Graphical abstract

Keywords: Immune evasion, In situ, Monoallelic, Polycistronic, Silencing, Trans-splicing
Highlights
-
•
Much of what we know about trypanosomatid biology has its origin in studies on VSGs.
-
•
Monotelomeric VSG expression and epigenetic switching are remarkable examples of allelic exclusion.
-
•
DNA repair processes allow a new VSG to be copied into the single transcribed locus.
Abstract
Studies on Variant Surface Glycoproteins (VSGs) and antigenic variation in the African trypanosome, Trypanosoma brucei, have yielded a remarkable range of novel and important insights. The features first identified in T. brucei extend from unique to conserved-among-trypanosomatids to conserved-among-eukaryotes. Consequently, much of what we now know about trypanosomatid biology and much of the technology available has its origin in studies related to VSGs. T. brucei is now probably the most advanced early branched eukaryote in terms of experimental tractability and can be approached as a pathogen, as a model for studies on fundamental processes, as a model for studies on eukaryotic evolution or often all of the above. In terms of antigenic variation itself, substantial progress has been made in understanding the expression and switching of the VSG coat, while outstanding questions continue to stimulate innovative new approaches. There are large numbers of VSG genes in the genome but only one is expressed at a time, always immediately adjacent to a telomere. DNA repair processes allow a new VSG to be copied into the single transcribed locus. A coordinated transcriptional switch can also allow a new VSG gene to be activated without any detectable change in the DNA sequence, thereby maintaining singular expression, also known as allelic exclusion. I review the story behind VSGs; the genes, their expression and switching, their central role in T. brucei virulence, the discoveries that emerged along the way and the persistent questions relating to allelic exclusion in particular.
1. Introduction
Variant Surface Glycoproteins (VSGs) are developmentally regulated genes that mediate immune evasion. Activated in the tsetse-fly salivary-gland [1], and inactivated upon return to the tsetse-fly mid-gut, they produce a protective cell coat throughout the mammalian infectious cycle. The coat must provide robust protection as Trypanosoma brucei occupy the bloodstream and tissue-spaces of their hosts and are fully exposed to immune surveillance in this hostile environment. Indeed, as an infection persists, the vast majority of the parasite population is periodically eliminated. The key features underlying successful immune evasion are clone-specific singular VSG expression combined with switching from one VSG to another. The metacyclic cells in the salivary gland are challenging to study since VSG expression is heterogeneous during this phase and the yield of T. brucei from flies is limiting. Most studies, therefore, have been conducted using bloodstream forms, more recently in axenic culture. Antigenic variation continues to operate in this environment [2] indicating that host antibodies are selective rather than a trigger for variation. An advantage here is that VSG switching operates at a frequency of approximately 1 switch/105 cells per population doubling, allowing the analysis of almost homogeneous but switchable populations.
Many seminal discoveries have emerged from studies on VSGs in T. brucei and the drive to understand VSGs and their expression has also led to the development of many of the tools and technologies now available for a range of other studies on trypanosomatids. Indeed, studies on VSG expression revealed much of what we now know about gene expression in trypanosomatids. Some features are specific to VSG gene expression sites, while others operate across the genome and are conserved in trypanosomatids that do not express VSGs. Thus, work on VSGs has informed studies on other important parasites, including Trypanosoma cruzi and Leishmania species. VSGs in T. b. gambiense, T. b. rhodesiense, T. equiperdum, T. congolense and T. vivax are not discussed in detail here but a similar system of gene expression and antigenic variation appears to operate in T. brucei brucei and in these other African trypanosomes.
What I present below is a somewhat historical perspective on antigenic variation in T. brucei and, in this regard, I recommend further reading of some of the older papers in particular, not often cited these days but often impressive when viewed in this historical context. It is also worth noting that few studies on antigenic variation in T. brucei have been or are currently specifically focussed on the prospect of a therapy in the short term. The central role of VSGs in virulence does mean that improved knowledge in this area is likely to present further opportunities for intervention, however.
2. A very brief early history – pre VSG gene-cloning
Sir David Bruce had read David Livingstone's reports on the tsetse fly diseases known as nagana in cattle and sleeping sickness in humans and, while searching for the cause more than 100 years ago, reported that“ a rapidly moving object was seen lashing about among the red blood corpuscles … probably a trypanosome” [3]. Bruce also noted “the parasites come and go in the blood” and Franke & Ehrlich had deduced in 1905 that T. brucei acquired properties that conferred resistance to host “defensive substances”. Ronald Ross and others then enumerated the relapsing parasitaemia in patients [4], albeit treated with several different drugs during monitoring in this case. A number of parasitic infections of mammals are now known to display relapses due to the emergence of new variants that are no longer susceptible to the latest host immune response (Fig. 1A).
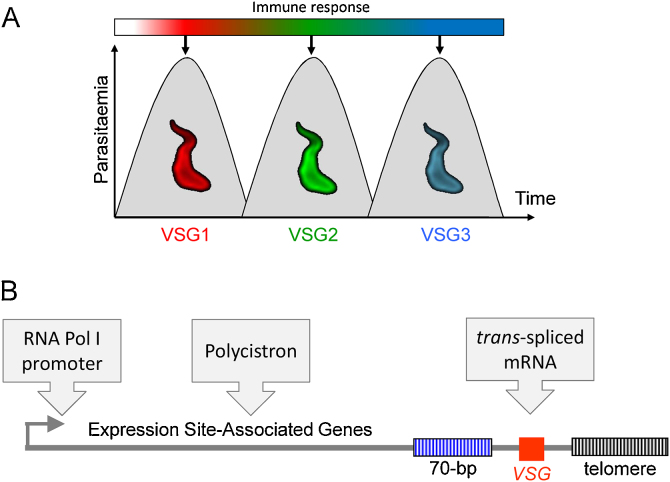
Fig. 1.
