Abstract
Purpose
The pain experience has multiple influences but little is known about how specific biological and psychological factors interact to influence pain responses. The current study investigated the combined influences of genetic (pro-inflammatory) and psychological factors on several pre-clinical shoulder pain phenotypes.
Methods
An exercise-induced shoulder injury model was used, and a priori selected genetic (IL1B, TNF/LTA region, IL6 single nucleotide polymorphisms, SNPs) and psychological (anxiety, depressive symptoms, pain catastrophizing, fear of pain, kinesiophobia) factors were included as the predictors of interest. The phenotypes were pain intensity (5-day average and peak reported on numerical rating scale), upper-extremity disability (5-day average and peak reported on the QuickDASH instrument), and duration of shoulder pain (in days).
Results
After controlling for age, sex, and race, the genetic and psychological predictors were entered separately as main effects and interaction terms in regression models for each pain phenotype. Results from the recruited cohort (n = 190) indicated strong statistical evidence for the interactions between 1) TNF/LTA SNP rs2229094 and depressive symptoms for average pain intensity and duration and 2) IL1B two-SNP diplotype and kinesiophobia for average shoulder pain intensity. Moderate statistical evidence for prediction of additional shoulder pain phenotypes included interactions of kinesiophobia, fear of pain, or depressive symptoms with TNF/LTA rs2229094 and IL1B.
Conclusion
These findings support the combined predictive ability of specific genetic and psychological factors for shoulder pain phenotypes by revealing novel combinations that may merit further investigation in clinical cohorts, to determine their involvement in the transition from acute to chronic pain conditions.
Keywords: Chronic pain, muscle pain, inflammation SNPs, IL1B, TNF/LTA region, IL6
Introduction
The body’s response to musculoskeletal injury involves a release of inflammatory mediators that drive the repair process by removing necrotic tissue and neutralizing associated enzymatic reactions that could cause additional cell death. However, when this response persists beyond a normal period of time, a chronic inflammatory reaction can cause excessive tissue damage at the site of injury, including secondary damage to uninjured tissue (23). The extent of tissue damage is one factor that accounts for the duration of the initial inflammatory reaction. Yet, many individuals experience robust inflammatory responses and substantial pain even in the absence of severe injury (23).
Individual genetic variation has been identified as a relevant factor for extending the length of the inflammatory response (15). In protein-coding regions of genes, single nucleotide polymorphisms (SNPs) have the potential to result in an alteration in protein function via residue changes, regulatory factor binding, and/or messenger RNA stability (38). Thus, we hypothesized that SNPs in genes involved with physiological processes that increase the inflammatory response could be associated with a heightened symptomatic response and prolonged recovery following musculoskeletal injury (3,34).
Studies have identified several pro-inflammatory genes, such as IL1B (15,26), IL6 (16,17), and TNF (34) in which SNPs are associated with altered inflammatory response. For example, in one study a promoter polymorphism in TNF, the Tumor Necrosis Factor gene, was associated with increased levels of the TNFα cytokine in the bloodstream, potentially related to a quicker progression in chronic obstructive pulmonary disease (34). Likewise, monocytes from individuals homozygous for the G alleles of the interleukin 6 gene (IL6) promoter SNPs -597 and -174 showed increased LPS-induced expression of IL6 (32). Studies in IL1B have been less definitive. For example, most studies have not been able to correlate specific polymorphisms with increases in gene expression or cytokine production (15,26), although a recent in vitro study of a promoter SNP showed altered transcription factor binding based on the alleles (20).
Inflammatory responses are important, but are not the only factors involved in the development of chronic pain conditions. Diatchenko et al (6) proposed a theoretical model that identified environmental, genetic, psychological, and pain amplification factors as relevant to the development of idiopathic pain conditions. Specific to the purpose of this paper are the pain-associated psychological factors included in the model, such as catastrophizing, fear of pain, anxiety, and depressive symptoms. These psychological factors are predictive of prolonged recovery following musculoskeletal injuries and have been associated with increased pain or disability in our previous studies of shoulder pain (11,28). Diatchenko et al (6) identified the importance of physiological and psychological interactions in their model, and similarly we are investigating genetic and pain-associated psychological risk factors for prediction of shoulder pain. In our previous work, based on high priority candidate genes (2) involved with direct or indirect modulation of nociception, we identified an interaction between elevated pain catastrophizing and the high pain sensitivity genotype of the catechol-O-methyltransferase gene (COMT) that was predictive of higher shoulder pain in exercise induced injury and surgical cohorts (10,12). In our most recent work we confirmed that the COMT genotype interacts with pain associated psychological factors, and identified additional interactions with variations in other pain modulatory genes - AVPR1A, KCNS1, and ADRB2 - that increase or prolong the pain experience (11).
These results provide support for the Diatchenko et al (6) model but further investigations are necessary to provide a better understanding of how heightened pain responses may occur. Interactions between genes involved with inflammatory responses and pain-associated psychological factors have not been previously studied. This is an important consideration because of potential for an additive effect for robust inflammatory responses being perpetuated by elevated levels of fear of pain, fear of movement, depressive symptoms, and pain catastrophizing. There are some precedents for inflammatory and psychological investigations in the literature, such as a study of the IL1A gene reporting discordance between gene expression and levels of perceived stress in individuals with chronic abdominal pain (29). Also, the degree of pain catastrophizing has been associated with increased IL6 responses to a painful challenge (8). While these studies investigated the association between inflammatory and psychological factors, they did not test for additive effects. Therefore, the purpose of the current study was to test for interactions between selected inflammatory gene variants and pain-associated psychological factors that predicted shoulder pain and disability phenotypes better than the individual genetic or psychological factor alone. This study used an exercise-induced muscle injury model that caused a standard injury of micro-trauma resulting in inflammation, muscular pain, and loss of physical function. This model was selected because it is a validated model of muscle-related shoulder pain and associated disability that lasts several days (9,28). We included several shoulder pain phenotypes as outcomes to represent different aspects of the pain experience. In this study we describe identification of interactions between genetic and psychological factors that are predictive of heightened symptomatic responses to a controlled muscle injury. This is a novel contribution to the understanding of processes underlying pain experiences
Methods
Procedures
All subjects underwent five testing sessions on consecutive days. During the first session, subjects 1) read and signed the informed consent approved by the University’s Institutional Review Board; 2) completed a series of brief questionnaires asking for demographic data; 3) filled out previously validated questionnaires; 4) had pre-injury (baseline) impairment measures taken; 5) had DNA collected via buccal swabs; and 6) performed a concentric-eccentric isokinetic exercise protocol on their dominant shoulder. Subjects were asked to return to the lab post-injury at 24 hour intervals for the next four days. If shoulder pain continued after the fifth study day, subjects were sent an email prompting them to report pain intensity via a web-based data collection tool. These procedures are explained in more detail in the subsequent sections.
