Abstract
Background
All site-specific interactions between HIV type-1 (HIV-1) subtype, human leukocyte anti-gen (HLA)-associated immune selection and integrase inhibitor resistance are not completely understood. We examined naturally occurring polymorphisms in HIV-1-integrase sequences from 342 antiretroviral-naive individuals from the Western Australian HIV Cohort Study and the Swiss HIV Cohort Study.
Methods
Standard bulk sequencing and sequence-based typing were used to generate integrase sequences and high-resolution HLA genotypes, respectively. Viral residues were examined with respect to drug resistance mutations and CD8+ T-cell escape mutations.
Results
In both predominantly subtype B cohorts, 12 of 38 sites that mediate integrase inhibitor resistance mutations were absolutely conserved, and these included the primary resistance mutations. There were 18 codons with non-primary drug resistance-associated substitutions at rates of up to 58.8% and eight sites with alternative polymorphisms. Five viral residues were potentially subject to dual-drug and HLA-associated immune selection in which both selective pressures either drove the same amino acid substitution (codons 72, 157 and 163) or HLA alleles were associated with an alternative polymorphism that would alter the genetic barrier to resistance (codons 125 and 193). The common polymorphism T125A, which was characteristic of non-subtype B and was also associated with carriage of HLA-B*57/*5801, increased the mutational barrier to the resistance mutation T125K.
Conclusions
Primary integrase inhibitor resistance mutations were not detected in the absence of drug exposure in keeping with sites of high constraint. Viral polymorphisms caused by immune selection and/or associated with non-subtype B might alter the genetic barrier to some non-primary resistance-associated mutations.
Introduction
HIV type-1 (HIV-1) integrase inhibitors are an important new addition to the antiretroviral armamentarium and bring renewed focus to the genetic and functional characteristics of the 32 kDa integrase enzyme, encoded by 864 bases within the 3′ HIV-1 pol. Integrase inhibitors block strand transfer and thus prevent the integration of the viral pre-integration complex into the host genome. The pyrimidinone analogue raltegravir is the first integrase strand transfer inhibitor to be in clinical use and has significant efficacy in integrase-inhibitor-naive patients when combined with other active agents, including those with significant triple antiretroviral class experience [1–3].
Integrase inhibitor resistance mutations
Major genetic pathways to integrase resistance, initially through N155H and shifting to Q148H/K/R-G140S or directly to Q148H/K/R-G140S, have been reported [4]. Another pathway through Y143C has been described more recently [5]. Elvitegravir, which remains in the final stages of clinical development, is additionally associated with T66I and E92Q in primary resistance [6]. In total, 38 unique integrase inhibitor resistance-associated mutations have been reported and cited consistently [7–10]. As resistance is generally mediated by reduced affinity in inhibitor binding to the integrase complex [11], the majority of resistance mutations lie within the presumed binding pocket [12]. Additionally, the major resistance mutations Q148H/RK and N155S have been shown to reduce replicative capacity [5,13]. The degree to which similar constraints operate at sites of secondary or minor mutations and their contribution to the overall tendency to develop resistance to integrase inhibitors remains less defined. The genetic barrier to phenotypic resistance in integrase might be lower than previously thought as a result of the selection of double and other secondary mutations, which confer marked fold reductions to drug susceptibility and restore viral infectivity and replication kinetics [14].
Interactive drug and immune selective pressures on HIV-1 integrase
The autologous viral sequence in the drug-naive patient is imprinted by a unique transmission history through all preceding hosts as well as the CD8+ T-cell selection pressure restricted by the patient’s own human leukocyte antigen (HLA) class I genotype. HIV-1 mutates within or near HLA-restricted epitopes to reduce epitope processing, HLA or T-cell receptor epitope binding, resulting in escape from CD8+ T-cell responses [15]. Similar to drug resistance, immune selection operates under the intrinsic constraints imposed by codon usage, amino acid physicochemistry, structure or function of viral residues and motifs, such that effects of individual immune escape mutations on replicative fitness are protein- and residue-specific [16–18]. HIV-1 Pol and integrase, in particular, is conserved relative to Nef, Env and all accessory proteins, although both Pol and integrase are more variable than Gag, suggesting that integrase has a higher degree of constraint in general. However, within integrase, naturally occurring polymorphisms (some of which were driven by T-cell selection) should act as a probe of genomic areas more vulnerable to mutate if targeted by antiretroviral drugs.
Studies of HIV-1 reverse transcriptase (RT), protease [19–22] and, more recently, hepatitis C virus (HCV) genes [23], have shown that drug selection and immune selection do intersect, leading to the potential for synergistic or antagonistic interactions between antiviral drugs and specific HLA alleles. In the case of HIV-1 RT and protease, these interactions conceivably account for some of the observed host- and ethnicity-specific variation in RT and protease inhibitor resistance and such interactions might be potentially very significant for novel agents, as illustrated by the challenges in developing Gag maturation inhibitors [24]. The genetic barrier to full resistance to integrase inhibitors and the role of HLA and the residue-specific effects on viral fitness remain incompletely defined, particularly for secondary sites and in natural isolates. We therefore sought to characterize polymorphism and immune selection in population-derived sequences of integrase, and to map areas of potential overlap with integrase inhibitor associated selection.
Methods
Patient cohorts
Study populations were derived from two large population-based cohorts in which the diversity of prevalent viral subtypes, host ethnicity and HLA for their respective regions would be captured. Patients were drawn from the Western Australian HIV Cohort Study (WAHCS) [25], a state-wide observational cohort study established in 1983, and the Swiss HIV Cohort Study (SHCS), a multicentre national research project initiated in 1988 [26,27]. Patients were selected on the basis of availability of stored pre-antiretroviral treatment plasma samples for viral sequencing and extracted DNA for high-resolution HLA class I genotyping. Plasma samples of the WAHCS were collected between 1997 and 2003 and of the SHCS between 1998 and 2005, well before any use of integrase inhibitors in either population. A final dataset of 179 and 163 patient-specific integrase sequences from the WAHCS and SHCS, respectively, were included for analysis based on the sequence inclusion criteria described below. Both cohort studies operate under institutional review board approvals, which require that patients provide written informed consent for collection and storage of samples, including for genetic studies.