VSG expression and switching. (A) VSG switching brings about antigenic variation. Combined with successive immune responses, this can generate a relapsing parasitaemia. Natural infections are more complex than this highly simplified schematic. (B) Studies on VSG expression revealed some unusual features. The single expressed VSG was found to be flanked by distinct repetitive sequences. Three further unusual features are indicated (boxes).
In the 1960s, Keith Vickerman's work using electron microscopy revealed the dense T. brucei coat responsible for clone-specific relapses [5]. The identification and purification of the coat proteins by George Cross followed in the 1970s [6] and then the cloning and sequencing of the corresponding cDNA in the late 1970s and early 1980s (detailed below). The VSG responsible for clone-specific immunity or antigenic variation were found to be Variant throughout much of their length, they were known to represent the major component of the trypanosome Surface coat and they were Glycoproteins, decorated with multiple sugar residues [6].
3. The variant surface glycoprotein coat
The 15 nm thick VSG coat covers the entire cell and is an essential virulence factor. Formed from approximately 10 million molecules of approximately 60 kDa, the coat represents up to 20% of total cell protein [6], facilitating the production of antisera that recognise distinct VSGs [7]. VSGs are present at the cell surface as homodimers and, despite extreme sequence divergence, display remarkably similar structures [8], partly due to a conserved arrangement of disulphide bonds. Coat exchange during a VSG-switch appears to be primarily by dilution during cell division since cells divide approximately every 6 h with shedding and turnover being relatively much slower [9]. Cytokinesis is in fact dependent upon VSG supply as demonstrated by knockdown experiments [10]. The VSG is not a transmembrane protein but is rather anchored in the membrane by glycosylphosphatidylinositol (GPI). Indeed, GPI was discovered in T. brucei and this yielded the first GPI structure [11] as well as a description of GPI biosynthesis [12]. GPI anchors were subsequently discovered in mammals and in other cells [13] but the biosynthetic pathways do display differences [14].
The fluid nature of the VSG coat allows for a remarkably high rate of recycling involving endocytosis at the flagellar pocket [15]. This allows the coat to be cleansed of antibodies, at low titre at least, and this requires vigorous directional cell motility mediated by the flagellum [16]. VSG coats are highly immunogenic however, so once antibody titre increases, the vast majority of parasites are eliminated and only cells with distinct VSG coats survive. As well as this variable function, the VSG coat also serves to protect less variable or even invariant surface proteins from immune effectors. Comparison of the genome sequences of T. brucei, T. congolense and T. vivax revealed not only how VSG repertoires evolved [17] but also allowed reconstruction of a surface phylome [17]. This revealed a diversity of potential non-VSG surface proteins, nutrient receptors and other ‘invariant’ surface glycoproteins. It appears that the densely packed and thick VSG coat can physically obstruct access to these proteins by conventional immunoglobulins while selectively allowing access to smaller molecules such as nutrients [18]. Nanobodies also have the potential to access the less variable epitopes usually hidden within the coat by the size exclusion limit [19].
4. VSG genes and their subtelomeric environment
Access to the first cloned VSG genes provided probes to explore copy number, location and diversity within trypanosome genomes, especially because the 3′ terminal regions of VSG mRNAs were found to be conserved [20]. It was soon recognised that the single expressed VSG was adjacent to a telomere, or a chromosome discontinuity that was subject to Bal31 exonuclease digestion [21], [22] and this strict association remains intact to date (Fig. 1B). This stimulated much interest in telomere biology in trypanosomes, which revealed the addition (and occasional large deletions) of TTAGGG/CCCTAA-repeats to growing telomeres [23], [24], [25]; the same hexameric repeats were later found at human telomeres. T. brucei telomeres were also found to terminate in t-loops [26] and this was followed by the identification and characterisation of telomerase reverse transcriptase, responsible for telomere growth, and other telomere-repeat-binding proteins [27].
The VSG-telomere association actually goes much further. Each T. brucei genome contains around 250 telomeres, almost all of which may be closely linked to non-transcribed VSG genes, with their 3′-ends closest to the telomere. Around 80% of these telomeres reside on 50–100 kbp long minichromosomes [22], [28], [29], which appear to be entirely dedicated to VSG archiving. Another ten or so telomeres reside on ‘intermediate’ chromosomes and the remaining 44 reside on the eleven pairs of megabase-chromosomes that comprise the diploid genome. With many additional arrays of subtelomeric VSGs, it is currently thought that up to 30% of an African trypanosome genome is dedicated to archiving up to 2000 VSG genes and gene-fragments.
Although the size of the VSG archive and the telomeric environment continue to present challenges for complete genome assembly and functional analyses, important insights into VSG gene evolution and diversification have emerged from genome sequencing [30] and the cloning and sequencing of large intact telomeric fragments [31]. Extensive hemizygous subtelomeric domains on the megabase chromosomes are dedicated to arrays of archival VSGs [32], meaning that many VSGs are present as a single copy even in a diploid genome. Most of these VSGs are pseudogenes in T. brucei (749/804 analysed) and these VSGs are flanked by 70-bp repeats (Fig. 1B) upstream [33] and highly conserved elements within the 3′-untranslated region (3′-UTR); both of these sequences facilitate recombination (see below). The 3′-UTR is also involved in specific stabilisation of the VSG mRNA at the bloodstream stage, contributing to a half-life of 4.5 h [34].
5. VSG gene expression – trans-splicing and polycistronic Pol I transcription
The ‘yeast to human’ view of eukaryotic diversity is very narrow so it is not surprising that several dogmas have been overturned by work on the divergent trypanosomes. Studies on VSGs revealed some unusual features underlying gene-expression in trypanosomatids for example (Fig. 1B). S1 nuclease protection and RNA blotting experiments revealed a spliced segment at the 5′ end of the VSG mRNA and reverse transcription then showed this “mini-exon” or “spliced leader” sequence to be 35 nt long [35]. Intriguingly, the same sequence was found at the 5′-end of two other VSG mRNAs [36]. Cis-splicing was initially considered to be the most likely explanation but it turned out that this same sequence was present on all mRNAs. In fact, discontinuous mRNA synthesis through bimolecular splicing or trans-splicing allows mature mRNA to be derived from precursor RNAs transcribed from two different chromosomes [37]. Trans-splicing was also found to operate in other trypanosomatids [38] and in nematodes.
