | Fold change DD post vs DD pre |
GO category |
| Common to both LD and DD | Common to both LD and DD | ||||||
| BCL3 | B-cell CLL/lymphoma 3 | 1.89 | Immune response | BCL3 | B-cell CLL/lymphoma 3 | 2.75 | Immune response |
| BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 1.42 | Inflammatory response | BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 1.73 | Inflammatory response |
| C5R1 | Complement component 5 receptor 1 (C5a ligand) | 1.89 | Immune response | C5R1 | Complement component 5 receptor 1 (C5a ligand) | 2.48 | Immune response |
| CALCA | Calcitonin/calcitonin-related polypeptide, alpha | 5.99 | Inflammatory response | CALCA | Calcitonin/calcitonin-related polypeptide, alpha | 4.78 | Inflammatory response |
| CCL2 | Chemokine (C-C motif) ligand 2 (MCP-1) | 11.76 | Inflammatory response | CCL2 | Chemokine (C-C motif) ligand 2 (MCP-1) | 4.37 | Inflammatory response |
| CCL4 | Chemokine (C-C motif) ligand 4 (MIP-1-beta) | 6.76 | Inflammatory response | CCL4 | Chemokine (C-C motif) ligand 4 (MIP-1-beta) | 3.22 | Inflammatory response |
| CCL18 | Chemokine (C-C motif) ligand 18 | 1.47 | Inflammatory response | CCL18 | Chemokine (C-C motif) ligand 18 (pulmonary and | 1.46 | Inflammatory response |
| CCL20 | Chemokine (C-C motif) ligand 20 (MIP-3 alpha) | 5.81 | Inflammatory response | CCL20 | Chemokine (C-C motif) ligand 20 (MIP-3 alpha) | 5.49 | Inflammatory response |
| IL1RL1 | Interleukin 1 receptor-like 1 | 8.77 | Immune response | IL1RL1 | Interleukin 1 receptor-like 1 | 2.49 | Immune response |
| IL1RN | Interleukin 1 | 7.25 | Inflammatory response | IL1RN | Interleukin 1 receptor antagonist | 4.88 | Inflammatory response |
| NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer | 1.18 | Immune response | NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer | 1.40 | Immune response |
| RIPK2 | Receptor-interacting serine-threonine kinase 2 | 1.67 | Inflammatory response | RIPK2 | Receptor-interacting serine-threonine kinase 2 | 1.62 | Inflammatory response |
| THBS1 | Thrombospondin 1 | 5.05 | Inflammatory response | THBS1 | Thrombospondin 1 | 4.67 | Inflammatory response |
| ELF3 | E74-like factor 3 | 2.79 | Inflammatory response | ELF3 | E74-like factor 3 | 1.94 | Inflammatory response |
| FCN1 | Ficolin (collagen/fibrinogen domain containing) 1 | 1.82 | Immune response | FCN1 | Ficolin (collagen/fibrinogen domain containing) 1 | 2.14 | Immune response |
| FCAR | Fc fragment of IgA, receptor for | 1.42 | Immune response | FCAR | Fc fragment of IgA, receptor for | 1.14 | Immune response |
| ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 |
1.29 | Immune response | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 |
1.25 | Immune response |
| Exclusive to LD | Exclusive to LD | ||||||
| CRP | C-reactive protein, pentraxin-related | 8.06 | Inflammatory response | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 11.36 | Inflammatory response |
| SAA1 /// SAA2 | Serum amyloid A1//A2 | 7.30 | Inflammatory response | IL8 | Interleukin | 6.06 | Inflammatory response |
| LBP | Lipopolysaccharide binding protein | 3.16 | Inflammatory response | SNF1LK | SNF1-like kinase | 5.59 | Immune response |
| RAB27A | RAB27A, member RAS oncogene family |
3.07 | Immune response | CCL3 | Chemokine (C–C motif) ligand 3 (MIP-1-alpha) | 5.08 | Inflammatory response |
| STAT3 | Signal transducer and activator of transcription 3 | 2.84 | Inflammatory response | NFIL3 | Nuclear factor, interleukin 3 regulated | 3.75 | Immune response |