Participants
Participants (n = 190) were otherwise healthy men and women of any racial/ethnic background. Participants were recruited from undergraduate and graduate courses at the local university and surrounding community. To meet the inclusion criteria, participants had to be 18–85 years old and not currently performing strength training exercises (no resistance exercise of the upper extremity during the previous six weeks). Participants were also excluded if they 1) were currently experiencing neck or shoulder pain, 2) had any neurological impairment of the upper extremity, such as loss of sensation, muscle weakness or reflex changes, 3) were currently taking pain medication or 4) had previous history of shoulder surgery. These eligibility criteria are the same as used in our previous studies (9,28).
Self-Report Measures
Negative Mood
Depressive symptoms were assessed through the Patient Health Questionnaire (PHQ). The PHQ is a 9-item measure that assesses both symptoms and severity of depression (19). The PHQ examines how often one has particular thoughts or feelings and is rated on a 4-point scale, where 0 means “not at all” and 3 means “nearly every day.” Anxiety symptoms were assessed with the 40-item State-Trait Anxiety Inventory (STAI) (13). Only the 20-item trait portion of the STAI was used in the data analysis to capture a dispositional construct.
Fear-Avoidance Model
Fear of pain, fear of re-injury/movement, and pain catastrophizing were the FAM-specific constructs of interest for this study. We used a shortened version of the Fear of Pain Questionnaire (FPQ-III). The shortened version contains nine items that correlated highly with the original 30-item scale in our previous study (28). The items assess fear of specific situations that would normally produce pain on a 5-point rating scale, with a score of 5 meaning “extremely painful” and a score of 1 meaning “not at all painful.” The Tampa Scale of Kinesiophobia (TSK) consists of 11 items and is used to measure the fear of movement/re-injury. It is rated on a 4-point scale, where a 4 represents “strongly agree” with the statement and a 1 represents a “strongly disagree.” Subjects were asked to fill out the TSK on each visit to the lab. The total score was used in the current study. The TSK has been deemed a valid and reliable method for determining fear of movement/re-injury in both clinical and nonclinical populations (40). The Pain Catastrophizing Scale (PCS) consists of 13 items and assesses different thoughts that may be associated with experiencing pain. It is rated on a 5-point scale, where a 4 means “you worry all the time about the pain” and a 0 means “not at all” (35). Subjects were instructed to rate the degree to which they have specified feelings when experiencing pain. Three dimensions of pain catastrophizing have been identified, but only the total score was used for the current study.
Genetic Data Generation
Gene and SNP Selection
Genetic predictors for this paper were selected a priori based on allele frequencies, status as tagging SNPs, functional data, and promising findings in human association studies involving experimental or clinical pain phenotypes. All eight SNPs chosen were bi-allelic. In this paper we report findings for 3 genes involved in inflammation (whose proteins are predominantly pro-inflammatory), and in a separate paper we recently reported findings for genes involved in peripheral or central pain modulation pathways (11). The specific SNPs selected for each gene (Table 1) had minor allele frequencies in Caucasian populations of European descent (the majority of our subjects) that ensured adequate power in statistical analyses. For two genes (IL1B, IL6), two-point diplotypes were available for use in statistical analysis due to high linkage disequilibrium between some adjacent SNP combinations, consistent with previously published data where there were only a few major haplotypes represented among the subjects (Table 2) (25). The haplotypes were combined into diplotypes for each subject, and these diplotypes were grouped together to represent putatively functionally-distinct groups (promoting more or less inflammation, based on prior evidence from the literature (15,16). However, at the TNF/LTA region, there was lower linkage disequilibrium and thus this locus was analyzed by individual SNPs (4). The SNPs included: IL1B (rs1143627, rs16944, rs1143634), IL6 (rs1800797, rs2069840), and the TNF/LTA region (rs1800629, rs229094, rs1800683).
Table 1.
Descriptive statistics for inflammatory genes and SNPs
| Gene | SNP | Genotype | Number, % | MAF Allele, Number, % |
|---|---|---|---|---|
| TNF/LTA | rs2229094* | CC | 15, 8.6% | C, 99, 28.4% |
| CT | 69, 99.7% | |||
| TT | 90, 51.7% | |||
| rs1800683* | AA | 21, 12.4% | A, 112, 33.1% | |
| AG | 70, 41.4% | |||
| GG | 78, 46.2% | |||
| TNF-308 | rs18000629* | AA | 3, 1.6% | A, 54, 14.5% |
| AG | 48, 25.8% | |||
| GG | 135, 72.6% | |||
| IL1B | rs1143627# | AA | 68, 36.8% | G, 145, 39.2% |
| GA | 89, 48.1% | |||
| GG | 28, 15.1% | |||
| rs16944# | AA | 26, 13.9% | A, 143, 38.2% | |
| GA | 91, 48.7% | |||
| GG | 70, 37.4% | |||
| rs1143634* | AA | 10, 5.6% | A, 57, 31.6% | |
| GA | 57, 32.0% | |||
| GG | 111, 62.4% | |||
| IL6 | rs1800797# | AA | 27, 14.3% | A, 125, 33.1% |
| AG | 71, 37.6% | |||
| GG | 91, 48.1% | |||
| rs2069840# | CC | 95, 50.3% | G, 120, 31.7% | |
| CG | 68, 36.0% | |||
| GG | 26, 13.8% |
Table Key:
- Included in data analysis as individual SNPs
- Included in data analysis as diplotypes
MAF = Minor Allele Frequency
Table 2.