The search for a VSG gene promoter extended further from the gene itself than expected. In most eukaryotes, each gene has its own promoter, while polycistrons are largely restricted to prokaryotes. Cloning and mapping upstream of an active VSG revealed a large expanse of imperfect 70-bp repeats [39] and then an Expression Site Associated Gene or ESAG [40] and then several more ESAGs [41] forming a polycistron. It was found that α-amanitin failed to inhibit ribosomal RNA (rRNA) transcription, as expected, but also VSG transcription [42] implicating RNA Pol I. These findings prompted a detailed analysis of RNA pol I and associated factors in T. brucei (see below). Polycistronic transcription was also found to operate within Pol II transcription units elsewhere in the genome [43] and this has proven to be a pervasive feature of trypanosomatid genomes. As for the VSG expression site promoter, the first one was eventually found around 60 kbp from the VSG [44] and was indeed, following mounting evidence, confirmed to recruit RNA Pol I in vitro [45]. Nuclear ‘run-on’ assays combined with inhibition of transcription elongation using UV exposure were instrumental in locating the VSG expression site promoter [46], the sequence of which was unrelated to the more conventional rDNA promoter [47]. The metacyclic promoters responsible for VSG transcription in the tsetse-fly salivary gland were also found to recruit RNA Pol I but were distinct from the bloodstream promoters and in this case were located only a short distance upstream of the telomeric VSGs [48]; these are among only a few monocistronic transcription units in trypanosomatids.
The result of all this work revealed a somewhat surprising situation whereby RNA Pol I and RNA Pol II are required to produce mature VSG expression site transcripts; transcription of the spliced leader sequence was confirmed to be RNA Pol II dependent [49]. Further surprises were in store as assessment of other potential Pol II promoters in T. brucei or in other trypanosomatids failed to yield any further conventional examples. Rather, around sixty RNA Pol II ‘transcription initiation sites’ appear to depend upon a particular chromatin structure [50] that is not readily reconstituted on reporter constructs.
Fifteen copies of the highly conserved subtelomeric VSG expression sites active in the bloodstream have now been identified, cloned by recombination in yeast and sequenced from the most widely studies T. brucei strain [31]. Present on the diploid and intermediate-chromosomes, these contain intact promoters and are competent transcription units but are typically reversibly repressed due to monoallelic VSG expression control. Many ESAGs found within these polycistronic units remain to be characterised but the evidence so far points to roles in host-parasite interactions. For example, a novel heterodimeric transferrin receptor, encoded by ESAG6 and ESAG7 and related to the N-terminal domain of the VSG, has the capacity to bind transferrin from different hosts with different affinities [51]. A human serum resistance associated gene, or SRA, found in T. brucei rhodesiense, also resembles a VSG and is also an ESAG [52] while another gene resembling a VSG, known as TgsGP, confers human-serum resistance to T. brucei gambiense [53]. ESAG4 genes are unrelated to VSGs, but also mediate host-parasite interactions. These genes encode adenylate cyclases which are released by lysed trypanosomes and inhibit the innate immune response [54]. Clearly, VSGs and their associated genes have been central to the ‘arms-race’ operating and evolving at the host-parasite interface. The vast reservoir available has allowed VSG genes to be co-opted to functions beyond classical antigenic variation. The relationships among these proteins could equally reflect an evolutionary origin of VSGs from ancient surface receptors.
6. Antigenic variation by VSG gene rearrangement
Subtelomeres are recombinogenic hotspots, plastic regions of genomes enriched in gene families that are most commonly involved in adaptation to different environments. Like many other cell types, African trypanosomes appear to have exploited these properties, in this case for the massive expansion and evolution of the VSG family and also for the ESAGs [55]. Antigenic variation in T. brucei involves switching to expression of a distinct VSG so the availability of VSG cDNA clones allowed researchers to look for changes associated with the activation and inactivation of those VSGs. Early analyses using Southern blotting [56], [57], northern blotting [58], DNAse I digestion [59] or DNA sequencing [60] revealed switching by duplicative transposition of a non-telomeric ‘basic copy’ VSG and replacement of the old VSG by the new ‘expression-linked copy’ at a single transcribed locus. Silent telomeric VSG cassettes appeared to be copied all the way to the end of the chromosome [61], [62] by a mechanism now known as ‘break-induced replication’ (BIR).
Subsequent analyses revealed a range of variations on the recombination theme and DNA homology emerged as the key driver of these VSG rearrangements [63] stimulating further studies on a range of T. brucei DNA-repair factors. Most VSGs are flanked upstream by common 70-bp repeats and even non-telomeric genes share sequences at the 3′-end [64], sometimes within the region encoding the VSG C-terminal domain and almost always in the conserved VSG 3′-UTR (Fig. 2). Replacement of larger parts of the expression site or non-duplicative telomere exchange [65] can bring about a switch but these events appear to be relatively rare and even actively suppressed [66]. Translocation to the active site is preferentially initiated by the long tracts of 70-bp repeats found upstream of the active VSG [67], [68] so the probability of being ‘selected’ as a template for repair depends upon these and other sequences shared with the active VSG locus [69]. Indeed, 70-bp repeat recombination is primarily responsible for the high-efficiency duplication of telomeric VSGs [61], [62], which can employ the BIR mechanism, while chromosome-internal VSGs must use a ‘gene-conversion’ mechanism, similar to BIR but with a second recombination junction, most often within the VSG 3′-UTR [39], [70]. Conversion of even shorter VSG segments can generate ‘mosaic’ VSGs that, because of their lower-frequency emergence, become increasingly important for parasite persistence in a chronic infection [64], [71], [72], [73], as immunity to more-frequently activated VSGs builds [67].