RNA quantitation analyses
Quantitative determination of plasma HIV-1 RNA levels (viral load [VL]) for all study patients was performed using the AmpliPrep/COBAS® TaqMan® HIV-1 assay version 1.5 (Roche Diagnostics, Castle Hill, New South Wales, Australia) according to manufacturer’s instructions and analysed using the COBAS® TaqMan® 48 Analyzer (Roche Diagnostics) with a linear dynamic range of 40–1×107 copies/ml.
HLA genotyping
HLA class I genotypes (HLA-A, -B and -C) were determined based on locus-specific PCR amplification of exons 2–3 and were all resolved to four-digit-level resolution using standard DNA sequence-based typing as previously described [28]. Ambiguities were resolved following sequencing with allele-specific subtyping primers. Sequence electropherograms were analysed using Assign™ (Conexio Genomics, Applecross, WA, USA).
HIV-1 integrase sequencing
Sequences from the WAHCS were obtained by amplification and sequencing of HIV-1 full-length genomes in multiple small overlapping fragments [29] and the data has been used in other published studies. Only integrase sequences were extracted from this pre-existing dataset for the purposes of this study. The sequencing method has been subsequently revised in our laboratory to long fragment amplification and SHCS samples were processed as follows: viral RNA for sequencing was isolated from stored plasma using lysis buffer (available separately in the Roche COBAS® Amplicor HCV sample preparation kit version 2.0; Roche Diagnostics). After reverse transcription (Superscript™ III Reverse Transcriptase; Invitrogen, Carlsberg, CA, USA), two approximately 6 kb overlapping fragments spanning the full genome were amplified by nested-PCR [30]. Direct sequencing was performed on PCR products using an ABI 3130XL Analyzer (Hitachi, Singapore) and electropherograms were analysed and edited using Assign™ (Connexio Genomics).
Integrase sequence analyses
HIV-1 integrase sequences generated from all individuals were aligned and numbered against the reference sequence HXB2 (amino acids 1–288, 864 base pairs; GenBank accession number K03455). Any sequences <50% complete because of amplification or sequencing failures or stop codons were excluded. The median level of sequence coverage from all included samples was 100% (interquartile range [IQR] 98.6–100). Subtype assignment was based on all gene sequences available for each individual using the National Center of Biotechnology Information genotyping tool [31]. Phylogenetic trees of the aligned sequences were visualized using the Neighbour-joining method based on the p-distance model with pairwise deletion within the MEGA software version 4.0 [32]. Mixtures in sequences and samples with stop codons were excluded from the calculation of prevalence of specific amino acids at single positions. Polymorphism rates were determined for each residue in integrase by calculating the proportions of non-consensus sequence relative to consensus in aligned sequences, after restriction of the dataset to subtype B viral sequences. Entropy, as an alternative measure of variability taking the diversity of all amino acid substitutions at each position into account, was scored at each codon for all clade B samples using −ΣP (s,i) logP (s,i), where P (s,i) is the probability of a given amino acid (s) appearing at a given position (i) [33].
Reference HIV-1 integrase inhibitor resistance mutations
The International AIDS Society–USA guidelines were the primary source of reference data on primary HIV-1 integrase inhibitor-associated resistance [7]. In addition, other mutations reported to be associated with resistance in published scientific literature up to December 2008 were considered [8–10].
Reference HLA allele-specific T-cell escape mutations and viral load analyses
A reference set of statistical associations between HLA alleles and HIV-1 polymorphisms was used to provide putative genotypic correlates of viral escape from HLA-restricted T-cell responses. As this population-based approach requires sufficient statistical power across many prevalent HLA-A, -B and -C genotypes and viral polymorphisms, associations computed in a large dataset (n=800) of predominantly clade B natural isolates from the USA and Australia as part of an independent study of HLA-associated viral adaptation were used [17]. These analyses used published methods in which associations between HLA alleles and HIV-1 polymorphisms were tested at single residues with adjustment for viral phylogenetic relatedness, linkage disequilibrium between major histocompatibility complex loci and multiple comparisons [34]. Adjustment for multiple comparisons utilized q-values, which estimate the false discovery rate among identified associations compared with a randomly permuted dataset. Only integrase-associations with q≤0.2 indicating 20% false discovery rates were used for this study. All HLA associations relevant to the regions of interest in this study were then examined separately using HLA and integrase sequence data from the study cohorts.
Associations between integrase polymorphisms and plasma VL were calculated in the WAHCS and the SHCS cohorts separately using ANOVA and Student’s t-tests with restriction of the dataset to subtype B viral sequences.