Descriptive statistics for inflammatory gene haplotypes and diplotypes
| Gene | Haplotype | Number | Frequency (%) |
|---|---|---|---|
| IL6(rs1800797, rs2069840) | 1 (GG) | 121 | 32.5% |
| 2 (AC) | 117 | 31.5% | |
| 3 (GC) | 134 | 36.0% | |
| IL1B(rs16944, rs1143627) | 1 (GA) | 211 | 60.3% |
| 2 (AG) | 139 | 39.7% | |
| Gene | Diplotype | Number | Frequency (%) |
| IL6 | A 11, 12, 22 | 84 | 45.2% |
| B 13, 23, 33 | 102 | 54.8% | |
| IL1B | 11 | 64 | 36.6% |
| 12 | 83 | 47.4% | |
| 22 | 28 | 16.0% |
IL1B was selected because its variants have been linked to common chronic pain conditions like knee (15) and hand osteoarthritis (26). More recently, IL1B has been examined as a candidate for susceptibility to depressive disorders (25). Peripheral IL1β communicates with the brain via neural and humoral pathways to induce brain expression of IL1β, which elicits mood changes (5), making it a potentially good candidate for interaction with pain associated psychological factors for influence on pain phenotypes. IL6 has also been a gene of interest for increased risk or severity of osteoarthritis (OA), although there is some debate on this link in the literature (37). Kamarainen et al (16) reported associations of IL6 with more severe forms of OA in the fingers, specifically with the presence of the minor G alleles of two promoter SNPs, including one in our study (rs1800797). Other SNPs for IL6 have been associated with pain related to sciatica (17), thereby providing evidence for IL6 in another common chronic pain condition and supporting its inclusion in our study. Finally, TNF was included as a gene of interest in this study because it has been associated with the modulation of pain severity in cancer patients (31) and increased inflammatory reaction related to sepsis syndrome (24). At the TNF locus, another pro-inflammatory gene encoding a TNF family member, LTA (lymphotoxin alpha), lies immediately upstream (3 kilobases) in the same orientation. Thus, this area is often referred to as the TNF/LTA gene region, and some SNPs in either gene theoretically could be influencing the other (34). To best cover this region given the parameters listed above, the chosen SNPs span 3.9 kb involving both genes. A well-studied SNP in the proximal TNF promoter (308 bp upstream of transcription start, rs1800629) is much less likely to impact LTA, thus we labeled this SNP TNF-308 (consistent with literature), whereas the other two SNPs (“TNF/LTA”) are in the LTA gene but could be part of the TNF promoter: rs1800683, in the LTA 5′ untranslated region, and rs2229094 in the coding region, encoding a cysteine-to-arginine substitution. Collectively, all three SNPs are in the “TNF/LTA region” for the purposes of this paper.
Genotyping
Genotyping of these eight SNPs was performed using standard methods as follows. Briefly, DNA was extracted from subject buccal swabs using the PureGene system (Qiagen). DNA quality and quantity was verified with spectrophotometry, and sample aliquots were diluted to 10 ng/ul. The DNA dilutions were genotyped in 96-well plate format using ABI/LifeTechnologies TaqMan SNP genotyping assays at the UF Pharmacogenomics Core, with an Applied Biosystems 7900 HT platform. The plates included several blanks and duplicates for quality control. In addition, genotyping of a few random samples was validated by DNA sequencing or restrictions digest of PCR products as a further quality control. Distribution of genotypes, haplotypes and diplotypes is shown in Tables 1 and 2.
Shoulder Pain Outcomes
Shoulder Pain Intensity
The Brief Pain Inventory (BPI) was used to measure pain intensity. The BPI is an abridged version used in non-clinical populations, consists of 4 questions, and is rated on an 11-point scale (0–10). The BPI has good test-retest reliability, especially over shorter intervals (18). The BPI asks subjects to rate their pain at worst, best and average over the past 24 hours and includes a rating for current pain. In this study we did not include the patient determined average pain rating due to it differing in nature from ratings with more discrete parameters (e.g. current pain intensity). For this study, the current, best, and worst ratings on the BPI were combined for a mathematical average for each day and this variable was used in our analysis. The average of these combined scores over 5 days represented the average pain intensity. Peak pain intensity was represented as the highest value of worst pain intensity recorded during the 5 day period.
Upper Extremity Disability
The QuickDASH (Disability of the Arm, Shoulder, and Hand) was completed and focused on the subject’s ability to use the affected arm in conditions of activity of daily living. The QuickDASH is an abridged version of the full DASH and has been found to be a valid and reliable tool for determining functional status (14). The QuickDASH provided a score from 0–100, which indicates a percentage of disability reported. The peak and average disability ratings for the QuickDASH were calculated in the same manner as BPI.
Duration of Shoulder Pain
Not all subjects were pain-free at the assessment on the 5th day. These subjects were sent an email each subsequent day prompting them to complete the BPI through a secure, web-based data collection system. Subjects continued to receive an email each day, for daily BPI assessment, until they rated their current pain at 0/10 and their worst pain was rated less than 2/10. The number of days it took to reach this criterion was recorded as the duration of shoulder pain.
Shoulder Injury Protocol
Muscle injury was induced using a Kin-Com isokinetic dynamometer (Chattanooga Group., Chattanooga, TN). Detailed methods for our exercise-induced injury model can be found in our previous work, however, a brief description will be provided here (9,28). Maximum voluntary isometric contraction (MVIC) was determined by having the participants perform three repetitions of maximal isometric shoulder external rotation. The highest torque value was recorded as their MVIC. After initial MVIC was determined, subjects completed maximal isokinetic concentric/eccentric external rotation repetitions to induce an experimental muscle injury. The speed was set at 60°/s for 3 sets of 10 repetitions. Subjects were given 30 seconds rest between sets. Following the isokinetic repetitions, MVIC was measured and if subjects could still generate greater than 50% of their initial MVIC, they performed an additional 1 to 8 sets of 10 repetitions at 60°/s. This was continued until their peak force was lower than 50% of the initial MVIC. Previous research has indicated the inability to achieve 50% of initial peak MVIC is a consistent indicator of muscle fatigue (39).
Data Analysis
All statistical analyses were conducted using SAS version 9.2 (SAS Institute, Cary, North Carolina). Summary statistics were calculated for all demographic, psychological, genetic, and shoulder pain outcome measures. For every inflammatory gene, a general linear model was fitted to assess its main effect (genotype level) and a series of expanded models were fitted to study its interaction with five psychological factors for each shoulder pain outcome. The inflammatory genes were included as diplotypes (IL1B and IL6), or the three individual TNF/LTA region SNPs in the respective regression models, representing 5 genetic factors for the primary analysis (Tables 1 and 2). The rs1143634 SNP for IL1B that was not included in the diplotype, was included as part of an exploratory analysis only. Psychological factors were kept in the original continuous metric and included as main effects in the regression models prior to being incorporated into the interaction term. Each model had the same structure with 4 increments including 1) demographic data (age, gender, and race), 2) genotype, 3) psychological factor, and 4) the gene by psychological factor interaction. In this approach the inflammatory gene by psychological interaction effect was determined individually after accounting for the other predictor variables, to identify its unique prediction of variability for the respective shoulder pain phenotypes.
In our linear modeling, we conducted a total of 25 independent tests to determine if interactions (5 genetic factors by 5 psychological factors) improved prediction for each shoulder pain outcome. Bonferroni correction would yield a threshold alpha level of 0.002 for each outcome. While this might be a conservative correction for genetic studies (1), the value of 0.002 provides a convenient benchmark in assessing the outcome of the analyses reported below. In what follows then, interaction terms with p values <0.002 were considered “strong” statistical evidence for predicting the pain phenotype of interest, while those with p values ≥0.002 but <0.05 as showing “moderate” statistical evidence for predicting the pain phenotype of interest. Interaction terms with p values ≥0.05 were not further considered.