Fig. 2.
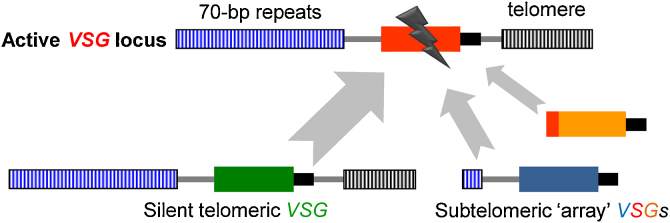
DNA recombination is central to VSG switching. The active subtelomeric VSG locus is prone to spontaneous DNA breaks. Three classes of repair templates are shown with VSGs represented as coloured boxes; those sharing more (flanking) homology with the active site are used more frequently (grey arrows). The homologous sequences indicated are the 70-bp repeats (blue stripes), the telomeric repeats (black stripes), the VSG 3′-UTRs (thick black bars) and a portion of the VSG coding sequence (red). A break in or around the active VSG is followed by DNA resection extending towards the 70-bp repeats, often initiating recombination in this region.
This understanding of the shared sequences that drive recombination can now explain why the expression of VSGs is ‘semi-ordered’ or somewhat predictable. Any order though will be isolate-dependent, less predictable as the infection progresses and will ultimately be highly dynamic and variable in different infections and also from one epidemic to the next, meaning that herd-immunity is unlikely to be achieved. The potential VSG repertoire is ultimately larger than a single genomic repertoire due to segmental gene conversion and inter-strain mating in the tsetse fly [74]. This genome plasticity also impacts the VSGs expressed in the tsetse fly salivary gland and presents stark challenges in terms of any vaccine strategy that targets VSGs.
VSG recombination does not appear to be naturally triggered by a site-specific nuclease but rather appears to depend upon the inherent instability of subtelomeres [75], [76]. Breaks were shown to arise naturally at telomeric VSG loci, probably due to replication fork collapse, and an artificial, meganuclease-induced DNA break at the active site can trigger a switch [75], [77]. These breaks initiate DNA resection producing ssDNA and triggering a homology search. Once a suitable template is found, DNA can be copied from the template to repair the lesion [76]. Non-homologous DNA repair does not appear to operate in T. brucei, placing an emphasis on RAD51-dependent homologous recombination [63], [78]. This has had a major impact on our ability to manipulate the T. brucei genome and also makes an important contribution to VSG gene rearrangements. There is an alternative form of microhomology-mediated end-joining, however. This end-joining is RAD51-independent and may be particularly effective within 70-bp repeats, thereby making a substantial contribution to the duplicative transposition of VSGs [76].
Nuclear positioning and the chromatin environment of VSGs may be important for VSG recombination [77]. In the bloodstream form, telomeres [79], active [80] and silent VSG expression sites [81] are distributed throughout the nuclear space rather than sequestered at the periphery and this may facilitate homology searching during DNA repair. Notably, the active VSG expression site specifically migrates to the nuclear periphery during differentiation to the insect stage [82].
Switching occurs in only approximately 0.001% of cells per cell division cycle in experimental in vitro culture or during frequent syringe passage but appears to be much higher naturally; switch rate returns to >0.2% following transmission through flies [83] and also apparently increases during growth in vivo in mammals [68], [84]. This transition is not understood but could involve the acquisition of a hyper-labile or hyper-recombinogenic state at the active VSG locus [85]. Rapid switching also operates in metacyclic cells obtained from tsetse-fly salivary glands [86] but in this case, switching does not involve recombination [87].
The vast and dynamic VSG gene family, the large number of subtelomeres in T. brucei and the incomplete sequence coverage currently available for these regions presents challenges but current techniques should now allow some longstanding questions to be addressed. For example, where in the genome are often entirely pseudogene-derived mosaic-VSGs assembled and presumably selected for? Do non-templated VSG mutations [88] naturally contribute to immune evasion? Are novel VSGs typically preserved or permanently lost once successfully used for immune evasion? In terms of this last question, the presence of intact VSGs on minichromosomes [29] could reflect an effective archiving mechanism for novel VSGs. It will also be of interest to determine whether VSG recombination requires or exploits dedicated or specifically modified DNA repair factors, such as BRCA2 with an expanded set of RAD51-interacting repeats [89].
7. Antigenic variation by VSG transcription (in)activation – allelic exclusion
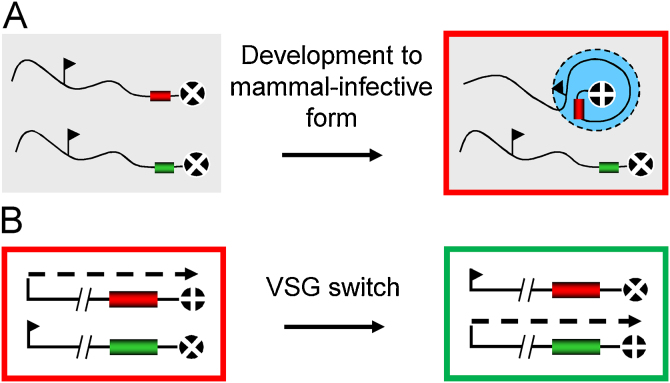
Studies on T. brucei clones with switched VSGs also revealed that some switching events were not associated with duplicative VSG transposition [90]. Pulsed-field gel-electrophoresis, combined with Southern blotting, which had also facilitated ‘mapping’ of the VSG rearrangements described above, provided confirmation of coordinated on and off switching, known as ‘in situ’ switching [28]. This showed that certain telomeric VSGs were reversibly active or repressed and could be subject to transcriptional switching as well as recombination [61], [91]. The in situ switches are classical epigenetic switches since they occur without changes in the DNA sequence [44]. Thus, although gene expression control in trypanosomatids is primarily post-transcriptional, VSG exclusion represents a prominent exception (Fig. 3). Although simultaneous expression of two VSGs from distinct telomeric sites has been reported in T. equiperdum [92], VSG double-expressors arose rarely and were unstable in T. brucei [93]. Thus, VSG allelic exclusion is generally strictly maintained.
