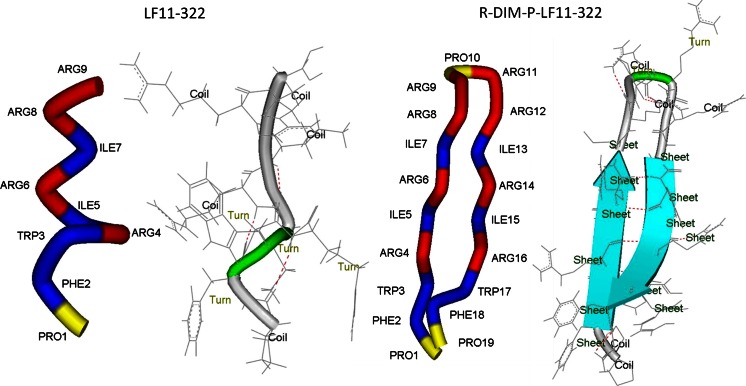
Fig. 6.
Secondary structure predictions for peptide LF11-322 and R-DIM-P-LF11-322 were performed by use of the program PEP-FOLD (Maupetit et al. 2009, 2010; Thévenet et al. 2012). The best model was plotted with DS ViewerPro 5.0 for Windows. On the particular left sides the amino acids are colored corresponding to their hydrophobicity according to the whole residue (octanol) hydrophobicity scale by Wimley and White (Wimley and White 1996). (hydrophilic amino acids are colored in red, hydrophobic amino acids are colored in blue, slightly charged amino acid Pro is colored in yellow). On the particular right sides predicted secondary structures are plotted and delineated, predicted hydrogen bonds are indicated as red dashed lines