Abstract
Summary: A number of technologies, including CRISPR/Cas, transcription activator-like effector nucleases and zinc-finger nucleases, allow the user to target a chosen locus for genome editing or regulatory interference. Specificity, however, is a major problem, and the targeted locus must be chosen with care to avoid inadvertently affecting other loci (‘off-targets’) in the genome. To address this we have created ‘Genome Target Scan’ (GT-Scan), a flexible web-based tool that ranks all potential targets in a user-selected region of a genome in terms of how many off-targets they have. GT-Scan gives the user flexibility to define the desired characteristics of targets and off-targets via a simple ‘target rule’, and its interactive output allows detailed inspection of each of the most promising candidate targets. GT-Scan can be used to identify optimal targets for CRISPR/Cas systems, but its flexibility gives it potential to be adapted to other genome-targeting technologies as well.
Availability and implementation: GT-Scan can be run via the web at: http://gt-scan.braembl.org.au.
Contact: t.bailey@uq.edu.au
1 INTRODUCTION
Targeted genome editing and targeted transcriptional control are essential tools of modern molecular biology. Currently popular approaches include CRISPR-Cas (Jinek et al., 2012), transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs) (Miller et al., 2007). Each of these technologies allows the user to design a system to target a specific genomic sequence. However, because of the limited or uncertain sequence specificity of technologies based on CRISPR/Cas (Hsu et al., 2013), TALENs and ZFNs (Gabriel et al., 2011), it is crucial that the intended genomic target be as unique as possible within the genome. Ensuring target uniqueness will reduce the number of ‘off-targets’ in the genome and minimize unintended genome editing or regulatory effects.
The first step in the effective use of any genome targeting technology is therefore to identify the possible target(s) within the region of interest (e.g. gene, exon or promoter) that have the fewest off-targets. The ideal targets are subsequences within the region of interest that have no exact copies elsewhere in the genome. Because targeting affinity is a continuum, near-exact copies that differ at one or a few positions from the intended target are also undesirable (Jinek et al., 2012). Furthermore, the positions of the mismatches in off-targets may also affect their affinity. For example, in the CRISPR/Cas system, mismatches in 5′ positions are generally less disruptive than those in 3′ positions (Jiang et al., 2013). An exception is a single nucleotide near the 3′ end of the target, which has little or no effect on specificity, regardless of the base pair present in that position (Mali et al., 2013). In ZFN targets, a central ‘spacer’ has little affect on affinity. Target selection should therefore consider both exact and near-exact matches to the target as off-targets, as well as the positions of mismatches in off-targets.
The particular genome editing technology being used may place other constraints on the ideal targets in addition to uniqueness. For example, CRISPR/Cas targets must contain a particular short sequence at the 3′ end called a protospacer adjacent motif (PAM) (Mali et al., 2013). Target selection must consider such constraints as well.
2 GT-SCAN
We have developed Genomic Target Scan (GT-Scan) to aid in the selection of optimal genomic targets for genome editing or transcriptional control via systems based on CRISPR/Cas and other systems. GT-Scan is a flexible web-based tool that scans a user-defined genomic region for candidate targets and ranks them in terms of the number of exact or approximate off-targets in the genome. GT-Scan allows the user to define a ‘target rule’ in a simple format that specifies target length, constrained positions and positions with high-, low- or no-target (and off-target) specificity. GT-Scan’s output is interactive, allowing the user to examine candidate targets and the characteristics of their potential off-targets (number of mismatches, positions of mismatches, genomic locations). The GT-Scan website currently supports target selection in >25 Ensembl genomes.
To use GT-Scan, the user selects the appropriate genome from a menu and provides the DNA sequence of the genomic region in which they wish to identify optimal targets. They then choose a ‘rule-pair’ consisting of a ‘target rule’ and an ‘off-target filter’, or they can create their own custom rule-pair. Candidate targets are positions in the given genomic region (on either strand) that match the target rule. For each candidate target, GT-scan reports all potential off-targets in the genome that have no more than three mismatches with the candidate target and match the off-target filter. The user can also separately limit the number of high-specificity mismatches in off-targets.