Fig. 3.
VSG allelic exclusion is not yet understood. (A) Only one of the telomeric VSGs (red and green) is active and only in the mammalian stages. This associates with a focus of extranucleolar RNA Pol I and other factors (light blue) and produces the VSG coat (outer red box). The ‘+’ and ‘x’ symbols indicate the active and silent telomeres, respectively. (B) Transcription can occasionally switch from one telomeric VSG to another; a coordinated epigenetic switch maintains allelic exclusion. The dashed arrow indicates transcription. Other details as in A.
The mechanism that distinguishes among almost identical VSG expression sites in the bloodstream form suppresses both transcription initiation [94] and elongation [95] and this results in a remarkable 10,000-fold abundance differential among active and silent VSG mRNAs [96]. Despite the distinct sequences of rDNA promoters, these promoters, inserted at VSG expression sites, adopt the transcriptional status of that site [97], [98]. A second VSG inserted at an active VSG expression site is similarly also active [99], [100]. In contrast to this locus-specific control in the bloodstream form, it seems that VSG promoter-specific elements, distinct from rDNA promoters, allow selective down-regulation of all VSG transcription during differentiation to the insect-stage [101].
A novel DNA base, the hydroxylated and glucosylated derivative of thymidine or base-J [102], was first discovered at silent VSG loci. Evidence of J-base came from blocked cleavage by certain restriction enzymes such as PstI as revealed on Southern-blots. Base-J is notably absent from insect-stage T. brucei but, although implicated for many years, no role in antigenic variation has been demonstrated [103]. What is established is that J-base is required for RNA Pol II transcription termination in L. major [104] so it remains possible that J-base also presents a barrier to certain RNA polymerases or DNA polymerases in T. brucei, potentially impacting transcription or DNA recombination and repair, respectively.
Transcription of the single active VSG by RNA Pol I, rather than RNA Pol II, suggested a potential ‘privileged domain’ model for activation based on association with the nucleolus. As it turned out, active VSG transcripts and the active VSG locus were found to be extranucleolar [81]. Like the nucleolus though, this region, known as the expression-site body, is associated with an accumulation of RNA Pol I [80], a trypanosomatid-specific transcription initiation complex known as class I transcription factor A [105] and also a pol I-associated high-mobility group factor [106].
It remains likely that the telomeric environment is important for VSG allelic exclusion as well as for VSG recombination. There is evidence of a role for the telomere-binding protein, RAP1, in VSG silencing [96] and this silencing is also dependent upon chromatin structure [107], [108]; since this topic is covered in a recent review [77] it is not covered in any detail here. Briefly, the histones, histone chaperones, chromatin remodelers, chromatin modifiers, cohesins and nuclear lamina, as well as other chromatin-associated factors, contribute to repression. DOT1B in particular, has a major role in establishing the silent state during in situ switching [109]. Thus, transcription is clearly repressed or attenuated at telomeric VSG loci. Understanding allelic exclusion will remain challenging until we know more about the selection of a single VSG for expression and how this is coordinated with the silencing of all other VSGs.
8. Concluding remarks
The abundance of the active VSG, quite stable yet reversible VSG repression and the ease of genetic manipulation and cell culture mean that T. brucei provides a highly tractable experimental system for the study of monoallelic expression and antigenic variation. There has been tremendous progress in our understanding in this area, how the VSGs are organised and expressed and how expression is switched, through recombination in particular. An ancient and ongoing ‘arms race’ between host immunity and parasite immune evasion has been illuminated through studies on VSGs. The set of monocistronic VSGs first expressed in the tsetse-fly salivary gland facilitates the establishment of a mammalian infection following a blood-meal. The multiplicity of telomeric VSGs with alternative collections of ESAGs may then provide an opportunity to select the optimal expression site for effective nutrition and growth in distinct host environments [51]. Recombination can then allow for VSG switching compatible with continued expression of a favoured set of ESAGs. The vast reservoir of VSG genes allows the presentation of constantly changing epitopes at the cell surface to counter the hosts’ capacity for adaptive immunity.
Access to genome sequence data changed the research landscape, allowing easy access to a vast number of VSG sequences, factors involved in transcription, telomere-binding, recombination and repair and chromatin-based control. There has been an inevitable focus on factors related to those with known functions in other eukaryotes, however, meaning that a lot of territory still remains uncharted in trypanosomatid research. Recent technical advances in areas such as forward genetics [110], proteomics [111], improved access to T. brucei developmental stages in in vitro culture [112] and new and improved technologies to come, will surely now help to deliver answers to some of the outstanding questions.
Acknowledgements
I thank many colleagues for VSG-related discussions over the years, in particular George Cross, Lucy Glover, Sebastian Hutchinson, Sam Alsford, Mark Field, Richard McCulloch, Gloria Rudenko, Miguel Navarro, Martin Taylor, Dave Barry, Piet Borst and John Boothroyd. Research in my laboratory is supported by a Wellcome Trust Senior Investigator Award [100320/Z/12/Z].