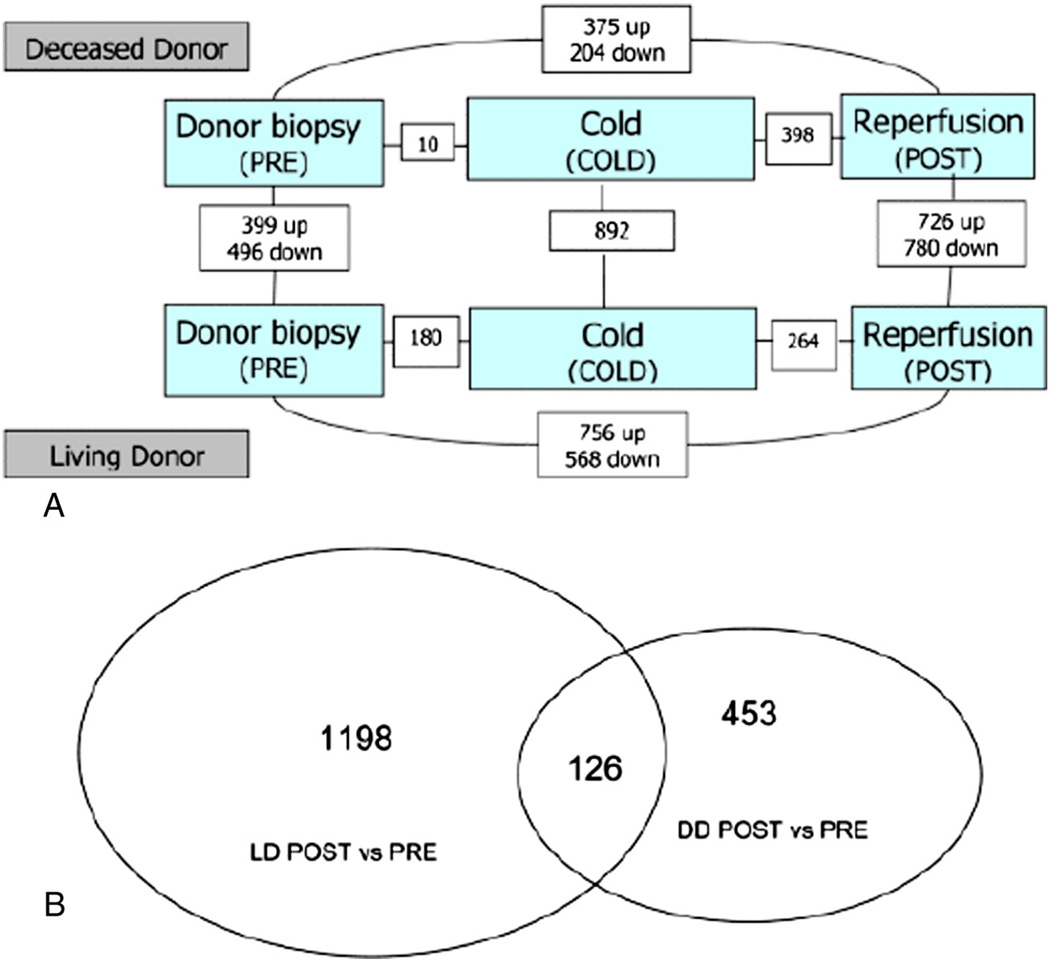
This study also showed that the molecular signatures of LD grafts differ significantly from those of DD grafts (Fig. 4). We demonstrated that molecular networks associated with regeneration are activated immediately in partial LD grafts and are associated with proinflammatory and cell-cycle pathways. The genes of the IL-6/STAT3 pathway, including SOCS3 (a feedback mediator the regulates STAT3 after partial hepatectomy), hepatocyte growth factor (HGF), and NF-kB1, all of which are involved in hepatoprotection and regeneration are up-regulated [23,52–54]. Furthermore, networks involving cell-cycle progression and de novo biosynthesis of polyamines and pyrimidines are also up-regulated [10]. The type of graft (LD vs. DD) also impacts gene expression associated with regeneration. By using EASE analysis to assign significance to biological functions as opposed to individual genes, we demonstrated that the 12 functions up-regulated in living donor grafts were mainly associated with cell proliferation and tissue regeneration, while the 17 functions down-regulated in living donor grafts were associated with metabolic liver functions. This indicates that the gene expression profile orchestrates a redistribution of energy to favor regeneration in partial LD grafts [10]. There were only 17 overlapping genes between LD and DD grafts. LD grafts showed differential up-regulation of interleukin-associated receptor genes (IL1R and 1L4R) and genes associated with innate immunity (e.g. lipopolysaccharide binding protein [LBP]). It is important to assess differences in mechanisms when using grafts from LD as opposed to DD in order to guide post-operative management and optimize patient outcomes.
Fig. 4.
A. Diagram of number of differentially expressed genes in each class comparison. The numbers of differentially expressed genes between groups are illustrated in the small boxes connecting the larger shaded boxes (at P-value of 0.005). B. Venn diagram of overlap of differentially expressed genes in LD and DD POST reperfusion, compared to PRE transplantation for each graft type. Copyright © 2009 The Authors Journal compilation © 2009 The American Society of Transplantation and the American Society of Transplant Surgeons [10]; Open access article distributed under the Creative Commons Attribution License.
Some of the differences between LD and DD may be due to brain death. A study by Weiss et al. [55] attempted to evaluate cytokine gene expression profiles in brain-dead (n = 32) and living (n = 26) donors, comparing the data to post-OLT organ function. Biopsies were performed at the time of donor laparotomy, before preservation, at the time of transplantation, and 1 hour post-reperfusion. Cytokine expression was assessed using real-time reverse transcriptase-PCR (RT-PCR). The inflammatory cytokines IL-6, IL-10, TNF-α, TGF-β, and MIP-1α were significantly up-regulated in brain-dead donors compared to living donors immediately post-laparotomy. Thus, the significant differential up-regulation of inflammatory cytokines by brain death may lead to worse I/R injury post-OLT [55]. Furthermore, pre-transplant transcriptome analysis indicated C3 gene expression differences between LD and DD that were directly correlated to cold ischemia duration [56], further suggesting that the length of cold ischemia time plays a critical role in I/R injury.
