ABSTRACT
Three Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamase (ESBL) were obtained from three dairy cows with clinical mastitis in two farms in western Japan. Two of the 3 isolates from cows in different farms were able to transfer plasmids carrying the blaCTX-M-2 gene to Escherichia coli recipient. Pulsed-field gel electrophoresis (PFGE) patterns of the 2 isolates were different from each other, although restricted-fragment patterns of the two conjugative plasmids were similar to each other. Additionally, PCR-based replicon typing revealed that both the plasmids belonged to type Inc.T. These results suggest that ESBL-encoding genes can be distributed in bacteria on dairy farms through the plasmids.
Keywords: bovine mastitis, ESBL, Klebsiella pneumonia
Bovine mastitis due to Klebsiella pneumoniae causes a significant drop in milk production and high mortality of the affected cows [9]. Recently, K. pneumoniae strains that produce extended-spectrum beta-lactamase (ESBL) were isolated from cases of bovine mastitis in Europe [6, 13]. ESBLs hydrolyze not only 1st- and 2nd-generation cephalosporins but also 3rd- and 4th-generation cephalosporins, and association of ESBL genes with transferable plasmids has often been reported [8]. The emergence of bacteria that produce ESBLs in domestic animals is a threat to animal hygiene and public health [16]. In the present study, we isolated K. pneumoniae from 20 dairy cows with clinical mastitis in multiple farms in western Japan. Three of the isolates were found to be resistant to ceftiofur, a 3rd-generation cephalosporin. Thus, the isolates were further characterized.
From April to September in 2011, milk samples were obtained from 20 dairy cows with clinical mastitis that were raised on 6 farms (farms A, B, C, E, F and G) located in western Japan. All the samples were collected before starting antimicrobial treatment. Rearing practices and the treatment for mastitis were reported from the farms. Farms A and C were located within 1.5 km of each other. The other 4 farms were more than 7 km west of farms A and C. Clinical symptoms observed included heat, induration and swelling of udders, fever, anorexia and reduced milk production. Aerobic culture on DHL agar plates yielded a heavy growth of pinkish red colonies in pure culture. These colonies were identified as K. pneumoniae by the API 20E test (bioMérieux, Marcy l’Etoile, France). A representative isolate was selected randomly from 1 sample, and a total of 20 isolates were used for further examinations. All the isolates were subtyped using pulsed-field gel electrophoresis (PFGE) with XbaI-digested chromosomal DNAs as described elsewhere [17].
Minimum inhibitory concentrations (MICs) were determined by the agar dilution method using Mueller-Hinton agar (Becton, Dickinson and Co., Sparks, MD, U.S.A.) plates according to the recommendations given in document M07-A8 of the Clinical and Laboratory Standards Institute (CLSI) [4] and interpreted using the resistance breakpoints defined in the previous study [15] as follows: ampicillin (AMP) and cefazolin (CEZ), 32 µg/ml; ceftiofur (CTF), 8 µg/ml; dihydrostreptomycin (DSM), 32 µg/ml; gentamicin (GEN), 16 µg/ml; kanamycin (KAN), 64 µg/ml; oxytetracycline (OTC), 16 µg/ml; chloramphenicol (CHL), 32 µg/ml; nalidixic acid (NAL), 32 µg/ml; enrofloxacin (ERFX), 2 µg/ml; and trimethoprim (TMP), 16 µg/ml. Each antibiotic reagent, except for CTF, was purchased from Sigma-Aldrich Co. LLC. (St. Louis, MO, U.S.A.). CTF was obtained from Pfizer Inc. (Tokyo, Japan). ESBL-production among CTF-resistant isolates was screened and confirmed using the disc diffusion method based on the criteria provided by the CLSI [5]. Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality control strains.
ESBL-producing strains were tested by PCR targeting the blaCTX-M gene using the specific primer set as follows: (CTX-M-1: 5′-GACGATGTCACTGGCTGAGC-3′ and 5′-AGCCGCCGACGCTAATACA-3′, CTX-M-2: 5′-GCGACCTGGTTAACTAACAATC-3′ and 5′-CGGTAGTATTGCCCTTAAGCC-3′, CTX-M-3: 5′-CGCTTTGCCATGTGCAGCACC-3′ and 5′-GCTCAGTACGATCGAGCC-3′, CTX-M-4: 5′-GCTGGAGAAAAGCAGCGGAG-3′ and 5′-GTAAGCTGACGCAACGTCTG-3′, CTX-M-5: 5′-ATGATGAGAAAAAGCGTAAGG-3′ and 5′-TTAATAACCGTCGGTGAC-3′). Amplified products were sequenced using the same primer pair.