References
- 1.Tetley L., Turner C.M., Barry J.D., Crowe J.S., Vickerman K. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J Cell Sci. 1987;87(Pt 2):363–372. doi: 10.1242/jcs.87.2.363. [DOI] [PubMed] [Google Scholar]
- 2.Doyle J.J., Hirumi H., Hirumi K., Lupton E.N., Cross G.A. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology. 1980;80:359–369. doi: 10.1017/s0031182000000810. [DOI] [PubMed] [Google Scholar]
- 3.Bruce D. The Croonian Lectures on trypanosomes causing disease in man and domestic animals in central Africa. Br Med J. 1915;2:5–10. doi: 10.1136/bmj.2.2844.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross R., Thomson D. A case of sleeping sickness showing regular periodical increase of the parasites disclosed. Br Med J. 1910;1:1544–1545. doi: 10.1136/bmj.1.2582.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969;5:163–193. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- 6.Cross G.A., Identification purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 7.Cross G.A. Cross-reacting determinants in the C-terminal region of trypanosome variant surface antigens. Nature. 1979;277:310–312. doi: 10.1038/277310a0. [DOI] [PubMed] [Google Scholar]
- 8.Blum M.L., Down J.A., Gurnett A.M., Carrington M., Turner M.J., Wiley D.C. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- 9.Seyfang A., Mecke D., Duszenko M. Degradation, recycling, and shedding of Trypanosoma brucei variant surface glycoprotein. J Protozool. 1990;37:546–552. doi: 10.1111/j.1550-7408.1990.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 10.Sheader K., Vaughan S., Minchin J., Hughes K., Gull K., Rudenko G. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci USA. 2005;102:8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson M.A., Homans S.W., Dwek R.A., Rademacher T.W. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 12.Masterson W.J., Doering T.L., Hart G.W., Englund P.T. A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson M.A., Williams A.F. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson M.A. Glycosylphosphatidylinositol biosynthesis validated as a drug target for African sleeping sickness. Proc Natl Acad Sci USA. 2000;97:10673–10675. doi: 10.1073/pnas.97.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field M.C., Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- 16.Engstler M., Pfohl T., Herminghaus S., Boshart M., Wiegertjes G., Heddergott N. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 17.Jackson A.P., Allison H.C., Barry J.D., Field M.C., Hertz-Fowler C., Berriman M. A cell-surface phylome for African trypanosomes. PLoS Negl Trop Dis. 2013;7:e2121. doi: 10.1371/journal.pntd.0002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehlert A., Wormald M.R., Ferguson M.A. Modeling of the N-glycosylated transferrin receptor suggests how transferrin binding can occur within the surface coat of Trypanosoma brucei. PLoS Pathog. 2012;8:e1002618. doi: 10.1371/journal.ppat.1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stijlemans B., Caljon G., Natesan S.K., Saerens D., Conrath K., Perez-Morga D. High affinity nanobodies against the Trypanosome brucei VSG are potent trypanolytic agents that block endocytosis. PLoS Pathog. 2011;7:e1002072. doi: 10.1371/journal.ppat.1002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder H.K., Boothroyd J.C., Weber H. Homologous 3′-terminal regions of mRNAs for surface antigens of different antigenic variants of Trypanosoma brucei. Nucleic Acids Res. 1981;9:4745–4753. doi: 10.1093/nar/9.18.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Lange T., Borst P. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature. 1982;299:451–453. doi: 10.1038/299451a0. [DOI] [PubMed] [Google Scholar]
- 22.Williams R.O., Young J.R., Majiwa P.A. Genomic environment of T. brucei VSG genes: presence of a minichromosome. Nature. 1982;299:417–421. doi: 10.1038/299417a0. [DOI] [PubMed] [Google Scholar]
- 23.Bernards A., Michels P.A., Lincke C.R., Borst P. Growth of chromosome ends in multiplying trypanosomes. Nature. 1983;303:592–597. doi: 10.1038/303592a0. [DOI] [PubMed] [Google Scholar]
- 24.Blackburn E.H., Challoner P.B. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell. 1984;36:447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- 25.Van der Ploeg L.H., Liu A.Y., Borst P. Structure of the growing telomeres of trypanosomes. Cell. 1984;36:459–468. doi: 10.1016/0092-8674(84)90239-3. [DOI] [PubMed] [Google Scholar]
- 26.Munoz-Jordan J.L., Cross G.A., de Lange T., Griffith J.D. t-loops at trypanosome telomeres. Embo J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreesen O., Li B., Cross G.A. Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol. 2007;5:70–75. doi: 10.1038/nrmicro1577. [DOI] [PubMed] [Google Scholar]
- 28.Van der Ploeg L.H., Schwartz D.C., Cantor C.R., Borst P. Antigenic variation in Trypanosoma brucei analyzed by electrophoretic separation of chromosome-sized DNA molecules. Cell. 1984;37:77–84. doi: 10.1016/0092-8674(84)90302-7. [DOI] [PubMed] [Google Scholar]
- 29.Wickstead B., Ersfeld K., Gull K. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 2004;14:1014–1024. doi: 10.1101/gr.2227704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson A.P., Berry A., Aslett M., Allison H.C., Burton P., Vavrova-Anderson J. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc Natl Acad Sci USA. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callejas S., Leech V., Reitter C., Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 34.Berberof M., Vanhamme L., Tebabi P., Pays A., Jefferies D., Welburn S. The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. Embo J. 1995;14:2925–2934. doi: 10.1002/j.1460-2075.1995.tb07292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Ploeg L.H., Liu A.Y., Michels P.A., De Lange T., Borst P., Majumder H.K. RNA splicing is required to make the messenger RNA for a variant surface antigen in trypanosomes. Nucleic Acids Res. 1982;10:3591–3604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boothroyd J.C., Cross G.A. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 37.Kooter J.M., De Lange T., Borst P. Discontinuous synthesis of mRNA in trypanosomes. Embo J. 1984;3:2387–2392. doi: 10.1002/j.1460-2075.1984.tb02144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borst P. Discontinuous transcription and antigenic variation in trypanosomes. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 39.Liu A.Y., Van der Ploeg L.H., Rijsewijk F.A., Borst P. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences. J Mol Biol. 1983;167:57–75. doi: 10.1016/s0022-2836(83)80034-5. [DOI] [PubMed] [Google Scholar]