Regeneration is a critical component of recovery after donation in LD liver transplantation, where often 60% or more of the liver is removed to transplant in another individual [57]. While most donors do well, there remains significant morbidity associated with the procedure. Many show incomplete regeneration in the first 3–6 months, with a significant incidence of complications and a small but present risk of death or liver failure [58]. A better understanding of factors influencing regeneration may provide possible targets for intervention, minimizing subsequent morbidity and mortality. We performed a pilot study investigating differences in hepatic gene expression between donors with complete regeneration compared to those with less successful regeneration in 24 right lobe donors. Using Affymetrix Human Gene 1.0 ST array chips, liver biopsies from these donors were analyzed for gene expression at baseline and in remnant left lobes immediately after resection. Data were analyzed using BRB-ArrayTools and pathway analysis was done using Ingenuity Pathway Analysis (IPA). Living donors with successful liver regeneration show differential expression of a high number of genes immediately post-resection compared to baseline, markedly different from those with deficient regeneration. Class comparison of baseline liver before and after resection in the good regeneration group yielded many more differentially expressed genes, than those that did not reconstitute their mass as well. Many genes were mainly related to cell proliferation, inflammation and metabolism, and metabolic pathways (aminoacyl tRNA synthesis), and stress pathways (acute phase response), were among the most significantly regulated pathways. Among biological functions, genes involved in cell growth and proliferation and cell death were most differentially expressed. In contrast, the poor regeneration group demonstrated very little change in expression before and after resection. The lack of significant change in genomic profile in the poorly regenerating livers suggests a possible inhibition or delay in initiation of recovery and regeneration molecular pathways, and may identify potential areas for intervention.
6. New directions: microRNA markers of liver injury, regeneration, and rejection
MicroRNAs are a specific class of single-stranded noncoding RNAs that act to negatively modulate genes implicated in cellular function and metabolism [59]. They act through translational repression of target mRNAs. miRNAs have been detected in biological fluids which makes them a valuable non-invasive diagnostic tool for distinguishing between diseased individuals and healthy controls [60]. Altered miRNA expression has been observed in malignant, infectious, autoimmune, metabolic and cardiovascular diseases [61]. The link between miRNAs and immunity and inflammatory diseases is well established [62,63].
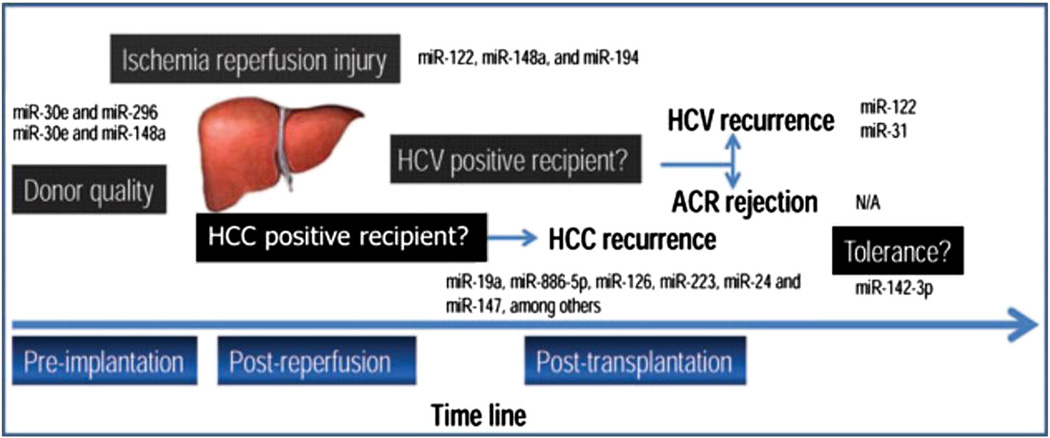
The identification of physiologic miRNA and their mRNA targets in liver regeneration was demonstrated in mice by Schug et al. [64] using a high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) assay. They hypothesized that miRNA activity would be more accurately described by quantifying the abundance of miRNA in the RNA-induced silencing complex (RISC) as opposed to using miRNA levels on their own. Nine miRNAs showed a significant increase and seven showed a significant decrease in RISC recruitment. Cell-cycle progression and checkpoint control genes showed maximal enrichment in the RISC at 36 and 48 hours; genes involved in amino acid metabolism, lipid metabolism and cell growth decreased in the RISC post-PH [64]. Furthermore, FGF1 and VEGFA are miRNA regulated [64,65]. Using a 70% partial hepatectomy mouse model, Zhou et al. [65] identified miRNA-26 as an important regulator of hepatocyte proliferation in liver regeneration. Fig. 5 shows the use of miRNAs as potential biomarkers for graft function and condition.
Fig. 5.
Potential applications of miRNAs as biomarkers in different conditions involved in graft function in liver transplantation. The liver transplantation model is used in the present figure to show the most important applications of miRNAs as biomarkers. The miRNAs included in the figure are the result of several publications and have been found to behave as disease-associated markers. This figure depicts the many opportunities for evaluating these markers (e.g. conditions likeAR [with or without HCV] have not yet been evaluated). Moreover, most of these results lack mechanistic studies exploring the role of the miRNAs in the disease. Copyright © 2012 Kilian Weigand et al [4]; Open access article distributed under the Creative Commons Attribution License.
Long noncoding RNA (lncRNA) have been postulated to play a role in regeneration and Xu et al. [66] conducted a genome-wide lncRNA analysis during liver regeneration after two-thirds partial hepatectomy in mice. A lncRNA associated with liver regeneration (lncRNA-LALR1) was shown to enhance hepatocyte proliferation through activation of Wnt-b-catenin signaling. Furthermore, a human analog of this lncRNA (hLALR1) was identified on chromose 16 by RACE analysis.