Results
One hundred ninety subjects completed the study, and the cohort had an average age of 23.0±6.0 (mean ±sd) with 61% female participants. The predominant race was white (81%) and 90% of the cohort was right hand dominant. The baseline score for the FPQ was 23.4±5.8, for the PCS was 9.9±7.7, for the PHQ was 2.7±3.2, for the STAI was 45.6±3.1, and for the TSK was 18.0±4.2. Exercise-induced muscle pain resulted in average shoulder pain intensity scores (mean±sd) of 0.4±0.8 (day of injury protocol), 2.0±1.4 (24 hours after injury protocol), 2.4±1.7 (48 hours after injury protocol), 2.1±1.7 (72 hours after injury protocol) and 1.2±1.2 (96 hours after injury protocol) respectively. The average peak pain intensity rating (mean±sd) was 5.0±2.4. The corresponding upper extremity disability reported (mean±sd) was 2.7±4.6 (day of injury protocol), 11.6±11.6 (24 hours after injury protocol), 16.5±13.8 (48 hours after injury protocol), 15.5±13.8 (72 hours after injury protocol) and 10.8±10.8 (96 hours after injury protocol). The average peak upper extremity disability was 19.6±15.0. The average duration of shoulder pain in days (mean±sd) was 6.1±1.8, with all subjects providing complete data on shoulder pain duration.
Correlations between the variables used as shoulder pain outcomes ranged from r = 0.25 to r = 0.95, with 8/10 of the correlations being below r = 0.60. The outcome measures that correlated below 0.60 were retained as different phenotypes since they shared less than 36% of variance. As expected, the highest correlations were between 5-day shoulder pain intensity and peak shoulder pain intensity (r = 0.86) and 5-day upper extremity disability and peak upper extremity disability (r = 0.95). Since we had planned a priori to consider these as separate measures on conceptual grounds, these phenotypes were also analyzed separately.
Regression models included age, sex, and race to control for these effects on the shoulder pain phenotypes, as well as the individual main effects for the genotype and psychological factor of interest. Models meeting our criteria for strong or moderate statistical evidence are summarized in Table 3 and highlighted below in more detail for each shoulder pain phenotype.
Table 3.
Regression analyses for prediction of shoulder pain phenotypes by genetic and psychological factors
| Model Description | R-Square Increment | R-Square Full Model | p-value Full Model | ||||
|---|---|---|---|---|---|---|---|
| Age, Sex, and Race | Add Genotype | Add Psychological Factor | Add Interaction Term | P-value Interaction Term | |||
| Prediction of 5-Day Average Pain Intensity | |||||||
| Age, sex, and race + TNF/LTA (rs2229094) + PHQ + TNF/LTA (rs2229094)*PHQ | 0.024 | 0.004 | 0.029 | 0.080 | 0.0018 | 0.136 | 0.0328 |
| Age, sex, and race + IL1B + TSK + IL1B*TSK | 0.024 | 0.004 | 0.024 | 0.081 | 0.0013 | 0.129 | 0.0412 |
| Prediction of Peak Pain Intensity | |||||||
| Age, sex, and race + TNF/LTA (rs2229094) + PHQ + TNF/LTA (rs2229094)*PHQ | 0.052 | 0.007 | 0.057 | 0.056 | 0.0155 | 0.172 | 0.0260 |
| Age, sex, and race + IL1B + TSK + IL1B*TSK | 0.052 | 0.007 | 0.052 | 0.052 | 0.0126 | 0.157 | 0.0412 |
| Prediction of 5-Day Average Upper Extremity Disability | |||||||
| Age, sex, and race + IL1B + FPQ + IL1B*FPQ | 0.080^ | 0.003 | 0.119^ | 0.037 | 0.0429 | 0.239 | 0.0014 |
| Age, sex, and race + TNF/LTA (rs2229094) + TSK + TNF/LTA (rs2229094)*TSK | 0.080^ | 0.003 | 0.103^ | 0.041 | 0.0404 | 0.227 | 0.0036 |
| Prediction of Shoulder Pain Duration | |||||||
| Age, sex, and race + TNF/LTA (rs2229094) + PHQ + TNF/LTA (rs2229094)*PHQ | 0.022 | 0.034 | 0.039 | 0.168 | <0.0005 | 0.263 | <0.0005 |
Table Key: Models in bold font meet criterion for strong statistical evidence of interaction term (p < 0.002);
indicates p-valued for R-Square main effect is < 0.05;
Gene without rs number is the associated diplotype; FPQ – Fear of Pain Questionnaire, PHQ - Patient Health Questionnaire (depressive symptoms), TSK – Tampa Scale of Kinesiophobia;
5-Day Shoulder Pain Intensity
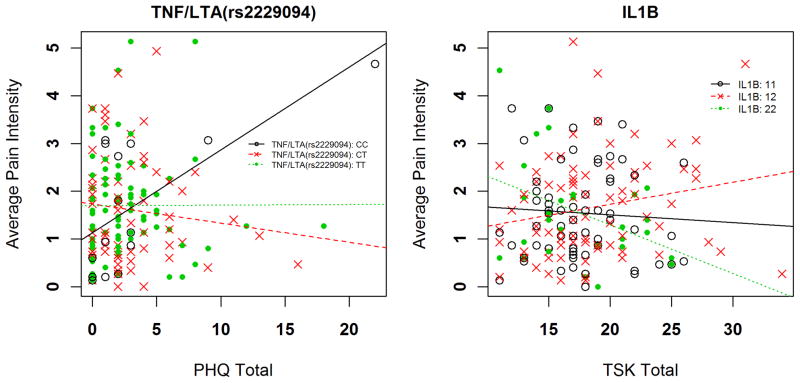
Two different genetic x psychological interactions had strong statistical evidence for explaining variance in 5-day average shoulder pain intensity ratings (Table 3). First, in the full regression model for the TNF/LTA SNP rs2229094 and the PHQ, total variance explained was an estimated 13.6%, with the interaction independently accounting for 8.0% of the overall variance (p = 0.0018 for the interaction term). Increasing PHQ scores were more strongly associated with increasing pain intensity for the CC genotype in comparison to the CT and TT genotypes (Figure 1a). The interaction resulted in an increased pain intensity rating of 0.56 and 0.73 for every 1 standard deviation increase in PHQ score among individuals with rs2229094 CC compared to TT and CT genotypes respectively. Second, in the full regression model for the IL1B diplotype and the TSK score, total variance explained was an estimated 12.9%, with the interaction term independently accounting for 8.1% of the overall variance (p = 0.0013 for the interaction term). Increasing TSK scores were associated with decreasing pain intensity for the IL1B “22” diplotype (homozygous for the AG haplotype), in comparison to the “11 and 12” diplotype (Figure 1b). The interaction resulted in a decreased pain intensity rating of 0.68 for every 1 standard deviation increase in TSK score among individuals for the IL1B “22” diplotype when compared to “12” diplotype.