Plasmid DNAs were extracted from ESBL-producing strains using Plasmid Midi Kit (Qiagen, Hilden, Germany) and electrophoresed in an agarose gel in Tris-acetate-EDTA buffer. For southern blot analysis, PCR-amplified ESBL gene fragments from one of the ESBL-producing strains, K. pneumoniae A46 (see below or Table 1), were labeled with digoxigenin using DIG High Prime Labeling and Detection Starter Kit (Roche, Indianapolis, IN, U.S.A.).
Table 1. Characteristics of K. pneumoniae isolates obtained from milk samples of mastitic dairy cows.
| Isolate* | Isolation date | MIC value
(µg/ml)** |
ESBL bla gene | Conjugation to E. coli | |||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | CEZ | CTF | DSM | KAN | OTC | ||||
| A45 | Aug 9, 2011 | 32 | 4 | 0.25 | 2 | 2 | 0.25 | n/a*** | n/a*** |
| A46 | Aug 17, 2011 | 512 | 256 | 8 | 128 | 2 | 2 | blaCTX-M-2 | + |
| A48 | Aug 22, 2011 | 32 | 1 | 0.5 | 128 | 2 | 2 | n/a | n/a |
| A49 | Aug 22, 2011 | 16 | 1 | 0.5 | 256 | 4 | 2 | n/a | n/a |
| B29 | Jul 1, 2011 | 16 | 4 | 0.5 | 128 | 2 | 2 | n/a | n/a |
| B30 | Jul 5, 2011 | 32 | 2 | 0.5 | 256 | 2 | 512 | n/a | n/a |
| B31 | Jul 13, 2011 | 32 | 1 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B32 | Jul 13, 2011 | 32 | 1 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B34 | Jul 14, 2011 | 32 | 2 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B35 | Jul 15, 2011 | 32 | 1 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B36 | Jul 15, 2011 | 32 | 1 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B40 | Jul 22, 2011 | 32 | 1 | 0.5 | 256 | 2 | >512 | n/a | n/a |
| B44 | Aug 16, 2011 | 32 | 2 | 0.5 | 128 | 2 | 512 | n/a | n/a |
| B50 | Sep 15, 2011 | 32 | 2 | 1 | 256 | 2 | 512 | n/a | n/a |
| B51 | Sep 15, 2011 | 32 | 2 | 1 | 256 | 2 | 512 | n/a | n/a |
| C22 | Apr 21, 2011 | 512 | 128 | 8 | 128 | 2 | 1 | blaCTX-M-2 | - |
| C52 | Sep 21, 2011 | >512 | 256 | 16 | 128 | 2 | 2 | blaCTX-M-2 | + |
| E33 | Jul 13, 2011 | 32 | 256 | 1 | 64 | 2 | 2 | n/a | n/a |
| F42 | Jul 26, 2011 | 256 | 1 | 0.5 | 512 | 512 | 256 | n/a | n/a |
| G53 | Sep 20, 2011 | 32 | 1 | 0.5 | 2 | 2 | 2 | n/a | n/a |
*The first letter (alphabet) indicates each of the farms where cows were raised. **AMP, ampicilin; CEZ, cefazolin; CTF, ceftiofur; DSM, dihydrostreptomycin; OTC, oxytetracycline; KAN, kanamycin, Underlined numbers in boldface represent resistance to each of the antimicrobials. ***Not applicable.
In the transfer experiments [17], ESBL-producing strains were used as donor strains, and E. coli ML1410 was used as a recipient. Overnight cultures of donor and recipient strains were co-cultured in freshly prepared broth and incubated at 37°C for 6 hr. Transconjugants were selected on DHL plates containing 50 µg/ml of AMP and 25 µg/ml of NAL. To compare restriction fragment length patterns (RFLPs) of plasmids conjugated into E. coli ML1410, the plasmid DNAs were extracted, digested with DraI or PstI and electrophoresed. These RFLPs were compared with that of the plasmid harboring the blaCTX-M-2 gene originated from E. coli strain D1633 that was isolated from a commercial broiler chicken with colibacillosis. Plasmids of parent ESBL-producing strains and the transconjugants were typed by PCR-based replicon typing [3].