In a unique model of human auxiliary liver transplant (ALT) where part of the native damaged organ was left in place, adjacent to the transplanted liver, specific miRNAs were shown to regulate the cell cycle, angiogenesis, and innate immunity in regeneration [9]. Liver biopsies from 11 patients undergoing ALT for acute liver failure were obtained during reperfusion at transplantation and during subsequent biopsies and miRNAs were assessed using microarray technologies. Initial miRNA expression distinguished between successful and unsuccessful regeneration as early as the first biopsy immediately after reperfusion. For successful regeneration, the network of down-regulated miRNAs consists of miRNA-200b, miRNA-183, ZEB1, and SP1, which promote apoptosis and inhibit cell proliferation. Furthermore, the network of up-regulated miRNA for successful regeneration includes miRNA-27a, miRNA-494, miRNA-1224, and miRNA-149, with up-regulation leading to inhibition of apoptosis, as well as increased cell proliferation and angiogenesis. Inhibition of miRNA-150, miRNA-503, miRNA-663 promotes proliferation in vitro [9].
Farid et al. [61] investigated serum hepatocyte derived miRNAs as markers of hepatic injury in liver transplantation. Serum samples from healthy controls and liver transplant recipients (n = 107) and peritransplant liver allograft biopsy samples (n = 45) were analyzed via the real-time PCR quantification of miR-122, miR-148a, and miR-194. The expression ofmiR-122 andmiR-148a in liver tissue was significantly reduced with prolonged ischemic times; however serum levels of these were elevated in patients with liver injury, correlating with amino-transferase levels. The authors suggest that graft injuries associated with longer warm ischemic times reduce the level of hepatocyte miRNAs, possibly due to release of miRNA from injured cells. In addition, miRNA levels in liver recipients in the early post-transplant period correlated with transaminase levels, with serum samples with high AST and ALT levels having miR-122 elevated over 100-fold andmiR-148a andmiR-194 elevated 30- and 40- fold compared to healthy controls (P < 0.005). This study demonstrated that liver injury can be reflected by the release of miRNAs into circulation, which may be early, stable, and sensitive biomarkers of injury following transplantation.
The role of miRNA as regulators of immune response and immune function in transplant is currently a focus for multiple investigations. Typically, studies have explored miRNA expression within the allograft and in the periphery to decipher pathways regulating immune response and rejection, and have addressed the use of miRNA as biomarkers informing allograft status. Most investigations performed in humans have been conducted in the kidney transplant setting where it has been shown that subsets of mature miRNAs can differentiate acutely rejecting kidney allografts from normal allografts [67–70]. Data relating to miRNA expression in the human liver rejection are very limited. In the study by Farid et al. [61], they tested a limited number of miRNAs in sera of recipients during biopsy-proven rejection, demonstrating significantly elevated serum miR-122 with similar kinetics to those observed for liver injury enzymes such as AST and ALT. Research in the rat model demonstrated similar findings where plasma miR-146 was associated with acute rejection, whereas miR-122 is associated with liver injury [71,72]. This suggests that miRNA measured in serum of transplant recipients may serve as ideal and novel biomarkers for identification of injury and management of the inflammatory and immune response in liver transplant patients.
7. Identifying future potential treatment strategies
We have discussed human genomic studies that demonstrate how I/R injury is mediated through proinflammatory and apoptotic pathways. Experimental strategies to reduce I/R injury in animal models have attempted to blockade cytokine and chemokine pathways, adhesion molecules, NF-κB, specific MAP kinases, and metalloproteinases [16]. Studies have also discussed induction of protective genes and modulation of the innate immune system [16,73–78]. Selective neutralization of chemokine TCA3 has been associated with reduction of injury in partial versus whole grafts [79]. Down-regulation of the MEK/ERK1/2 pathway through CO administration renders hepatoprotective effects through HO-1 activation in rats [19,80]. Reduced tissue damage in models of I/R injury is also observed with the overexpression of Bcl-2 [73]. While none of these interventions have been implemented in human studies, they all have potential for future therapeutic strategies.
An area of increasing research is the use of ischemic preconditioning (IP) where a brief period of ischemia generates a protective response against harmful effects of longer durations of ischemia. IP down-regulates genes involved in cell death, inflammation, immune responses, stress, and cell-cycle modulation – all up-regulated by I/R injury, notably [81]. IP-induced overexpression of glutathione S-transferase mu transcripts could contribute to decreased oxidative stress. However, the overexpression of fatty acid synthase may increase oxidative stress and the TNF ligand superfamily member 10 may enhance apoptotic pathways [82]. Thus, the clinical effectiveness of IP is still under discussion. Lastly, hypothermic machine perfusion is another technique which may reduce preservation injury and improve graft function, and was shown to significantly reduce proinflammatory cytokine expression, thereby reducing downstream activation of adhesion molecules and migration of leukocytes that induce apoptosis [83].
Manipulation of the lipid pathways may also improve outcomes. Animal models show that PPARα promotes resolution of I/R injury of the liver, and is important for liver regeneration [84–86]. It is possible that either enhancing or inhibiting these pathways using small molecules that are in clinical use may increase regeneration or minimize inflammation. Synthetic PPARα ligands such as Wy-14653, GW7647 or fibrates increase PPARα half-life by preventing ubiquitination and degradation and the PPARα agonist (Wy-14643) has decreased ischemic injury in liver and heart IR models [87–89].