Figure 1.
Interaction of genetic and psychological factors for average shoulder pain intensity
Peak Shoulder Pain Intensity
The same genetic and psychological interactions provided moderate statistical evidence for prediction of peak shoulder pain intensity ratings (Table 3). First, in the full regression model for the TNF/LTA SNP rs2229094 and the PHQ, total variance explained was an estimated 17.2%, with the interaction term independently accounting for 5.6% of the overall variance (p = 0.0155 for the interaction term). Increasing PHQ scores were more strongly associated with pain intensity for the CC genotype in comparison to the CT and TT genotypes, consistent with the 5-day pain intensity data. The interaction resulted in an increased pain intensity rating of 1.25 and 1.12 for every 1 standard deviation increase in PHQ score among individuals with the TNF/LTA rs2229094 CC genotype compared to TT and CT genotypes respectively. Second, in the full regression model for the IL1B diplotype and TSK, total variance explained was an estimated 15.7%, with the interaction term independently accounting for 5.2% of the overall variance (p = 0.0126 for the interaction term). Increasing TSK scores were inversely associated with pain intensity for the IL1B “22” diplotype, in comparison to the “12” diplotype, also consistent with 5-day pain intensity results. The interaction resulted in a decreased pain intensity rating of 1.19 for every 1 standard deviation increase in TSK score among individuals with the IL1B “22” diplotype compared to the “12” diplotype.
5-Day Upper Extremity Disability
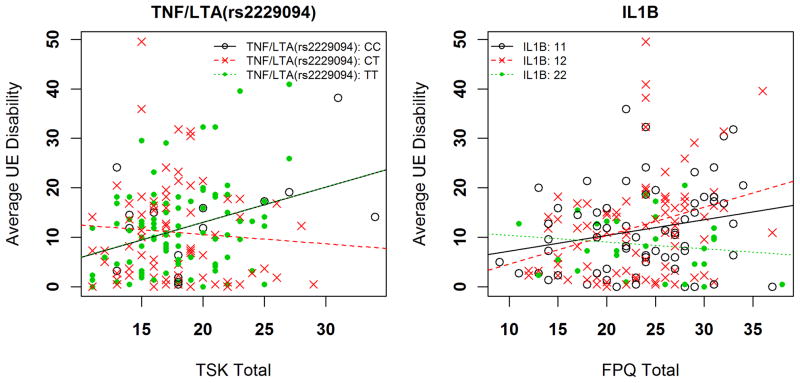
Two genetic x psychological interactions had moderate statistical evidence for the 5-day average upper-extremity disability phenotype (Table 3). First, the full model with the TNF/LTA rs2229094 and TSK scores accounted for an estimated 22.7% of the overall variance, with the interaction term independently contributing 4.1% (p = 0.0404) variance. Increasing TSK scores were associated with increased disability scores for the CC and TT genotypes (Figure 2a; lines representing CC and TT genotypes overlap). The interaction resulted in increased disability ratings of 5.91 for every 1 standard deviation increase in TSK score among individuals with the TT and CC genotype compared to CT genotype. Second, the full model including interaction between the IL1B diplotype and FPQ score explained an estimated 23.9% of the overall variance in this pain phenotype, with the interaction term independently accounting for 3.7% of the overall variance (p=0.0429 for the interaction term). Increasing FPQ scores were associated with increased disability in individuals with the IL1B “12” and “11” diplotypes (Figure 2b). The interaction resulted in decreased disability ratings of 7.0 for every 1 standard deviation increase in FPQ score among individuals with the IL1B “12” diplotype compared to “22” diplotype.
Figure 2.
Interaction of genetic and psychological factors for average upper extremity disability
Peak Upper Extremity Disability
No genetic-psychological interactions met our criterion for providing statistical evidence of improving the prediction of peak upper-extremity disability phenotype.
Duration of Shoulder Pain
One genetic x psychological interaction emerged for prediction of pain duration that met our criterion for strong statistical evidence (Table 3). The full model with TNF/LTA rs2229094 and PHQ scores explained an estimated 26.3% of the variance, with the interaction term accounting for 16.8% of the overall variance (p < 0.0005 for the interaction term). Increasing PHQ scores were associated with increased disability scores for the CC genotype (Figure 3). The interaction resulted in an increased duration of 1.19 and 1.75 for every 1 standard deviation increase in PHQ scores among individuals with the CC genotype compared to TT and CT genotypes respectively.
Figure 3.
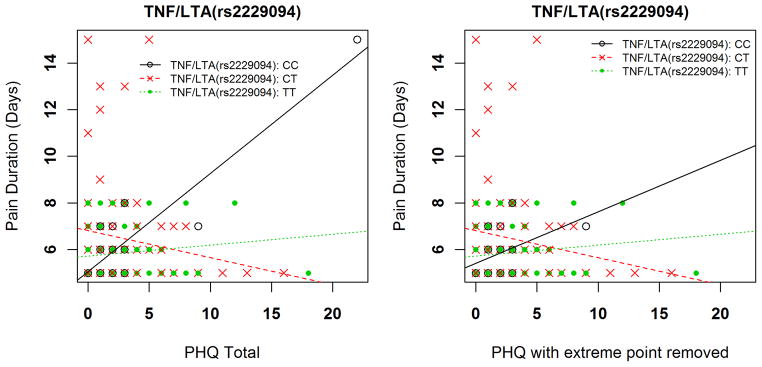
Interaction of genetic and psychological factor for shoulder pain duration
Influence of One Extreme PHQ Data Point
Visual inspection of the data provided an indication that there was one extreme PHQ score. This point was confirmed to be more than 3 standard deviations beyond the mean PHQ scores for this sample. There was consideration of dropping this point from the analyses, but we had made no a priori decision rules for eliminating extreme points. Furthermore, although this point was “extreme” for this sample it was not at the end range of the PHQ scale (observed value = 20, maximum value = 27) and is potentially more representative of PHQ scores from clinical pain samples. Therefore we decided to keep the data point in the primary analyses and report separately on the influence of this point. When this point was removed, only the interaction with the TNF/LTA SNP rs2229094 remained for the peak pain intensity pain phenotype (p = 0.0040 for the interaction term with point removed). No other statistical support for genetic interactions with the PHQ remained if the extreme PHQ data point was removed from the analysis. Figure 3 provides an example of how removal of the extreme PHQ point altered the findings for the shoulder pain duration phenotype.
Exploratory Analysis
The IL1B rs1143634 SNP was analyzed separately as it could not be included in the haplotype. This SNP interacted with the PHQ for the average pain intensity phenotype (p = 0.0114) with the combined effect resulting in increased pain scores for genotype AG. This SNP also interacted with the PHQ for the shoulder pain duration phenotype (p = 0.0032) with the combined effects resulting in decreased days for the same genotype. However, neither of these interactions remained if the extreme data point was removed.