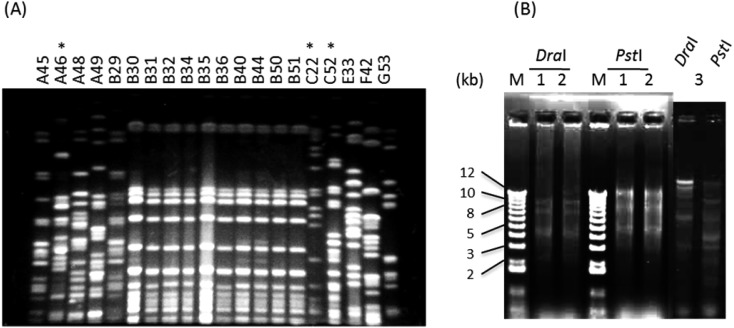
Among the 20 isolates, a total of 11 PFGE patterns were distinguished by more than 6-band differences (Fig. 1A). Ten of 11 isolates obtained from cows on farm B had identical PFGE patterns. Rates of resistance to AMP and DSM were 90% and 90%, respectively (Table 1). All the isolates were susceptible to GEN (0.5–1), CHL (2–8), NAL (4–16), ERFX (<0.13) and TMP (0.5–2) with the MIC ranges (µg/ml) described in parentheses. Three isolates (A46, C22 and C52) that were resistant to CTF (MICs: 8, 8 and 16 µg/ml, respectively) were confirmed to be ESBL producers and harbored the ESBL-encoding gene blaCTX-M-2. A southern blot analysis confirmed that the gene was located on plasmids with more than 50 kb plasmids in size (data not shown). Transferability tests demonstrated that 2 of the 3 ESBL-producing K. pneumoniae isolates (A46 and C52) were able to transfer the R plasmids to the E. coli recipient (Table 1). DraI- and PstI-digested patterns of the plasmids obtained from 2 transconjugants were apparently similar to each other and were distinguishable from those of the plasmid obtained from E. coli strain D1633 (Fig. 1B). The plasmids from the two transconjugants, which were derived from A46 and C52, and those from the parental strains were confirmed to be type Inc.T. On the other hand, the plasmid from strain C22 belonged to type Inc.FIC.
Fig. 1.
(A) PFGE patterns of XbaI-digested chromosomal DNA of Klebsiella pneumoniae isolates obtained from milk samples of mastitic dairy cows. The isolates are described in Table 1. Asterisks above the isolate number indicate ESBL-producing isolates. (B) Restriction profiles of DraI- and PstI-digested plasmids from blaCTX-M-2-positive transconjugants. Lanes 1 and 2, plasmids from K. pneumoniae A46 and C52, respectively, conjugated into E. coli. Lane 3, plasmids from E. coli strain D1633, conjugated into ML1410 laboratory strain.
In the present study, three isolates of ESBL-producing K. pneumoniae were obtained from cows with clinical mastitis that were reared on 2 farms (farms A and C). The blaCTX-M-2 gene was detected in transferable plasmids in 2 of the 3 ESBL-producing K. pneumoniae. The PFGE patterns of the 2 isolates were distinguishable, although the 2 conjugative plasmids carrying the blaCTX-M-2 gene had similar RFLP profiles and belonged to an identical replicon type. These results suggest that ESBL genes can be distributed in bacteria on dairy farms through the plasmids. According to the reports from the farms, animals in farm A were administered cefazolin and cefuroxime for 6 and 9 days, respectively, and those in farm C were administered cefazolin, cefuroxime and cefapirin for 3 days. The 3 cows infected with ESBL-producing K. pneumoniae were not recovered. Prescriptions or information in the other farms were unavailable. The farmers and employees in farms A and C did not visit each other, and no common veterinarians attended animals in both the farms. Companies supplied cattle feed to these farms were different from each other. Other environmental factors may contribute to occurrence of the R-plasmid carrying the blaCTX-M-2 gene in K. pneumoniae, because the geographical distance between farms A and C was relatively close. Dahmen et al. [6] showed low molecular diversity of ESBL-producing strains causing cattle mastitis in France with respect to ESBL genes, plasmids and clones. In a study of ESBL-producing K. pneumoniae and E. coli isolates obtained from rectal samples of broiler chickens from different farms in Japan [10], it was suggested that transferable plasmids carrying the blaCTX-M-2 or blaCTX-M-14 gene are distributed among those species.
In the present study, 10 of 11 isolates obtained from cows in farm B had identical PFGE patterns, suggesting that the strain with this PFGE profile had spread clonally. Munoz et al. [14] reported that one K. pneumoniae mastitis outbreak on a single dairy farm was attributed to contagious transmission or to exposure of multiple cows to a common source, because the isolates had identical random amplified polymorphic DNA (RAPD) types.
E. coli harboring the blaCTX-M-14 gene was detected in milk from mastitic cows in Switzerland [7]. The blaSHV-12 and blaCTX-M-15 genes were reported in Enterobacter and Klebsiella isolates from clinical bovine mastitis in Egypt [1]. According to reports of monitoring of ESBL-producing bacteria from food-producing animals in Japan, K. pneumoniae harboring the blaSHV-60 gene were isolated from raw milk from a healthy cow [10], and bovine isolates of E. coli with the blaCTX-M-2 gene were obtained from 1.5% (6/396) of fecal samples and surfaces of carcasses of cattle (0.7%, 2/270) [16] and animals with colibacillosis (2.8%, 2/72) [2]. In Japan, cephalosporins including ceftiofur are approved for parenteral use in cattle. Although the association of the use of ceftiofur with the emergence of EBSL producers in cattle is controversial [11, 12, 16, 18], continuous monitoring and molecular characterization of ESBL producers remain important to reveal the factors involved in introducing these strains into farms and the genes responsible for the resistance.