Post-operative liver damage with prolonged hyperbilirubinemia during liver regeneration is a major problem in patients receiving partial grafts, diseased livers which require excess hepatectomy, or small-for-size grafts. Changes in the expression of ATP-binding cassette transporters have been implicated as critical events in liver failure during regeneration [90]. Down-regulation of the multidrug resistance protein-2 (MRP-2) is a direct cause of conjugated hyperbilirubinemia. Thus, regulating MRP-2 may treat cholestasis which negatively impacts regeneration of small grafts [90].
8. Conclusion
The use of genomic profiling and biomarkers for the identification and management of liver transplant recipients is a rapidly evolving field. We continue to see the application of new technologies to persistent clinical processes and problems. There is ample evidence that these new platforms can provide an over-abundance of critical and informative data. The key will be how best to analyze, interpret, and utilize these data. Further investigation of genomic signatures and mechanisms through new methods offers great promise, but continues to pose significant challenges. Addressing these challenges will require unique collaborative efforts between surgeons, hepatologists, basic scientists, geneticists, epidemiologists, biostatisticians, bioengineers, and bioinformaticians.
Footnotes
No Conflicts of Interest.
References
- 1.Zhao K, Zhao GM, Wu D, et al. Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J Biol Chem. 2004;279:34682–34690. doi: 10.1074/jbc.M402999200. [DOI] [PubMed] [Google Scholar]
- 2.Anderson CD, Pierce J, Nicoud I, Belous A, Knox CD, Chari RS. Modulation of mitochondrial calcium management attenuates hepatic warm ischemia–reperfusion injury. Liver Transpl. 2005;11:663–668. doi: 10.1002/lt.20407. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell CC, Okaya T, Martignoni A, Husted T, Schuster R, Lentsch AB. Divergent functions of CD4+ T lymphocytes in acute liver inflammation and injury after ischemia–reperfusion. Am J Physiol Gastrointest Liver Physiol. 2005;289:G969–G976. doi: 10.1152/ajpgi.00223.2005. [DOI] [PubMed] [Google Scholar]
- 4.Weigand K, Brost S, Steinebrunner N, Buchler M, Schemmer P, Muller M. Ischemia/Reperfusion injury in liver surgery and transplantation: pathophysiology. HPB Surg. 2012;2012:176723. doi: 10.1155/2012/176723. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins GM, Anderson RM. Experimental pathology of the liver, 1: Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 6.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olthoff KM. Hepatic regeneration in living donor liver transplantation. Liver Transpl. 2003;9:S35–S41. doi: 10.1053/jlts.2003.50229. [DOI] [PubMed] [Google Scholar]
- 8.Borozan I, Chen L, Sun J, et al. Gene expression profiling of acute liver stress during living donor liver transplantation. Am J Transplant. 2006;6:806–824. doi: 10.1111/j.1600-6143.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 9.Salehi S, Brereton HC, Arno MJ, et al. Human liver regeneration is characterized by the coordinated expression of distinct microRNA governing cell cycle fate. Am J Transplant. 2013;13:1282–1295. doi: 10.1111/ajt.12183. [DOI] [PubMed] [Google Scholar]
- 10.de Jonge J, Kurian S, Shaked A, et al. Unique early gene expression patterns in human adult-to-adult living donor liver grafts compared to deceased donor grafts. Am J Transplant. 2009;9:758–772. doi: 10.1111/j.1600-6143.2009.02557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinbroum AA. N-acetyl-l-cysteine mitigates aortic tone injury following liver ischemia–reperfusion. J Cardiovasc Pharmacol. 2005;45:509–515. doi: 10.1097/01.fjc.0000159640.36900.5d. [DOI] [PubMed] [Google Scholar]
- 12.Hines IN, Hoffman JM, Scheerens H, et al. Regulation of postischemic liver injury following different durations of ischemia. Am J Physiol Gastrointest Liver Physiol. 2003;284:G536–G545. doi: 10.1152/ajpgi.00400.2002. [DOI] [PubMed] [Google Scholar]
- 13.Fondevila C, Busuttil RW, Kupiec-Weglinski JW. Hepatic ischemia/reperfusion injury—a fresh look. Exp Mol Pathol. 2003;74:86–93. doi: 10.1016/s0014-4800(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 14.Jaeschke H, Lemasters JJ. Apoptosis versus oncotic necrosis in hepatic ischemia/reperfusion injury. Gastroenterology. 2003;125:1246–1257. doi: 10.1016/s0016-5085(03)01209-5. [DOI] [PubMed] [Google Scholar]
- 15.Peralta C, Fernandez L, Panes J, et al. Preconditioning protects against systemic disorders associated with hepatic ischemia–reperfusion through blockade of tumor necrosis factor-induced P-selectin up-regulation in the rat. Hepatology. 2001;33:100–113. doi: 10.1053/jhep.2001.20529. [DOI] [PubMed] [Google Scholar]
- 16.Lutz J, Thurmel K, Heemann U. Anti-inflammatory treatment strategies for ischemia/reperfusion injury in transplantation. J Inflamm (Lond) 2010:7. doi: 10.1186/1476-9255-7-27. http://dx.doi.org/10.1186/1476-9255-7-27 [27-9255-7-27]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryter SW, Morse D, Choi AM. Carbon monoxide: to boldly go where NO has gone before. Sci STKE. 2004;2004:RE6. doi: 10.1126/stke.2302004re6. [DOI] [PubMed] [Google Scholar]