Results
HIV-1 subtype and HLA distributions
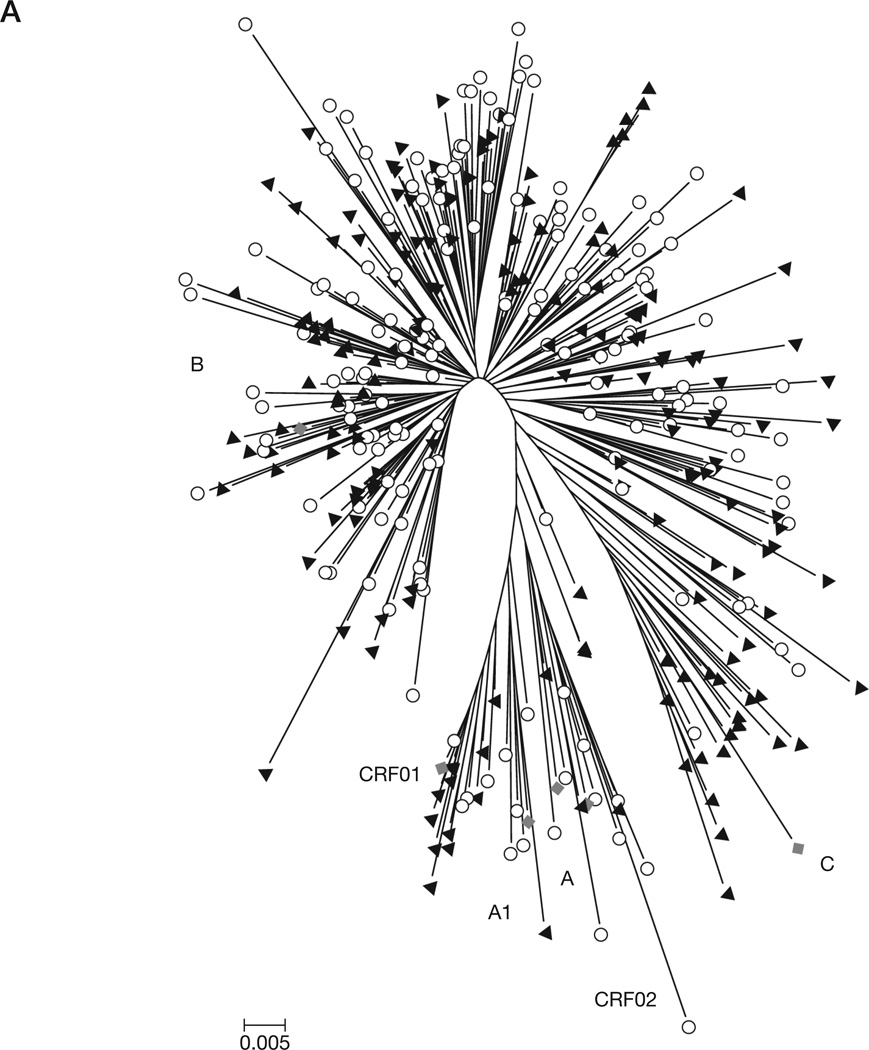
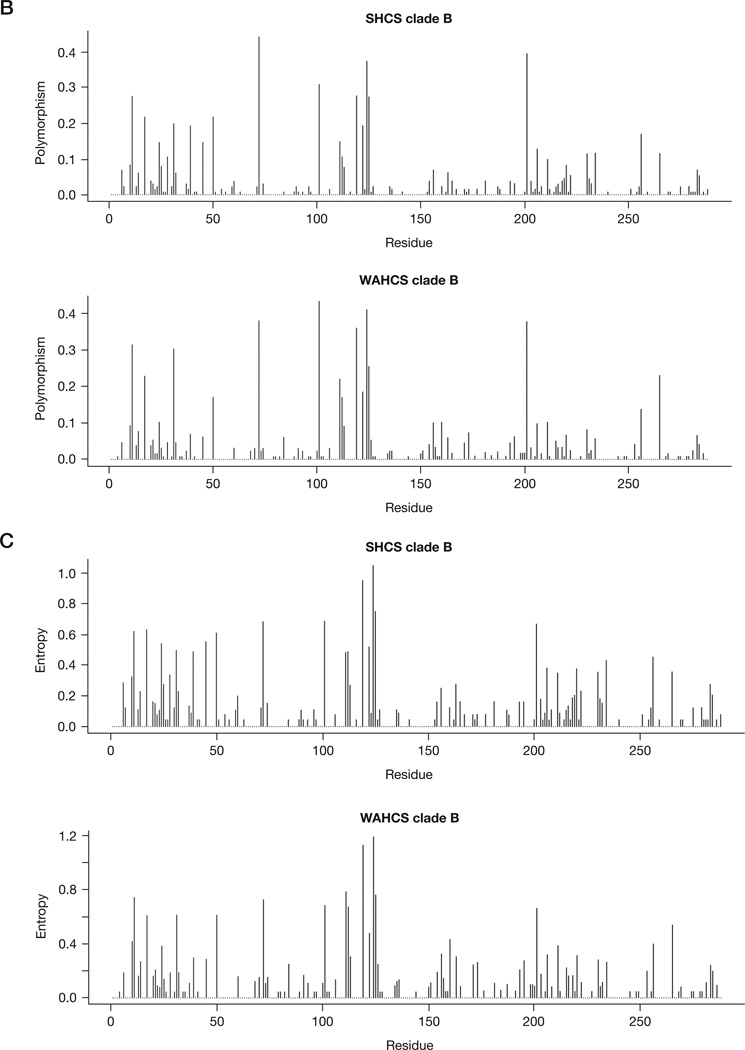
A phylogenetic tree of all sequences from both cohorts combined indicated the predominance of subtype B (82.8% in SHCS and 74.9% in WAHCS; Figure 1A). The subtype distribution among non-subtype B sequences in the SHCS was 3.7% A, 0.6% C, 0.6% CRF01, 1.8% CRF02, 0.6% CRF06, 0.6% CRF14 and 9.2% intersubtype recombinants and in the WAHCS was 4.5% C, 3.9% CRF01, 0.6% CRF02, 0.6% CRF06 and 15.6% intersubtype recombinants. Among all B subtypes, we did not detect significant clustering of sequences based on cohort origin, such that WAHCS and SHCS integrase sequences interdigitated within the subtype B phylogenetic space (Figure 1A). Similarly, the population polymorphism rates and entropy scores per residue were not significantly different at most positions between the two cohorts when restricting to subtype B samples only (Figure 1B and 1C).
Figure 1. Subtype distribution and viral diversity in the WAHCS and SHCS populations.
(A) Neighbour-joining phylogenetic analysis of HIV type-1 integrase sequences used in the study. The Swiss HIV Cohort Study (SHCS; circles) and the Western Australian HIV Cohort Study (WAHCS; black triangles) reference strains (diamonds; the GenBank submission numbers are indicated in brackets) included are A (M62320), A1 (AF069670), B (K03455), C (U46016), CRF01 (U54771) and CRF02 (L39106). (B) Integrase polymorphism profiles. (C) Integrase entropy profiles.
The most prevalent HLA types for HLA-A, -B, -C were also similar in both cohorts (data not shown) and were comparable to HLA distributions seen in published population studies, reflecting the predominance of White (Caucasian) ethnic background in both source populations [35].