- 40.Cully D.F., Ip H.S., Cross G.A. Coordinate transcription of variant surface glycoprotein genes and an expression site associated gene family in Trypanosoma brucei. Cell. 1985;42:173–182. doi: 10.1016/s0092-8674(85)80113-6. [DOI] [PubMed] [Google Scholar]
- 41.Pays E., Tebabi P., Pays A., Coquelet H., Revelard P., Salmon D. The genes and transcripts of an antigen gene expression site from T. brucei. Cell. 1989;57:835–845. doi: 10.1016/0092-8674(89)90798-8. [DOI] [PubMed] [Google Scholar]
- 42.Kooter J.M., Borst P. α-Amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 1984;12:9457–9472. doi: 10.1093/nar/12.24.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imboden M.A., Laird P.W., Affolter M., Seebeck T. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res. 1987;15:7357–7368. doi: 10.1093/nar/15.18.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zomerdijk J.C., Ouellette M., ten Asbroek A.L., Kieft R., Bommer A.M., Clayton C.E. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. Embo J. 1990;9:2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson P.J., Kooter J.M., Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 47.Zomerdijk J.C., Kieft R., Shiels P.G., Borst P. α-Amanitin-resistant transcription units in trypanosomes: a comparison of promoter sequences for a VSG gene expression site and for the ribosomal RNA genes. Nucleic Acids Res. 1991;19:5153–5158. doi: 10.1093/nar/19.19.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alarcon C.M., Son H.J., Hall T., Donelson J.E. A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol Cell Biol. 1994;14:5579–5591. doi: 10.1128/mcb.14.8.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilinger G., Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegel T.N., Hekstra D.R., Kemp L.E., Figueiredo L.M., Lowell J.E., Fenyo D. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitter W., Gerrits H., Kieft R., Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 52.Xong H.V., Vanhamme L., Chamekh M., Chimfwembe C.E., Van Den Abbeele J., Pays A. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 53.Uzureau P., Uzureau S., Lecordier L., Fontaine F., Tebabi P., Homble F. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 2013;501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 54.Salmon D., Vanwalleghem G., Morias Y., Denoeud J., Krumbholz C., Lhomme F. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science. 2012;337:463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- 55.Horn D., McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol. 2010;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoeijmakers J.H., Frasch A.C., Bernards A., Borst P., Cross G.A. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature. 1980;284:78–80. doi: 10.1038/284078a0. [DOI] [PubMed] [Google Scholar]
- 57.Williams R.O., Young J.R., Majiwa P.A. Genomic rearrangements correlated with antigenic variation in Trypanosoma brucei. Nature. 1979;282:847–849. doi: 10.1038/282847a0. [DOI] [PubMed] [Google Scholar]
- 58.Pays E., Delronche M., Lheureux M., Vervoort T., Bloch J., Gannon F. Cloning and characterization of DNA sequences complementary to messenger ribonucleic acids coding for the synthesis of two surface antigens of Trypanosoma brucei. Nucleic Acids Res. 1980;8:5965–5981. doi: 10.1093/nar/8.24.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pays E., Lheureux M., Steinert M. The expression-linked copy of a surface antigen gene in Trypanosoma is probably the one transcribed. Nature. 1981;292:265–267. doi: 10.1038/292265a0. [DOI] [PubMed] [Google Scholar]
- 60.Borst P., Frasch A.C., Bernards A., Van der Ploeg L.H., Hoeijmakers J.H., Arnberg A.C. DNA rearrangements involving the genes for variant antigens in Trypanosoma brucei. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):935–943. doi: 10.1101/sqb.1981.045.01.110. [DOI] [PubMed] [Google Scholar]
- 61.Bernards A., De Lange T., Michels P.A., Liu A.Y., Huisman M.J., Borst P. Two modes of activation of a single surface antigen gene of Trypanosoma brucei. Cell. 1984;36:163–170. doi: 10.1016/0092-8674(84)90085-0. [DOI] [PubMed] [Google Scholar]
- 62.Scholler J.K., Myler P.J., Stuart K.D. A novel telomeric gene conversion in Trypanosoma brucei. Mol Biochem Parasitol. 1989;35:11–19. doi: 10.1016/0166-6851(89)90137-0. [DOI] [PubMed] [Google Scholar]
- 63.McCulloch R., Barry J.D. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcello L., Barry J.D. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pays E., Guyaux M., Aerts D., Van Meirvenne N., Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- 66.Kim H.S., Cross G.A. TOPO3α influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrison L.J., Majiwa P., Read A.F., Barry J.D. Probabilistic order in antigenic variation of Trypanosoma brucei. Int J Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 68.Timmers H.T., de Lange T., Kooter J.M., Borst P. Coincident multiple activations of the same surface antigen gene in Trypanosoma brucei. J Mol Biol. 1987;194:81–90. doi: 10.1016/0022-2836(87)90717-0. [DOI] [PubMed] [Google Scholar]
- 69.Laurent M., Pays E., Van der Werf A., Aerts D., Magnus E., Van Meirvenne N. Translocation alters the activation rate of a trypanosome surface antigen gene. Nucleic Acids Res. 1984;12:8319–8328. doi: 10.1093/nar/12.22.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernards A., Van der Ploeg L.H., Frasch A.C., Borst P., Boothroyd J.C., Coleman S. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]
- 71.Hall J.P., Wang H., Barry J.D. Mosaic VSGs and the scale of Trypanosoma brucei antigenic variation. PLoS Pathog. 2013;9:e1003502. doi: 10.1371/journal.ppat.1003502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pays E., Van Assel S., Laurent M., Darville M., Vervoort T., Van Meirvenne N. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell. 1983;34:371–381. doi: 10.1016/0092-8674(83)90371-9. [DOI] [PubMed] [Google Scholar]