- 18.Coito AJ, Buelow R, Shen XD, et al. Heme oxygenase-1 gene transfer inhibits inducible nitric oxide synthase expression and protects genetically fat Zucker rat livers from ischemia–reperfusion injury. Transplantation. 2002;74:96–102. doi: 10.1097/00007890-200207150-00017. [DOI] [PubMed] [Google Scholar]
- 19.Kaizu T, Ikeda A, Nakao A, et al. Protection of transplant-induced hepatic ischemia/reperfusion injury with carbon monoxide via MEK/ERK1/2 pathway downregulation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G236–G244. doi: 10.1152/ajpgi.00144.2007. [DOI] [PubMed] [Google Scholar]
- 20.Tsung A, Stang MT, Ikeda A, et al. The transcription factor interferon regulatory factor-1 mediates liver damage during ischemia–reperfusion injury. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1261–G1268. doi: 10.1152/ajpgi.00460.2005. [DOI] [PubMed] [Google Scholar]
- 21.Kim KH, Dhupar R, Ueki S, et al. Donor graft interferon regulatory factor-1 gene transfer worsens liver transplant ischemia/reperfusion injury. Surgery. 2009;146:181–189. doi: 10.1016/j.surg.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 23.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol. 2012;57:692–694. doi: 10.1016/j.jhep.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 25.Fujiyoshi M, Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J Hepatobiliary Pancreat Sci. 2011;18:13–22. doi: 10.1007/s00534-010-0304-2. [DOI] [PubMed] [Google Scholar]
- 26.Miyaoka Y, Miyajima A. To divide or not to divide: revisiting liver regeneration. Cell Div. 2013;8 doi: 10.1186/1747-1028-8-8. 8-1028-8-8. http://dx.doi.org/10.1186/1747-1028-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taub R. In: Hepatology: a textbook of liver disease. Zakim DZ, Boyer WB, editors. Ch 2. Philadelphia, USA: Saunders; 2003. [Google Scholar]
- 28.Cressman DE, Greenbaum LE, DeAngelis RA, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficientmice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci U S A. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 31.Leu JI, Crissey MA, Craig LE, Taub R. Impaired hepatocyte DNA synthetic response posthepatectomy in insulin-like growth factor binding protein 1-deficient mice with defects in C/EBP beta and mitogen-activated protein kinase/extracellular signal-regulated kinase regulation. Mol Cell Biol. 2003;23:1251–1259. doi: 10.1128/MCB.23.4.1251-1259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiaffonati L, Cairo G, Tacchini L, et al. Protein synthesis and gene expression in transplanted and postischemic livers. Transplantation. 1993;55:977–982. doi: 10.1097/00007890-199305000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Damrauer SM, Studer P, da Silva CG, et al. A20 modulates lipid metabolism and energy production to promote liver regeneration. PLoS One. 2011;6:e17715. doi: 10.1371/journal.pone.0017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann DV, Lam WW, Hjelm NM, et al. Human liver regeneration: hepatic energy economy is less efficient when the organ is diseased. Hepatology. 2001;34:557–565. doi: 10.1053/jhep.2001.27012. [DOI] [PubMed] [Google Scholar]
- 35.Arai M, Yokosuka O, Chiba T, et al. Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol Chem. 2003;278:29813–29818. doi: 10.1074/jbc.M212648200. [DOI] [PubMed] [Google Scholar]
- 36.White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J Biol Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 37.Juskeviciute E, Vadigepalli R, Hoek JB. Temporal and functional profile of the transcriptional regulatory network in the early regenerative response to partial hepatectomy in the rat. BMC Genomics. 2008:9. doi: 10.1186/1471-2164-9-527. http://dx.doi.org/10.1186/1471-2164-9-527 [527-2164-9-527]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olthoff KM. Molecular pathways of regeneration and repair after liver transplantation. World J Surg. 2002;26:831–837. doi: 10.1007/s00268-002-4060-6. [DOI] [PubMed] [Google Scholar]
- 39.Debonera F, Aldeguer X, Shen X, et al. Activation of interleukin-6/STAT3 and liver regeneration following transplantation. J Surg Res. 2001;96:289–295. doi: 10.1006/jsre.2001.6086. [DOI] [PubMed] [Google Scholar]
- 40.Kiuchi T, Kasahara M, Uryuhara K, et al. Impact of graft size mismatching on graft prognosis in liver transplantation from living donors. Transplantation. 1999;67:321–327. doi: 10.1097/00007890-199901270-00024. [DOI] [PubMed] [Google Scholar]
- 41.Debonera F, Krasinkas AM, Gelman AE, et al. Dexamethasone inhibits early regenerative response of rat liver after cold preservation and transplantation. Hepatology. 2003;38:1563–1572. doi: 10.1016/j.hep.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 42.Francavilla A, Polimeno L, Barone M, Azzarone A, Starzl TE. Hepatic regeneration and growth factors. J Surg Oncol Suppl. 1993;3:1–7. doi: 10.1002/jso.2930530503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabibian JH, Girotra M, Yeh HC, et al. Sirolimus based immunosuppression is associated with need for early repeat therapeutic ERCP in liver transplant patients with anastomotic biliary stricture. Ann Hepatol. 2013;12:563–569. [PubMed] [Google Scholar]