Naturally occurring integrase inhibitor resistance and natural polymorphisms at resistance-associated codons
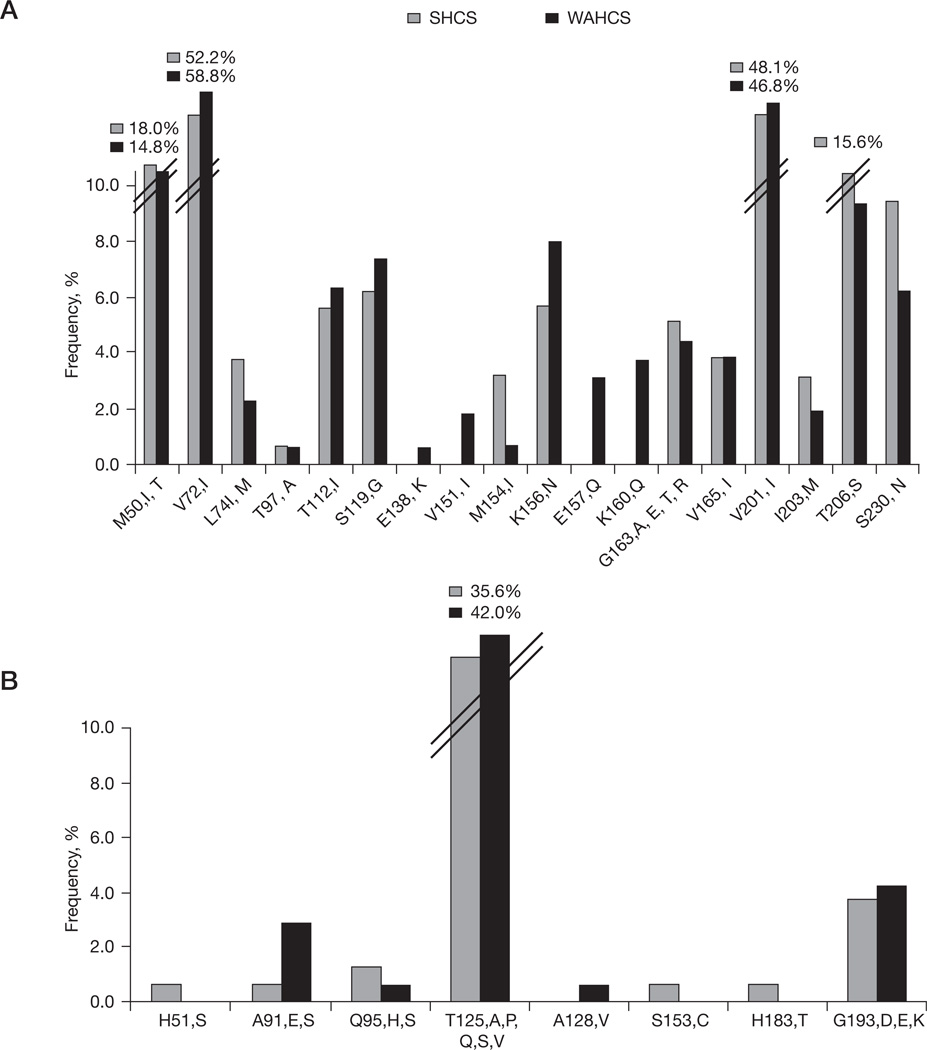
Of the 38 major and minor integrase inhibitor resistance associated codons, 44, 61, 66, 92, 121, 140, 143, 147, 148, 155, 226 and 263 were the only positions absolutely conserved across all 342 sequences and these included six primary resistance-associated mutations for raltegravir and elvitegravir (T66I, E92Q, G140S, Y143C/H/R Q148H/R/K and N155S/H). Q148K-G140S double mutants were not detected. There were 18 codons at which at least one sequence contained a (non-primary) drug resistance-associated mutation and in nine of these codons, the prevalence of the resistance mutation exceeded 5% in either the SHCS or WAHCS (M50I prevalence 18.0% and 14.8%, V72I 52.2% and 58.8%, T112I 5.6% and 6.3%, S119G 6.2% and 7.3%, K156N 5.7% and 8.0%, G163A/E/Q/T/R 5.1% and 4.4%, V201I 48.1% and 46.8%, T206S 15.6% and 9.3%, and S230N 9.4% and 6.2% for the SHCS and the WAHCS, respectively; Figure 2A).
Figure 2. Naturally occurring resistance-associated and alternative polymorphisms in the WAHCS and SHCS viral sequences.
(A) Frequency distribution histogram of 18 naturally occurring integrase inhibitor resistance mutations. (B) Frequency distribution histogram of six polymorphisms at integrase inhibitor resistance-associated sites. SHCS, Swiss HIV Cohort Study; WAHCS, Western Australian HIV Cohort Study.
There were eight resistance-associated sites that exhibited some polymorphism caused by alternative substitutions and mixtures than those associated with drug resistance; however, for six of these (51, 91, 95, 128, 153 and 183), alternative polymorphisms were rare (prevalence <3%) over all subtypes (Figure 2B). Codon 193 was polymorphic in 4.2% of WAHCS and 3.8% of SHCS sequences and codon 125 was highly polymorphic with rates of 42.0% and 35.6% in the WAHCS and SHCS, respectively. Notably, codons 50, 72, 112, 119, 125 and 201, in which either baseline drug resistance or alternative polymorphisms were seen, were among the most entropic positions within subtype B integrase, which suggests relatively low levels of mutational constraint at these sites (Figure 1B and 1C). Furthermore, three of these variable sites (112, 125 and 201) had specific polymorphisms in non-subtype B versus subtype B viruses (P<0.001 in all cases), whereas codons 95 and 230, which were more conserved within subtype B, were significantly different between B and non-B subtypes (P=0.004 and P=0.008, respectively).