- 73.Roth C.W., Longacre S., Raibaud A., Baltz T., Eisen H. The use of incomplete genes for the construction of a Trypanosoma equiperdum variant surface glycoprotein gene. Embo J. 1986;5:1065–1070. doi: 10.1002/j.1460-2075.1986.tb04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peacock L., Bailey M., Carrington M., Gibson W. Meiosis and haploid gametes in the pathogen Trypanosoma brucei. Curr Biol. 2014;24:181–186. doi: 10.1016/j.cub.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boothroyd C.E., Dreesen O., Leonova T., Ly K.I., Figueiredo L.M., Cross G.A. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glover L., Alsford S., Horn D. DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog. 2013;9:e1003260. doi: 10.1371/journal.ppat.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glover L., Hutchinson S., Alsford S., McCulloch R., Field M.C., Horn D. Antigenic variation in African trypanosomes: the importance of chromosomal and nuclear context in VSG expression control. Cell Microbiol. 2013;15:1984–1993. doi: 10.1111/cmi.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glover L., McCulloch R., Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perez-Morga D., Amiguet-Vercher A., Vermijlen D., Pays E. Organization of telomeres during the cell and life cycles of Trypanosoma brucei. J Eukaryot Microbiol. 2001;48:221–226. doi: 10.1111/j.1550-7408.2001.tb00306.x. [DOI] [PubMed] [Google Scholar]
- 80.Navarro M., Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 81.Chaves I., Zomerdijk J., Dirks-Mulder A., Dirks R.W., Raap A.K., Borst P. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95:12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Landeira D., Navarro M. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol. 2007;176:133–139. doi: 10.1083/jcb.200607174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turner C.M. The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol Lett. 1997;153:227–231. doi: 10.1111/j.1574-6968.1997.tb10486.x. [DOI] [PubMed] [Google Scholar]
- 84.Lee M.G., Van der Ploeg L.H. Frequent independent duplicative transpositions activate a single VSG gene. Mol Cell Biol. 1987;7:357–364. doi: 10.1128/mcb.7.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hovel-Miner G.A., Boothroyd C.E., Mugnier M., Dreesen O., Cross G.A., Papavasiliou F.N. Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog. 2012;8:e1002900. doi: 10.1371/journal.ppat.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Esser K.M., Schoenbechler M.J. Expression of two variant surface glycoproteins on individual African trypanosomes during antigen switching. Science. 1985;229:190–193. doi: 10.1126/science.3892689. [DOI] [PubMed] [Google Scholar]
- 87.Lenardo M.J., Esser K.M., Moon A.M., Van der Ploeg L.H., Donelson J.E. Metacyclic variant surface glycoprotein genes of Trypanosoma brucei subsp. rhodesiense are activated in situ, and their expression is transcriptionally regulated. Mol Cell Biol. 1986;6:1991–1997. doi: 10.1128/mcb.6.6.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu Y., Alarcon C.M., Hall T., Reddy L.V., Donelson J.E. A strand bias occurs in point mutations associated with variant surface glycoprotein gene conversion in Trypanosoma rhodesiense. Mol Cell Biol. 1994;14:3971–3980. doi: 10.1128/mcb.14.6.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hartley C.L., McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young J.R., Donelson J.E., Majiwa P.A., Shapiro S.Z., Williams R.O. analysis of genomic rearrangements associated with two variable antigen genes of Trypanosoma brucei. Nucleic Acids Res. 1982;10:803–819. doi: 10.1093/nar/10.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myler P., Nelson R.G., Agabian N., Stuart K. Two mechanisms of expression of a predominant variant antigen gene of Trypanosoma brucei. Nature. 1984;309:282–284. doi: 10.1038/309282a0. [DOI] [PubMed] [Google Scholar]
- 92.Baltz T., Giroud C., Baltz D., Roth C., Raibaud A., Eisen H. Stable expression of two variable surface glycoproteins by cloned Trypanosoma equiperdum. Nature. 1986;319:602–604. doi: 10.1038/319602a0. [DOI] [PubMed] [Google Scholar]
- 93.Chaves I., Rudenko G., Dirks-Mulder A., Cross M., Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. Embo J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nguyen T.N., Muller L.S., Park S.H., Siegel T.N., Gunzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res. 2014;42:3164–3176. doi: 10.1093/nar/gkt1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vanhamme L., Poelvoorde P., Pays A., Tebabi P., Van Xong H., Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 96.Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Horn D., Cross G.A. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 98.Rudenko G., Blundell P.A., Dirks-Mulder A., Kieft R., Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 99.Munoz-Jordan J.L., Davies K.P., Cross G.A. Stable expression of mosaic coats of variant surface glycoproteins in Trypanosoma brucei. Science. 1996;272:1795–1797. doi: 10.1126/science.272.5269.1795. [DOI] [PubMed] [Google Scholar]
- 100.Smith T.K., Vasileva N., Gluenz E., Terry S., Portman N., Kramer S. Blocking variant surface glycoprotein synthesis in Trypanosoma brucei triggers a general arrest in translation initiation. PLoS ONE. 2009;4:e7532. doi: 10.1371/journal.pone.0007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horn D., Cross G.A. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. Embo J. 1997;16:7422–7431. doi: 10.1093/emboj/16.24.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Borst P., Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu Rev Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 103.Cliffe L.J., Siegel T.N., Marshall M., Cross G.A., Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Luenen H.G., Farris C., Jan S., Genest P.A., Tripathi P., Velds A. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brandenburg J., Schimanski B., Nogoceke E., Nguyen T.N., Padovan J.C., Chait B.T. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. Embo J. 2007;26:4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narayanan M.S., Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41:2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Figueiredo L.M., Cross G.A. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9:148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stanne T.M., Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot Cell. 2010;9:136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Figueiredo L.M., Janzen C.J., Cross G.A. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K.F. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Urbaniak M.D., Guther M.L., Ferguson M.A. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS ONE. 2012;7:e36619. doi: 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kolev N.G., Ramey-Butler K., Cross G.A., Ullu E., Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]