- 44.Toso C, Patel S, Asthana S, et al. The impact of sirolimus on hepatocyte proliferation after living donor liver transplantation. Clin Transplant. 2010;24:695–700. doi: 10.1111/j.1399-0012.2009.01159.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu YX, Jin LM, Zhou L, et al. Sirolimus attenuates reduced-size liver ischemia–reperfusion injury but impairs liver regeneration in rats. Dig Dis Sci. 2010;55:2255–2262. doi: 10.1007/s10620-009-1002-2. [DOI] [PubMed] [Google Scholar]
- 46.Palmes D, Zibert A, Budny T, et al. Impact of rapamycin on liver regeneration. Virchows Arch. 2008;452:545–557. doi: 10.1007/s00428-008-0604-y. [DOI] [PubMed] [Google Scholar]
- 47.Berberat PO, Friess H, Schmied B, et al. Differentially expressed genes in postperfusion biopsies predict early graft dysfunction after liver transplantation. Transplantation. 2006;82:699–704. doi: 10.1097/01.tp.0000233377.14174.93. [DOI] [PubMed] [Google Scholar]
- 48.Conti A, Scala S, D'Agostino P, et al. Wide gene expression profiling of ischemia–reperfusion injury in human liver transplantation. Liver Transpl. 2007;13:99–113. doi: 10.1002/lt.20960. [DOI] [PubMed] [Google Scholar]
- 49.Conti A, Scala S, Romano M, et al. Gene expression profile in liver transplantation and the influence of gene dysregulation occurring in deceased donor grafts. Open Surg J. 2011;5:1–12. [Google Scholar]
- 50.Friedman BH, Wolf JH, Wang L, et al. Serum cytokine profiles associated with early allograft dysfunction in patients undergoing liver transplantation. Liver Transpl. 2012;18:166–176. doi: 10.1002/lt.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olthoff KM, Kulik L, Samstein B, et al. Validation of a current definition of early allograft dysfunction in liver transplant recipients and analysis of risk factors. Liver Transpl. 2010;16:943–949. doi: 10.1002/lt.22091. [DOI] [PubMed] [Google Scholar]
- 52.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 53.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest. 2003;112:978–980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuboki S, Okaya T, Schuster R, et al. Hepatocyte NF-kappaB activation is hepatoprotective during ischemia–reperfusion injury and is augmented by ischemic hypothermia. Am J Physiol Gastrointest Liver Physiol. 2007;292:G201–G207. doi: 10.1152/ajpgi.00186.2006. [DOI] [PubMed] [Google Scholar]
- 55.Weiss S, Kotsch K, Francuski M, et al. Brain death activates donor organs and is associated with a worse I/R injury after liver transplantation. Am J Transplant. 2007;7:1584–1593. doi: 10.1111/j.1600-6143.2007.01799.x. [DOI] [PubMed] [Google Scholar]
- 56.Roedder S, Vitalone M, Khatri P, Sarwal MM. Biomarkers in solid organ transplantation: establishing personalized transplantation medicine. Genome Med. 2011;3:37. doi: 10.1186/gm253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fouraschen S, Kurian S, Emond J, et al. Living donors with successful liver regeneration show distinct changes in hepatic genomic profiles compared to donors with incomplete regeneration. Hepatology. 2011;54:386A–387A. [Google Scholar]
- 58.Abecassis MM, Fisher RA, Olthoff KM, et al. Complications of living donor hepatic lobectomy—a comprehensive report. Am J Transplant. 2012;12:1208–1217. doi: 10.1111/j.1600-6143.2011.03972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Murad M, Cui YY, et al. miRNA regulation of liver growth after 50% partial hepatectomy and small size grafts in rats. Transplantation. 2011;91:293–299. doi: 10.1097/TP.0b013e318204756c. [DOI] [PubMed] [Google Scholar]
- 60.Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13:11–19. doi: 10.1111/j.1600-6143.2012.04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farid WR, Pan Q, van der Meer AJ, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18:290–297. doi: 10.1002/lt.22438. [DOI] [PubMed] [Google Scholar]
- 62.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 64.Schug J, McKenna LB, Walton G, et al. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genomics. 2013:14. doi: 10.1186/1471-2164-14-264. http://dx.doi.org/ 10.1186/1471-2164-14-264 [264-2164-14-264]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Ju W, Wang D, et al. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One. 2012;7:e33577. doi: 10.1371/journal.pone.0033577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu D, Yang F, Yuan JH, et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 67.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of microRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008;19:81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lorenzen JM, Volkmann I, Fiedler J, et al. UrinarymiR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11:2221–2227. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 70.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011;11:2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei L, Gong X, Martinez OM, Krams SM. Differential expression and functions of microRNAs in liver transplantation and potential use as non-invasive biomarkers. Transpl Immunol. 2013;29:123–129. doi: 10.1016/j.trim.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu J, Wang Z, Tan CJ, et al. Plasma microRNA, a potential biomarker for acute rejection after liver transplantation. Transplantation. 2013;95:991–999. doi: 10.1097/TP.0b013e31828618d8. [DOI] [PubMed] [Google Scholar]