HLA allele-specific polymorphism at resistance associated codons
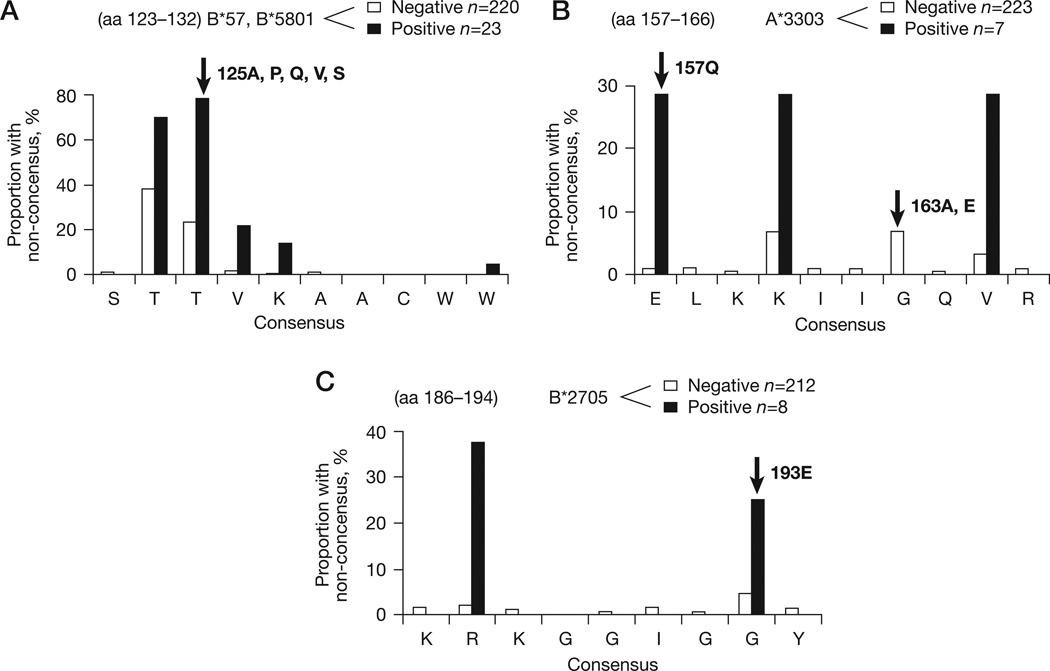
We examined the relationship between sites of integrase inhibitor resistance, HLA-restricted epitopes in integrase and evident CD8+ T-cell selection (Figure 3). Five codons shared associations with integrase inhibitor resistance and carriage of particular HLA alleles at the population level in the reference dataset (Table 1). At three of these positions (72, 157 and 163), immune and drug selection was associated with the same amino acid substitution, suggesting these sites were potentially subject to synergistic drug and immune pressure by virtue of their HLA genotype (Figure 4). V72I is associated with carriage of HLA-A*0206 alleles in population analyses and an HLA-A*0206 epitope (HLEGKVILV) is predicted to span positions 67–75 by the Epi-pred prediction programme [36]. HLA-A*0206 was rare in our study cohorts; however, the only individual carrying this allele had V72I in their autologous integrase sequence. E157Q at the p1 of the known HLA-A*3303-restricted ER9 epitope (ELKKIIGQVR) [37] is strongly associated with HLA-A*3303 in the reference associations dataset and this was also a significant association within the study dataset (P<0.005). Although G163E is at the p6 position of the same epitope and is associated with drug resistance and HLA-A*3303 carriage in the reference associations dataset, none of the HLA-A*3303-positive individuals within the study population had G163E (Figure 4).
Figure 3. Overlapping sites of integrase inhibitor resistance and HLA-restricted T-cell selection.
Map of integrase inhibitor resistance-associated sites and overlapping known (solid line) and predicted (dashed line) human leukocyte antigen (HLA)-restricted CD8+ T-cell epitopes are indicated with horizontal lines above the amino acid sequences. Known epitopes have been sourced from the Los Alamos National Laboratory HIV Immunology database and predicted epitopes are based on the Epi-pred prediction programme [35]. All amino acids are indicated by single-letter code. Highly conserved motives, the HHCC motif (coordinates zinc-binding), the DDE motif, the N-terminal domain (amino acids 1–50), the catalytic core domain (amino acids 51–212) and the C-terminal domain (amino acids 213–288) are indicated.
Table 1.
Amino acid substitutions at sites associated with synergistic or antagonistic drug and HLA-restricted immune selection
| HLA allele | Epitope | Epitope position in integrase |
Amino acid position in integrase |
Non-adapted | Adapted | Drug resistance- associated mutation |
|---|---|---|---|---|---|---|
| A*0206 | HLEGKVILV | 67–75 | 72 | V | I | Ia |
| B*5701, B*5703, B*5801 | STTVKAACWW | 123–132 | 125 | T | Ab | K |
| A*3303 | ELKKIIGQVR | 157–166 | 157 | E | Q | Qa |
| A*3303 | ELKKIIGQVR | 157–166 | 163 | G | E/A | Aa, Ea, Q, T, R |
| B*2705 | KRKGGIGGY | 186–194 | 193 | G | E | R |
Resistance associated substitution.
Mechanism of escape through loss of T-cell receptor recognition. HLA, human leukocyte antigen.
Figure 4. HLA allele-specific epitope variation at drug resistance sites.
(A) T125A is associated with HLA-B*57/*5801. (B) E157Q and G163A,E are associated with HLA-A*3303. (C) G193R is assoiciated with HLA-B*2705. Amino acid (aa) distribution at four codons (125, 157, 163 and 193, indicated with arrows) known to be integrase inhibitor resistance sites and associated with human leukocyte antigen (HLA) allele-specific polymorphism in reference data are shown. The polymorphism rate within and flanking the codons are shown in the presence (black bars) and absence (white bars) of the associated HLA allele. Polymorphism at 163 was associated with HLA-A*3303 in the reference associations dataset but not in the study population. Codon 72 was associated with HLA-A*0206 but is not included as only one individual carried this allele. HLA-B57/*5801-restricted SW10 and HLA-A*3303-restricted ER9 are published epitopes (Los Alamos National Laboratory), and KY9 is a predicted HLA-B*2705-restricted epitope based on the Epi-pred prediction programme [35]. Note the scale of the y-axis varies between panels.
The remaining dual immune/drug resistance-associated codons were 125 and 193. These were also the two most polymorphic drug resistance-associated residues in integrase (Figure 2B). Notably, G193E is an HLA-B*2705-associated polymorphism at p8 of the Epi-pred predicted epitope KRKGGIGGY (Figure 4) and distinct from the resistance change G193R. In the study population, 193G was present in six individuals carrying HLA-B*2705 versus 207 not carrying HLA-B*2705 and two patients had 193E versus five with other HLA alleles (P=0.022; Figure 4). Although both changes are single nucleotide step mutations, G193E predicts a reduced mutational barrier to the resistance change according to the Blosum62 scoring matrix [38], which is based on observed frequencies of different amino acid substitutions over short evolutionary periods in closely related proteins across all natural proteins (E193R score =0 versus G193R score =−2). Alterations of genetic and mutational barrier to resistance were observed for the most common polymorphism and HLA-B57/*5801-associated T125A as well, described in more detail below.