Discussion
Our purpose was to identify inflammatory genes that interact with established pain associated psychological factors for predicting heightened shoulder pain responses. An exercise-induced injury model was selected because it produces damage to muscle tissue, causing local inflammation, pain, disability, and functional deficits (9,28). This study is the first that we are aware of to examine interactions involving inflammatory genes in a way that is consistent with a Diatchenko et al’s (6) theoretical model for how heightened pain responses may lead to the development of chronic pain. This study identified several novel interactions that may contribute to predicting pain responses following acute muscle injury, thereby also having the potential to improve the prediction of chronic musculoskeletal pain conditions in future studies. The combinations identified in this study are noteworthy because the additive effect of the genetic and psychological factor was above and beyond the predictive value of either factor individually. These results extend our previous work that identified psychological interactions with pain modulation genes (11). These results provide further support for the importance of interactions between genetic and psychological risk factors in the prediction of pre-clinical shoulder pain phenotypes.
Our previous gene association studies for pain phenotypes have focused on pain candidate genes like catechol-O-methyltransferase (COMT) (12). The current study aimed to increase the scope of genetic association studies for pain responses by examining genes involved with inflammatory processes with an established influence on pain and disability outcomes. For example, Licciardone et al (21) found strong associations between IL1B and IL6 concentrations and the number of key osteopathic lesions in relation to chronic low back pain. In addition, Dinarello et al (7) found that by blocking IL1 activity in inflammatory diseases the severity of the condition was reduced. The current study’s approach, however, was to consider more than just the main effect of the genetic influence by testing statistically for interactions with relevant pain associated psychological factors. A precedent for such investigations comes from work like Peace et al (29) and Edwards et al (8), but our study is the first we are aware of to test for additive effects.
A hallmark characteristic of chronic pain conditions is elevated pain and disability, as well as pain duration that lasts longer than what is normally expected. Collectively, the results from this study identified additive effects of inflammatory genes and psychological factors that were predictive of heightened pain responses. Results indicated that the TNF/LTA locus and IL1B interacted with depressive symptoms and kinesiophobia respectively for predicting average and peak pain intensity. In these regression models the predictors explained from 12.9% to 17.2% of the variance in pain intensity outcomes. The test for interaction between TNF/LTA region SNPs and depressive symptoms revealed that depression was positively associated with pain intensity only in one of the genotype groups (CC at rs2229094) and in the expected direction of association, as the C allele has been associated with increased risk of inflammation-related traits (22,36). The C allele encodes a predicted amino acid substitution in a transmembrane domain and thus could feasibly alter protein function (33). However the interaction for IL1B and kinesiophobia indicated that increased kinesiophobia predicted decreased pain for another genotype group: the “22” diplotype at rs16944 and rs1143627 promoter SNPs. This suggests that homozygosity for the AG haplotype (“2”) is protective for these pain measures, which is consistent with the “1” haplotype being part of a risk haplotype for osteoarthritis (4). This intriguing finding may merit further confirmatory investigation, including direct measurement of inflammatory mediators, because of the finding implicating an inflammatory pathway that reversed the adverse effects of a well-established psychological risk factor. One potential explanation worth future exploration is that subjects with this combination had a severe protective response to the muscle inflammation and kinesiophobia, resulting in lower pain scores due to immobilization. Confirming this explanation would require direct measures of movement in future studies (e.g. accelerometer), which we did not incorporate in this study.
The regression models predicting average upper-extremity disability were the most robust for this cohort, explaining over 20% of variance each. This pain phenotype had an IL1B diplotype and a TNF/LTA rs2229094 genotype that interacted with 2 related psychological factors (fear of pain and kinesiophobia respectively). The direction of the interaction was as expected for these genotypes, with specific variations in those genes resulting in higher disability scores for increasing levels of pain associated psychological distress. For example the CC and TT genotypes of the TNF/LTA SNP rs2229094 had higher disability with increasing kinesiophobia in comparison to the CT genotype. For IL1B the “12” diplotype had higher disability with increasing fear of pain scores, which was a complementary finding for the average pain intensity scores since different haplotypes from this gene had interactions with different psychological factors. The pain and disability results with IL1B suggest a potentially complex association between this genetic factor and relevant psychological factors, but future study is necessary to confirm or refute these findings. Exercise-induced muscle injury typically resolves within 96 hours but several studies have shown variation among individuals, including our own previous work with this model (31). In this study we identified only one interaction that explained additional variability in the duration following exercise induced pain. The combination of the CC TNF/LTA rs2229094 genotype and increasing depressive symptoms predicted risk for longer duration of pain. The duration phenotype was the only variable that had a single psychological factor implicated; indicating depressive symptoms may have a primary role in perpetuating inflammatory factors involved with the length of response following controlled muscle injury.
We believe that this muscle injury paradigm provides a valid pre-clinical model because it generates comparable levels of pain and disability scores to those reported in lower quartile clinical cohorts. Therefore, the specific combined factors identified in this study might make good candidates for future clinical studies predicting individuals at risk to develop chronic pain following acute musculoskeletal injury. It was beyond the intent of this study to identify mechanisms involved in these processes, so we can only speculate as to the mechanisms mediating the interactions between genetic and psychological factors. Increasing evidence suggests that the psychological factors included in our study can affect inflammation (8,27,30). Hence, it seems plausible that in the presence of a pro-inflammatory genotype, psychological processes that may themselves promote inflammation may be more strongly related to pain phenotypes. Alternatively, if a genotype represents a marker of risk for inflammation and a psychological factor confers risk for enhanced central pain processing, then these two independent risk factors for pain, mediated by divergent underlying mechanisms, would understandably produce additive effects on the pain phenotype. Another finding of this study that merits further investigation is evidence of a protective effect for certain genotypes when combined with elevated psychological scores. This was an interesting finding because it suggests that significant interactions between genetic and psychological factors can bi-directional (related to increased and decreased risk).
Notable strengths of this study include the use of a muscle injury model that allows for generation of different pain phenotypes and the ability to follow individuals from a pain-free to a painful state. Such observations are not available in clinical cohorts. Additionally we used a priori identified genetic and psychological factors with theoretical and empirical rationale to support their inclusion in our prediction models. There are however several limitations with this study that should be considered when interpreting the results. Our sample consisted of healthy, young individuals with lower levels of pain associated psychological distress. While the levels of psychological distress here are consistent with previous reports in the literature for healthy individuals (9,28), they are not representative of the levels expected in patients with chronic pain. The inflammatory genes selected for this study were not intended to be comprehensive, so conclusions are only relevant for the genes included in this study. Another limitation of this study is that by using only a few SNPs in each region, we cannot make inferences for contributions to these pain phenotypes from gene variants not in linkage disequilibrium with the selected SNPs, although the exploratory analysis with the single IL1B SNP rs1143634 suggested that other regions of this gene may play a role. Further, consistent with the notion that the inflammatory process is complex, IL6 is reported to have both pro- and anti-inflammatory effects depending on the type of injury, location, and duration; such properties could significantly complicate genetic analysis and interpretation, and might underlie the lack of significant findings for IL6 in our model (4). Also, since the majority of our participants were white (non-Hispanic) and we controlled for race in our regression analyses; we may have missed subtle influences of race on the pain phenotypes. Finally, because this study was designed as a pre-clinical association study we did not include any direct measures of the inflammatory process therefore we cannot make direct mechanistic or biological conclusions from this study.