REFERENCES
- 1.Ahmed A. M., Shimamoto T.2011. Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 55: 318–327. doi: 10.1111/j.1348-0421.2011.00323.x [DOI] [PubMed] [Google Scholar]
- 2.Asai T., Masani K., Sato C., Hiki M., Usui M., Baba K., Ozawa M., Harada K., Aoki H., Sawada T.2011. Phylogenetic groups and cephalosporin resistance genes of Escherichia coli from diseased food-producing animals in Japan. Acta Vet. Scand. 53: 52. doi: 10.1186/1751-0147-53-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J.2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63: 219–228. doi: 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2010. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Eighth Edition (M07-A8). CLSI, Wayne, PA, U.S.A. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute2010. Performance Standards for Antimicrobial Susceptibility Testing. Twentieth Informational (Supplement): M100–S20 CLSI, Wayne, PA, U.S.A. [Google Scholar]
- 6.Dahmen S., Metayer V., Gay E., Madec J. Y., Haenni M.2013. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 162: 793–799. doi: 10.1016/j.vetmic.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 7.Geser N., Stephan R., Hachler H.2012. Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8: 21. doi: 10.1186/1746-6148-8-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gniadkowski M., Palucha A., Grzesiowski P., Hryniewicz W.1998. Outbreak of ceftazidime-resistant Klebsiella pneumoniae in a pediatric hospital in Warsaw, Poland: clonal spread of the TEM-47 extended-spectrum beta-lactamase (ESBL)-producing strain and transfer of a plasmid carrying the SHV-5-like ESBL-encoding gene. Antimicrob. Agents Chemother. 42: 3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gröhn Y. T., Wilson D. J., Gonzalez R. N., Hertl J. A., Schulte H., Bennett G., Schukken Y. H.2004. Effect of pathogen-specific clinical mastitis on milk yield in dairy cows. J. Dairy Sci. 87: 3358–3374. doi: 10.3168/jds.S0022-0302(04)73472-4 [DOI] [PubMed] [Google Scholar]
- 10.Hammad A. M., Ahmed A. M., Ishida Y., Shimamoto T.2008. First characterization and emergence of SHV-60 in raw milk of a healthy cow in Japan. J. Vet. Med. Sci. 70: 1269–1272. doi: 10.1292/jvms.70.1269 [DOI] [PubMed] [Google Scholar]
- 11.Hiroi M., Yamazaki F., Harada T., Takahashi N., Iida N., Noda Y., Yagi M., Nishio T., Kanda T., Kawamori F., Sugiyama K., Masuda T., Hara-Kudo Y., Ohashi N.2012. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 74: 189–195. doi: 10.1292/jvms.11-0372 [DOI] [PubMed] [Google Scholar]
- 12.Kojima A., Ishii Y., Ishihara K., Esaki H., Asai T., Oda C., Tamura Y., Takahashi T., Yamaguchi K.2005. Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49: 3533–3537. doi: 10.1128/AAC.49.8.3533-3537.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locatelli C., Scaccabarozzi L., Pisoni G., Moroni P.2010. CTX-M1 ESBL-producing Klebsiella pneumoniae subsp. pneumoniae isolated from cases of bovine mastitis. J. Clin. Microbiol. 48: 3822–3823. doi: 10.1128/JCM.00941-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz M. A., Welcome F. L., Schukken Y. H., Zadoks R. N.2007. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York State. J. Clin. Microbiol. 45: 3964–3971. doi: 10.1128/JCM.00795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozaki H., Esaki H., Takemoto K., Ikeda A., Nakatani Y., Someya A., Hirayama N., Murase T.2011. Antimicrobial resistance in fecal Escherichia coli isolated from growing chickens on commercial broiler farms. Vet. Microbiol. 150: 132–139. doi: 10.1016/j.vetmic.2010.12.020 [DOI] [PubMed] [Google Scholar]
- 16.Shiraki Y., Shibata N., Doi Y., Arakawa Y.2004. Escherichia coli producing CTX-M-2 beta-lactamase in cattle, Japan. Emerg. Infect. Dis. 10: 69–75. doi: 10.3201/eid1001.030219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Someya A., Otsuki K., Murase T.2005. Antimicrobial susceptibilities of Salmonella isolates obtained from layer chicken houses on a commercial egg-producing farm in Japan, 1997 to 2002. J. Food Prot. 68: 2030–2034 [DOI] [PubMed] [Google Scholar]
- 18.Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V.2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45: 2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]