Integrase codon 125: interactions with subtype, HLA-B57/*5801 and genetic barrier to drug resistance
T125A was the most common natural polymorphism at a drug resistance-associated codon in the study. The frequency of T125A differed significantly between subtype B and non-subtype B (21.5% in subtype B and 89.9% in non-subtype B sequences; P<0.001) and was present among a range of non-subtype B subtypes, including A1, C, CRF01, CRF02, CRF06, CRF14 and intersubtype recombinants.
T125A association with HLA-B57/*5801 has been observed in several studies [39,40] and within our subtype-B-infected individuals, 17 individuals carried the HLA-B*5701, 1 carried the HLA-B*5703 and 6 carried the HLA-B*5801 allele. The T125A polymorphism was present in 11 (48%) of these 23 individuals compared with 38 (17%) in 220 not carrying the HLA-B57/* 5801 alleles. By contrast, 125T was present in 5 (22%) individuals carrying HLA-B57/*5801 versus 169 (77%) not carrying these alleles (P<0.001). In keeping with T-cell escape mutation, 125 was at p3 of the known HLA-B*5701, -B*5703 and -B*5801-restricted SW10 epitope (STTVKAACWW). Although the drug resistance change T125K is a minimum single nucleotide exchange, A125K is at least a two-step mutation. As 125A predominates in non-subtype B viruses and in individuals with HLA-B57/*5801 alleles, these individuals potentially have a greater genetic barrier to resistance mediated by this position. Furthermore, the change required for drug resistance from alanine to lysine (for example, A125K) is scored as a rarely tolerated mutational event (score =−1) in the blosum62 scoring matrix compared with the relatively frequent and therefore permissible exchange between threonine and alanine (for example, T125A; score =0).
Effect of resistance mutations on HLA-restricted CD8+ T-cell epitope recognition
We identified seven sites where drug resistance-associated mutations could potentially affect T-cell recognition of known CD8+ T-cell epitopes in integrase through alteration of anchor sites important for HLA–peptide binding (Figure 3). These seven sites were L74I/M at the C terminus of HLA-B*1510-restricted THLEGKIIL, 91 at p9 of YIEAEVIPA with unknown restriction, T97A at p2 of HLA-A*2601-restricted ETGQETAY, 126 at p9 of HLA-A*3002-restricted KIQNFRYY, 128 at p2 of HLA-A2-restricted AKAACWWAGI, 143 at p9 of HLA-B*1503-restricted KQEFGIPY and 165 at p2 of HLA-A*0205-restricted QVRDQAEHL.
Natural integrase polymorphisms and viral load
We tested the significance of amino acid–VL associations to determine if there were strong site-specific effects on viral fitness in vivo within integrase. In the combined subtype B WAHCS and SHCS dataset, six natural occurring polymorphisms were significantly associated with VL differences (Student’s t-test P<0.05) and also showed a consistent direction of association when stratified by cohort. Two of these polymorphisms, V72I and V201I, were the most common integrase inhibitor resistance mutations present in pretreatment sequences (Figure 2A) and in both cases, isoleucine was associated with a significantly higher VL than valine (P=0.025, mean delta log10 VL=0.21 at codon 72; P=0.00003, mean delta log10 VL=0.39 at codon 201). Furthermore, the putatively ‘more fit’ isoleucine was the consensus amino acid at codon 72 in both the WAHCS and SHCS populations, rather than a minority mutant. The four remaining codons with significant VL associations did not have any apparent relationship to drug resistance (136K versus 136E/Q/R/T, P=0.02, mean delta log10 VL=0.78; 265A versus 265 V, P=0.04, mean delta log10 VL=0.26; 106 G versus A, P=0.002, mean delta log VL= −0.91; and 208I versus 208L, P=0.03, mean delta log10 VL=−0.71). Notably, codons 72, 106, 136 and 201 are all within the catalytic domain of HIV-1 integrase.
Discussion
In this population-based review of all sites associated with integrase inhibitor resistance in 342 natural isolates of HIV-1 obtained from two geographically distinct cohorts in Switzerland and Australia, we observed that the primary raltegravir and elvitegravir resistance mutations T66I, E92Q, G140S, Y143C/H/R, Q148H/R/K and N155H were absent. Because the sequences pre-date use of integrase inhibitors in both populations, transmitted drug resistance would not be expected. However, the strong conservation of these residues is consistent with either a high genetic barrier to change or functional or structural constraint at these sites under endogenous selection pressures. This in turn predicts the high levels of antiviral efficacy seen in clinical trials of integrase inhibitors to date. Beyond these primary sites, integrase can vary significantly at certain residues, including changes known to contribute to phenotypic drug resistance and which might, based on our plasma VL analyses, have particular effects on viral fitness in vivo. These data suggest that the increase in phenotypic integrase inhibitor resistance mediated by codons 72 and 201 in particular, in which the resistant forms approach consensus frequencies, is related to effects on increasing replicative fitness.
Five sites in integrase are subject to baseline changes and interactions with drug resistance in an HLA allele-dependant manner, reflecting the overlap of HLA-restricted T-cell epitopes targeted by the hosts’ immune response and residues important for integrase inhibitor activity. Even if the immune driven changes are not those associated with drug resistance, such alternative polymorphisms alter the mutational barriers to particular resistance-associated changes. We focused on T125A as the most prevalent of these mutations. Codon 125 has been shown to vary by subtype in other studies [41,42]; however, we additionally observed that T125A was predicted by HLA-B57/5801 genotypes in subtype B viruses, and therefore showed that a component of the tendency to develop integrase inhibitor resistance is conditioned by host genetics. Notably, the closely related HLA-B*5701, HLA-B*5703, HLA-B*5801 and HLA-B*2705 alleles are associated with natural control of HIV-1 and favourable disease progression [43], and this is likely due (at least in part) to targeting of epitopes that are conserved and confer replicative costs in the event of T-cell escape mutations [44]. In the case of HLA-B57/5801 alleles, the greater barriers to immune escape and effect of T125A predict an added protection against integrase inhibitor resistance. Notably, the other HLA alleles (HLA-A*3303 and HLA-A*0206) associated with overlapping immune and drug selection are more common in populations of African and Asian origin; therefore, they might exert greater influences on baseline polymorphisms in developing countries and in increasingly diverse developed countries yet to be exposed to integrase inhibitors, and arguably also less studied in trials and observational studies to date.