Worth consideration is the strong influence that the extreme value for the PHQ had on the results. We reported the results including that point as we did not have a priori decision guidelines to remove data. Furthermore, this point is extreme for the sample but it did not represent the end scale value for the PHQ. Readers that do not agree with this decision will have a different interpretation of the study, based on our description of the outcomes with that point removed. A total of 7 genetic by psychological interactions were identified in these analyses, with 3 providing strong statistical evidence and the other 4 providing moderate statistical evidence. Overall, 3 interactions involved the PHQ, 3 involved the TSK, and 1 involved the FPQ. If the extreme PHQ point is removed from the analyses, only one interaction remains and it meets our criterion for moderate statistical evidence (the interaction with TNF/LTA SNP rs2229094 for peak pain intensity). Therefore, a more conservative interpretation of these analyses is that 5 interaction terms were identified with only one meeting the criterion for strong statistical evidence (IL1B and TSK for average pain intensity).
The results from this current study and our previous study (11) have identified multiple genetic and psychological factors that when considered simultaneously, are predictive of pain responses that may be relevant in the transition from acute to chronic pain conditions. Collectively these findings lend further support for a theoretical model of chronic pain development (6) and indicate that future research in clinical cohorts should consider the combination of genetic and psychological factors when predicting pain outcomes. Extending this approach to clinical cohorts is important in order to determine the ecological validity of these predictors for clinical pain phenotypes and to identify potential treatment targets that could be used to develop tailored pain management strategies. Future studies in this area should also consider examining additional inflammatory markers and examining in vivo implications of gene polymorphisms. Finally, combining prediction of inflammatory genes with previously identified pain modulatory genes (e.g. COMT) would be of interest to examine how additive genetic effects influence the experience of musculoskeletal pain.
Acknowledgments
Alberto Bursian, Brianna Castillo, Lauren Hardin, Andy Hogan, Kelly Larkin Kaiser, Natalie Martinez, Pamela McCurdy, Rachel Montgomery, Hannah Spilker, and Nhi Thieu assisted with exercise-induced injury protocol and data collection. We thank Will Eaton and Michelle Burch for assisting with genetic analyses.
Footnotes
Conflict of Interest: The authors of this manuscript have no competing or conflicting interests to declare.
The results of the present study do not constitute endorsement by the ACSM.
Funding Disclosure: This study was completed with funding from the National Institutes of Health - NIAMS (AR055899) and NINDS (NS045551)
References
- 1.Beaton DE, Wright JG, Katz JN Upper Extremity Collaborative G. Development of the QuickDASH: comparison of three item-reduction approaches. The Journal of bone and joint surgery American volume. 2005;87(5):1038–46. doi: 10.2106/JBJS.D.02060. [DOI] [PubMed] [Google Scholar]
- 2.Belfer I, Wu T, Kingman A, Krishnaraju RK, Goldman D, Max MB. Candidate gene studies of human pain mechanisms: methods for optimizing choice of polymorphisms and sample size. Anesthesiology. 2004;100(6):1562–72. doi: 10.1097/00000542-200406000-00032. [DOI] [PubMed] [Google Scholar]
- 3.Ben Aleya W, Sfar I, Habibi I, et al. Interleukin-18 gene polymorphisms in tunisian patients with inflammatory bowel disease. Digestion. 2011;83(4):269–74. doi: 10.1159/000319755. [DOI] [PubMed] [Google Scholar]
- 4.Cenit MC, Simeon CP, Vonk MC, et al. Influence of the IL6 gene in susceptibility to systemic sclerosis. The Journal of rheumatology. 2012;39(12):2294–302. doi: 10.3899/jrheum.120506. [DOI] [PubMed] [Google Scholar]
- 5.Dantzer R. Cytokine, sickness behavior, and depression. Neurologic clinics. 2006;24(3):441–60. doi: 10.1016/j.ncl.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diatchenko L, Nackley AG, Slade GD, Fillingim RB, Maixner W. Idiopathic pain disorders--pathways of vulnerability. Pain. 2006;123(3):226–30. doi: 10.1016/j.pain.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nature reviews Drug discovery. 2012;11(8):633–52. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards RR, Kronfli T, Haythornthwaite JA, Smith MT, McGuire L, Page GG. Association of catastrophizing with interleukin-6 responses to acute pain. Pain. 2008;140(1):135–44. doi: 10.1016/j.pain.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George SZ, Dover GC, Fillingim RB. Fear of pain influences outcomes after exercise-induced delayed onset muscle soreness at the shoulder. The Clinical journal of pain. 2007;23(1):76–84. doi: 10.1097/01.ajp.0000210949.19429.34. [DOI] [PubMed] [Google Scholar]
- 10.George SZ, Dover GC, Wallace MR, et al. Biopsychosocial influence on exercise-induced delayed onset muscle soreness at the shoulder: pain catastrophizing and catechol-o-methyltransferase (COMT) diplotype predict pain ratings. The Clinical journal of pain. 2008;24(9):793–801. doi: 10.1097/AJP.0b013e31817bcb65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George SZ, Parr JJ, Wallace MR, et al. Biopsychosocial Influence on Exercise-Induced Injury: Genetic and Psychologcial Combinationa Are Predictive of Shoulder Pain Phenotypes. The journal of pain : official journal of the American Pain Society. 2013 doi: 10.1016/j.jpain.2013.09.012. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George SZ, Wallace MR, Wright TW, et al. Evidence for a biopsychosocial influence on shoulder pain: pain catastrophizing and catechol-O-methyltransferase (COMT) diplotype predict clinical pain ratings. Pain. 2008;136(1–2):53–61. doi: 10.1016/j.pain.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gros DF, Antony MM, Simms LJ, McCabe RE. Psychometric properties of the State-Trait Inventory for Cognitive and Somatic Anxiety (STICSA): comparison to the State-Trait Anxiety Inventory (STAI) Psychological assessment. 2007;19(4):369–81. doi: 10.1037/1040-3590.19.4.369. [DOI] [PubMed] [Google Scholar]
- 14.Gummesson C, Ward MM, Atroshi I. The shortened disabilities of the arm, shoulder and hand questionnaire (QuickDASH): validity and reliability based on responses within the full-length DASH. BMC musculoskeletal disorders. 2006;7:44. doi: 10.1186/1471-2474-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jotanovic Z, Etokebe GE, Mihelic R, et al. IL1B -511(G>A) and IL1RN (VNTR) allelic polymorphisms and susceptibility to knee osteoarthritis in Croatian population. Rheumatology international. 2012;32(7):2135–41. doi: 10.1007/s00296-011-1946-3. [DOI] [PubMed] [Google Scholar]