- 73.Cooke DT, Hoyt EG, Robbins RC. Overexpression of human Bcl-2 in syngeneic rat donor lungs preserves posttransplant function and reduces intragraft caspase activity and interleukin-1beta production. Transplantation. 2005;79:762–767. doi: 10.1097/01.tp.0000153368.08861.15. [DOI] [PubMed] [Google Scholar]
- 74.Wang DS, Li Y, Dou KF, Li KZ, Song ZS. Utility of adenovirus-mediated Fas ligand and bcl-2 gene transfer to modulate rat liver allograft survival. Hepatobiliary Pancreat Dis Int. 2006;5:505–510. [PubMed] [Google Scholar]
- 75.Huang J, Nakamura K, Ito Y, et al. Bcl-xL gene transfer inhibits Bax translocation and prolongs cardiac cold preservation time in rats. Circulation. 2005;112:76–83. doi: 10.1161/CIRCULATIONAHA.105.535740. [DOI] [PubMed] [Google Scholar]
- 76.Blydt-Hansen TD, Katori M, Lassman C, et al. Gene transfer-induced local heme oxygenase-1 overexpression protects rat kidney transplants from ischemia/reperfusion injury. J Am Soc Nephrol. 2003;14:745–754. doi: 10.1097/01.asn.0000050760.87113.25. [DOI] [PubMed] [Google Scholar]
- 77.Chauveau C, Bouchet D, Roussel JC, et al. Gene transfer of heme oxygenase-1 and carbon monoxide delivery inhibit chronic rejection. Am J Transplant. 2002;2:581–592. doi: 10.1034/j.1600-6143.2002.20702.x. [DOI] [PubMed] [Google Scholar]
- 78.Braudeau C, Bouchet D, Tesson L, et al. Induction of long-term cardiac allograft survival by heme oxygenase-1 gene transfer. Gene Ther. 2004;11:701–710. doi: 10.1038/sj.gt.3302208. [DOI] [PubMed] [Google Scholar]
- 79.Xie JF, Wang G, Debonera F, et al. Selective neutralization of the chemokine TCA3 reduces the increased injury of partial versus whole liver transplants induced by cold preservation. Transplantation. 2006;82:1501–1509. doi: 10.1097/01.tp.0000243167.11566.eb. [DOI] [PubMed] [Google Scholar]
- 80.Matsumi J, Morimatsu H, Matsusaki T, et al. Heme breakdown and ischemia/reperfusion injury in grafted liver during living donor liver transplantation. Int J Mol Med. 2012;29:135–140. doi: 10.3892/ijmm.2011.821. [DOI] [PubMed] [Google Scholar]
- 81.Jassem W, Fuggle S, Thompson R, et al. Effect of ischemic preconditioning on the genomic response to reperfusion injury in deceased donor liver transplantation. Liver Transpl. 2009;15:1750–1765. doi: 10.1002/lt.21936. [DOI] [PubMed] [Google Scholar]
- 82.Raza A, Dikdan G, Desai KK, et al. Global gene expression profiles of ischemic preconditioning in deceased donor liver transplantation. Liver Transpl. 2010;16:588–599. doi: 10.1002/lt.22049. [DOI] [PubMed] [Google Scholar]
- 83.Henry SD, Nachber E, Tulipan J, et al. Hypothermic machine preservation reduces molecular markers of ischemia/reperfusion injury in human liver transplantation. Am J Transplant. 2012;12:2477–2486. doi: 10.1111/j.1600-6143.2012.04086.x. [DOI] [PubMed] [Google Scholar]
- 84.Okaya T, Lentsch AB. Peroxisome proliferator-activated receptor-alpha regulates postischemic liver injury. AmJ Physiol Gastrointest Liver Physiol. 2004;286:G606–G612. doi: 10.1152/ajpgi.00191.2003. [DOI] [PubMed] [Google Scholar]
- 85.Anderson SP, Yoon L, Richard EB, Dunn CS, Cattley RC, Corton JC. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36:544–554. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 86.Ezaki H, Yoshida Y, Saji Y, et al. Delayed liver regeneration after partial hepatectomy in adiponectin knockout mice. Biochem Biophys Res Commun. 2009;378:68–72. doi: 10.1016/j.bbrc.2008.10.176. [DOI] [PubMed] [Google Scholar]
- 87.Blanquart C, Barbier O, Fruchart JC, Staels B, Glineur C. Peroxisome proliferator-activated receptor alpha (PPARalpha) turnover by the ubiquitin–proteasome system controls the ligand-induced expression level of its target genes. J Biol Chem. 2002;277:37254–37259. doi: 10.1074/jbc.M110598200. [DOI] [PubMed] [Google Scholar]
- 88.el Azzouzi H, Leptidis S, Bourajjaj M, et al. Peroxisome proliferator-activated receptor (PPAR) gene profiling uncovers insulin-like growth factor-1 as a PPARalpha target gene in cardioprotection. J Biol Chem. 2011;286:14598–14607. doi: 10.1074/jbc.M111.220525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teoh NC, Williams J, Hartley J, Yu J, McCuskey RS, Farrell GC. Short-term therapy with peroxisome proliferation-activator receptor-alpha agonist Wy-14,643 protects murine fatty liver against ischemia–reperfusion injury. Hepatology. 2010;51:996–1006. doi: 10.1002/hep.23420. [DOI] [PubMed] [Google Scholar]
- 90.Kimura N, Hakamada K, Ikenaga SK, Umehara Y, Toyoki Y, Sasaki M. Gene expression of ATP-binding cassette transporters during liver regeneration after 90% hepatectomy in rats. Int J Mol Med. 2012;30:28–34. doi: 10.3892/ijmm.2012.972. [DOI] [PubMed] [Google Scholar]