Specific subtype drug resistance interactions have been described in other drug contexts, such as the more rapid selection of RT K65R on tenofovir in subtype C viruses [45] and the low prevalence of nelfinavir-associated D30N in subtype C viruses [46], among others [47]. The general efficacy and primary mutational patterns of raltegravir appear comparable in subtype B and non-subtype B viruses in available studies [48], but more data on non-subtype B viruses and more genetically diverse patient groups are still needed. The synergistic and antagonistic selective interactions between subtype and HLA-restricted T-cell responses and other antiretroviral drugs [19–22] have not been a major barrier to the broad uptake and use of available agents in many diverse populations. It is not yet clear how much these interactions contribute to variable aspects of RT inhibitor or protease inhibitor resistance, such as discordance between in vitro and in vivo genotypic resistance patterns, alternative mutational pathways and variable rates of emergence of drug resistance mutations not explained by drug dosing, plasma drug levels or adherence. Furthermore, the opposing set of interactions in which drug resistance mutations affect HLA–integrase epitope binding, as shown for other antiretroviral drugs [49,50], might influence the replication of drug-resistant variants subject to T-cell responses in emergent treatment failure.
As the use of integrase inhibitors expands to use within more diverse populations, different antiretroviral combinations, and to different clinical settings, a more detailed understanding of the overall genetic barrier to integrase inhibitor resistance afforded by these drugs should help guide their use. In particular, baseline integrase polymorphisms, which modestly reduce genetic barrier, might become clinically relevant if considering using integrase inhibitors without concurrent boosted protease inhibitors in treatment-experienced individuals [51]. We do not have a sufficient number of cases of integrase inhibitor treatment or failures across different combination regimens within the WAHCS or the SHCS to examine this question; however, pretreatment data are needed to plan studies of treated individuals. Our population-based data on natural constraint, overlapping immune targeting and site-specific effects on replicative capacity in HIV-1 integrase provides a rational basis for post hoc analyses of virological failures in clinical trials, new prospective studies of HLA-genotyped individuals commencing treatment and ongoing research on integrase interactions with covarying codons in the long-terminal repeats, RT and with other relevant host molecules.
Acknowledgements
AC, TMM and IFA generated all viral sequence data. MT and MJ led study implementation and manuscript preparation. IJ contributed to statistical analyses. MJ and SM are investigators of the WAHCS. AR, HF and HFG are SHCS investigators. All authors contributed to data interpretation and critical reading of the manuscript and have approved the final version of the manuscript. All authors wish to thank Anthony Fordham, Shay Leary and Silvana Gaudieri for assistance with preparation of the manuscript, all clinical and laboratory staff of the Centre for Clinical Immunology and Biomedical Statistics (CCIBS) and the Department of Clinical Immunology and Immunogenetics (DCII), Royal Perth Hospital and all participants of the WAHCS and the SHCS (Additional file 1).
This study was financially supported by the Australian National Health & Medical Research Council programme grant identification 384702. The SHCS is supported by the Swiss National Science Foundation (grant number 3345-062041) and by the SHCS research foundation. This project was in part supported by SHCS project number 471.
Footnotes
Disclosure statement
The authors declare no competing interests.
Additional file
Additional file 1: A list of the members of the SHCS can be found at http://
References
- 1.Markowitz M, Morales-Ramirez JO, Nguyen BY, et al. Antiretroviral activity, pharmacokinetics, and tolerability of MK-0518, a novel inhibitor of HIV-1 integrase, dosed as monotherapy for 10 days in treatment-naive HIV-1-infected individuals. J Acquir Immune Defic Syndr. 2006;43:509–515. doi: 10.1097/QAI.0b013e31802b4956. [DOI] [PubMed] [Google Scholar]
- 2.Cooper DA, Steigbigel RT, Gatell JM, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 3.Grinsztejn B, Nguyen BY, Katlama C, et al. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a Phase II randomised controlled trial. Lancet. 2007;369:1261–1269. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- 4.Delelis O, Malet I, Na L, et al. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 2009;37:1193–1201. doi: 10.1093/nar/gkn1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fransen S, Gupta S, Danovich R, et al. Loss of raltegravir susceptibility in treated patients is conferred by multiple non-overlapping genetic pathways. 17th International HIV Drug Resistance Workshop; 10–14 June 2008; Sitges, Spain. Abstract 7. [Google Scholar]
- 6.Shimura K, Kodama E, Sakagami Y, et al. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137) J Virol. 2008;82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–145. [PubMed] [Google Scholar]
- 8.Lataillade M, Chiarella J, Kozal MJ. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir Ther. 2007;12:563–570. [PubMed] [Google Scholar]
- 9.Xu L, Anderson J, Ferns B, et al. Genetic diversity of integrase (IN) sequences in antiretroviral treatment-naive and treatment-experienced HIV type 2 patients. AIDS Res Hum Retroviruses. 2008;24:1003–1007. doi: 10.1089/aid.2007.0303. [DOI] [PubMed] [Google Scholar]
- 10.Myers RE, Pillay D. Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase. J Virol. 2008;82:9228–9235. doi: 10.1128/JVI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dicker IB, Terry B, Lin Z, et al. Biochemical analysis of HIV-1 integrase variants resistant to strand transfer inhibitors. J Biol Chem. 2008;283:23599–23609. doi: 10.1074/jbc.M804213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee DJ, Robinson WE., Jr Preliminary mapping of a putative inhibitor-binding pocket for human immunodeficiency virus type 1 integrase inhibitors. Antimicrob Agents Chemother. 2006;50:134–142. doi: 10.1128/AAC.50.1.134-142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahara K, Wakasa-Morimoto C, Kobayashi M, et al. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 2009;81:141–146. doi: 10.1016/j.antiviral.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, Nakahara K, Seki T, et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brumme ZL, Tao I, Szeto S, et al. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- 17.John M, Heckerman D, Park L, et al. Genome-wide HLA-associated selection in HIV-1 and protein-specific correlations with viral load: an ACTG5142 study. 15th Conference on Retroviruses and Opportunistic Infections; 3–6 February 2008; Boston, MA, USA. Abstract 312. [Google Scholar]