- 16.Kamarainen OP, Solovieva S, Vehmas T, et al. Common interleukin-6 promoter variants associate with the more severe forms of distal interphalangeal osteoarthritis. Arthritis research & therapy. 2008;10(1):R21. doi: 10.1186/ar2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karppinen J, Daavittila I, Noponen N, et al. Is the interleukin-6 haplotype a prognostic factor for sciatica? European journal of pain. 2008;12(8):1018–25. doi: 10.1016/j.ejpain.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Keller S, Bann CM, Dodd SL, Schein J, Mendoza TR, Cleeland CS. Validity of the brief pain inventory for use in documenting the outcomes of patients with noncancer pain. The Clinical journal of pain. 2004;20(5):309–18. doi: 10.1097/00002508-200409000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. Journal of general internal medicine. 2001;16(9):606–13. doi: 10.1046/j.1525-1497.2001.016009606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landvik NE, Hart K, Haugen A, Zienolddiny S. Functional analysis of a lung cancer risk haplotype in the IL1B gene regulatory region. Journal of human genetics. 2012;57(11):747–52. doi: 10.1038/jhg.2012.106. [DOI] [PubMed] [Google Scholar]
- 21.Licciardone JC, Kearns CM, Hodge LM, Bergamini MV. Associations of cytokine concentrations with key osteopathic lesions and clinical outcomes in patients with nonspecific chronic low back pain: results from the OSTEOPATHIC Trial. The Journal of the American Osteopathic Association. 2012;112(9):596–605. doi: 10.7556/jaoa.2012.112.9.596. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Sheng H, Lu L, et al. Haplotype-based association of four lymphotoxin-alpha gene polymorphisms with the risk of coronary artery disease in Han Chinese. The Tohoku journal of experimental medicine. 2011;224(2):119–25. doi: 10.1620/tjem.224.119. [DOI] [PubMed] [Google Scholar]
- 23.Mann CJ, Perdiguero E, Kharraz Y, et al. Aberrant repair and fibrosis development in skeletal muscle. Skeletal muscle. 2011;1(1):21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menges T, Konig IR, Hossain H, et al. Sepsis syndrome and death in trauma patients are associated with variation in the gene encoding tumor necrosis factor. Critical care medicine. 2008;36(5):1456–62. e1–6. doi: 10.1097/CCM.0B013E318170ABB6. [DOI] [PubMed] [Google Scholar]
- 25.Misener VL, Gomez L, Wigg KG, et al. Tagging SNP association study of the IL-1beta gene (IL1B) and childhood-onset mood disorders. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2009;150B(5):653–9. doi: 10.1002/ajmg.b.30885. [DOI] [PubMed] [Google Scholar]
- 26.Moxley G, Han J, Stern AG, Riley BP. Potential influence of IL1B haplotype and IL1A-IL1B-IL1RN extended haplotype on hand osteoarthritis risk. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2007;15(10):1106–12. doi: 10.1016/j.joca.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 27.O’Donovan A, Slavich GM, Epel ES, Neylan TC. Exaggerated neurobiological sensitivity to threat as a mechanism linking anxiety with increased risk for diseases of aging. Neuroscience and biobehavioral reviews. 2013;37(1):96–108. doi: 10.1016/j.neubiorev.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parr JJ, Borsa PA, Fillingim RB, et al. Pain-related fear and catastrophizing predict pain intensity and disability independently using an induced muscle injury model. The journal of pain : official journal of the American Pain Society. 2012;13(4):370–8. doi: 10.1016/j.jpain.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peace RM, Majors BL, Patel NS, et al. Stress and gene expression of individuals with chronic abdominal pain. Biological research for nursing. 2012;14(4):405–11. doi: 10.1177/1099800412458350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penninx BW, Milaneschi Y, Lamers F, Vogelzangs N. Understanding the somatic consequences of depression: biological mechanisms and the role of depression symptom profile. BMC medicine. 2013;11:129. doi: 10.1186/1741-7015-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Gibby CC, Spitz MR, Yennurajalingam S, et al. Role of inflammation gene polymorphisms on pain severity in lung cancer patients. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(10):2636–42. doi: 10.1158/1055-9965.EPI-09-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O’Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20(3):218–23. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas J, Fernandez I, Pastor JC, et al. A strong genetic association between the tumor necrosis factor locus and proliferative vitreoretinopathy: the retina 4 project. Ophthalmology. 2010;117(12):2417–23. e1–2. doi: 10.1016/j.ophtha.2010.03.059. [DOI] [PubMed] [Google Scholar]
- 34.Sapey E, Wood AM, Ahmad A, Stockley RA. Tumor necrosis factor-{alpha} rs361525 polymorphism is associated with increased local production and downstream inflammation in chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 2010;182(2):192–9. doi: 10.1164/rccm.200912-1846OC. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan M, Bishop S, Pivik J. The Pain Catastrophizing Scale: Development and Validation. Psychol. 1995;7:524–32. [Google Scholar]
- 36.Takei K, Ikeda S, Arai T, Tanaka N, Muramatsu M, Sawabe M. Lymphotoxin-alpha polymorphisms and presence of cancer in 1,536 consecutive autopsy cases. BMC cancer. 2008;8:235. doi: 10.1186/1471-2407-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdes AM, Arden NK, Tamm A, et al. A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2010;18(5):699–704. doi: 10.1016/j.joca.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Varela MA, Amos W. Heterogeneous distribution of SNPs in the human genome: microsatellites as predictors of nucleotide diversity and divergence. Genomics. 2010;95(3):151–9. doi: 10.1016/j.ygeno.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Voight ML, Hardin JA, Blackburn TA, Tippett S, Canner GC. The effects of muscle fatigue on and the relationship of arm dominance to shoulder proprioception. The Journal of orthopaedic and sports physical therapy. 1996;23(6):348–52. doi: 10.2519/jospt.1996.23.6.348. [DOI] [PubMed] [Google Scholar]
- 40.Woby SR, Roach NK, Urmston M, Watson PJ. Psychometric properties of the TSK-11: a shortened version of the Tampa Scale for Kinesiophobia. Pain. 2005;117(1–2):137–44. doi: 10.1016/j.pain.2005.05.029. [DOI] [PubMed] [Google Scholar]