- 18.Rousseau CM, Daniels MG, Carlson JM, et al. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John M, Moore CB, James IR, Mallal SA. Interactive selective pressures of HLA-restricted immune responses and antiretroviral drugs on HIV-1. Antivir Ther. 2005;10:551–555. [PubMed] [Google Scholar]
- 20.Karlsson AC, Deeks SG, Barbour JD, et al. Dual pressure from antiretroviral therapy and cell-mediated immune response on the human immunodeficiency virus type 1 protease gene. J Virol. 2003;77:6743–6752. doi: 10.1128/JVI.77.12.6743-6752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 22.Mueller SM, Schaetz B, Eismann K, et al. Dual selection pressure by drugs and HLA class I-restricted immune responses on human immunodeficiency virus type 1 protease. J Virol. 2007;81:2887–2898. doi: 10.1128/JVI.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudieri S, Rauch A, Pfafferott K, et al. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new anti-viral therapy. Hepatology. 2009;49:1069–1082. doi: 10.1002/hep.22773. [DOI] [PubMed] [Google Scholar]
- 24.Adamson CS, Ablan SD, Boeras I, et al. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat) J Virol. 2006;80:10957–10971. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallal SA. The Western Australian HIV Cohort Study, Perth, Australia. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17(Suppl 1):S23–S27. doi: 10.1097/00042560-199801001-00008. [DOI] [PubMed] [Google Scholar]
- 26.Ledergerber B, von Overbeck J, Egger M, Luthy R. The Swiss HIV Cohort Study: rationale, organization and selected baseline characteristics. Soz Praventivmed. 1994;39:387–394. doi: 10.1007/BF01299670. [DOI] [PubMed] [Google Scholar]
- 27.Swiss HIV Cohort Study. [Accessed December 52008]; Available from www.shcs.ch. [Google Scholar]
- 28.Witt CS, Price P, Kaur G, et al. Common HLA-B8-DR3 haplotype in Northern India is different from that found in Europe. Tissue Antigens. 2002;60:474–480. doi: 10.1034/j.1399-0039.2002.600602.x. [DOI] [PubMed] [Google Scholar]
- 29.Oelrichs RB, Lawson VA, Coates KM, Chatfield C, Deacon NJ, McPhee DA. Rapid full-length genomic sequencing of two cytopathically heterogeneous Australian primary HIV-1 isolates. J Biomed Sci. 2000;7:128–135. doi: 10.1007/BF02256619. [DOI] [PubMed] [Google Scholar]
- 30.Chopra A, Degenaar J, Rushton B, et al. High throughput sequencing of full length HIV-1 genomes. 4th International AIDS Conference on HIV Pathogenesis, Treatment and Prevention; 22–25 July 2007; Sydney, Australia. Abstract WEPEA 049. [Google Scholar]
- 31.National Center for Biotechnology Information. Genotyping tool. [Accessed September 2008]; Available from http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi.
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 33.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 34.Carlson J, Kadie C, Mallal S, Heckerman D. Leveraging hierarchical population structure in discrete association studies. PLoS ONE. 2007;2:e591. doi: 10.1371/journal.pone.0000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.EMBL-EBI. Human leukocyte antigen allele ethnicity tool. [Updated October 2008. Accessed November 2008]; Available from http://www.ebi.ac.uk/imgt/hla/ethnicity.html. [Google Scholar]
- 36.Heckerman D, Kadie C, Listgarten J. Leveraging information across HLA alleles/supertypes improves epitope prediction. J Comput Biol. 2007;14:736–746. doi: 10.1089/cmb.2007.R013. [DOI] [PubMed] [Google Scholar]
- 37.Hossain MS, Tomiyama H, Inagawa T, Ida S, Oka S, Takiguchi M. Identification and characterization of HLA-A* 3303-restricted, HIV type 1 Pol- and Gag-derived cytotoxic T cell epitopes. AIDS Res Hum Retroviruses. 2003;19:503–510. doi: 10.1089/088922203766774559. [DOI] [PubMed] [Google Scholar]
- 38.Organization. The Blosum clustered scoring matrix. [Updated 26 August 1997. Accessed November 2008]; Available from ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62.
- 39.Allen TM, Altfeld M, Geer SC, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feeney ME, Tang Y, Pfafferott K, et al. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J Immunol. 2005;174:7524–7530. doi: 10.4049/jimmunol.174.12.7524. [DOI] [PubMed] [Google Scholar]
- 41.Rhee SY, Liu TF, Kiuchi M, et al. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology. 2008;5:74. doi: 10.1186/1742-4690-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malet I, Soulie C, Tchertanov L, et al. Structural effects of amino acid variations between B and CRF02-AG HIV-1 integrases. J Med Virol. 2008;80:754–761. doi: 10.1002/jmv.21169. [DOI] [PubMed] [Google Scholar]
- 43.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner BG, Oliveira M, Doualla-Bell F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20:F9–F13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 46.Grossman Z, Paxinos EE, Averbuch D, et al. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob Agents Chemother. 2004;48:2159–2165. doi: 10.1128/AAC.48.6.2159-2165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danovich R, Ke Y, Wan H, et al. Raltegravir has similar in vitro antiviral potency, clinical efficacy, and resistance patterns in B subtype and non-B subtype HIV-1. XVII International AIDS Conference; 3–8 August 2008; Mexico City, Mexico. Abstract TUAA 0302. [Google Scholar]
- 49.Mason RD, Bowmer MI, Howley CM, Gallant M, Myers JC, Grant MD. Antiretroviral drug resistance mutations sustain or enhance CTL recognition of common HIV-1 Pol epitopes. J Immunol. 2004;172:7212–7219. doi: 10.4049/jimmunol.172.11.7212. [DOI] [PubMed] [Google Scholar]
- 50.Schmitt M, Harrer E, Goldwich A, et al. Specific recognition of lamivudine-resistant HIV-1 by cytotoxic T lymphocytes. AIDS. 2000;14:653–658. doi: 10.1097/00002030-200004140-00004. [DOI] [PubMed] [Google Scholar]
- 51.Eron JAJ, Zajdenverg R, et al. Switching from stable lopinavir/ritonavir-based to raltegravir-based combination ART resulted in a superior lipid profile at week 12 but did not demonstrate non-inferior virologic efficacy at week 24. 16th Conference on Retroviruses and Opportunistic Infections; 8–11 February 2009; Montreal, QC. Canada. Abstract 70aLB. [Google Scholar]