The target rule defines high-, low- and no-specificity positions in candidate targets, using upper- and lower-case letters from the alphabet ACGTNX. To illustrate, consider the 23 nt rule-pair
which is appropriate for CRISPR/Cas9 systems. This target rule specifies that candidate targets must be 23 nt long and end with the -NGG PAM. It also encodes the information that the 10 most 5′ positions are low-specificity positions, and the following 13 are high-specificity positions, except for the special character N, which denotes a ‘no-specificity’ position. The off-target filter is composed of letters in the standard IUPAC alphabet for DNA, and the letter case has no effect. With the given off-target filter, GT-Scan ignores any potential off-target not ending with either an -NGG or -NAG PAM.
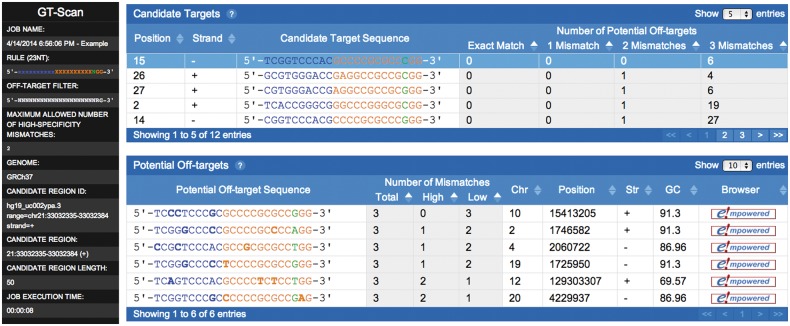
An example of the interactive HTML output produced by GT-Scan is shown in Figure 1. The job details on the left show that the user provided GT-Scan with a 50 bp region of DNA and specified scanning of the Ensembl human genome version GRCh37. GT-Scan detected that the candidate region matches the reference strand of chromosome 21 from position 33 032 335 to 33 032 384. GT-Scan ran for 8 s and found and evaluated 12 candidate targets within the candidate region that match the 23 bp rule chosen by the user. The best candidate target found by GT-Scan is unique in the human genome (0 in the ‘Exact Match’ column), and no other loci exist that differ from it by either one or two mismatches (0 in the ‘1 Mismatch’ and ‘2 Mismatches’ columns) Furthermore, only six loci in the human genome differ from the candidate by as few as three mismatches (six in the ‘3 Mismatches’ column). The second best candidate target present in the candidate region is similarly unique in the human genome allowing up to one mismatch; however, one locus differs from it by only two mismatches.
Fig. 1.
Sample GT-Scan output
Clicking on a candidate target in the upper table selects it and displays its potential off-targets in the lower table. The first column of the lower table shows the chosen candidate’s potential off-targets with mismatches highlighted in bold. High-, low- and no-specificity positions are denoted using orange, blue and green, respectively. The remaining columns show the number of mismatches in the potential off-target, its chromosomal location, its GC-content and a clickable link to its locus in a genome browser.
The GT-Scan algorithm extends the approach of Mali et al. (2013) to generalized target rules. Its first step is to convert the target rule into a regular expression by replacing X, x and N with wildcards. GT-Scan then finds all the candidate targets in the user-specified genomic region by scanning the input sequence for matches to the regular expression. Next, for each candidate target, GT-Scan generates a list of all possible off-targets by listing all possible words that match the candidate target except (possibly) at positions where the rule contains the no-specificity character (N). Note that the running time of GT-Scan therefore scales exponentially with the number of no-specificity characters in the rule (time proportional to 4n, where n is the number of no-specificity characters). GT-Scan then combines the possible off-targets for all targets into a single list of words and uses the Bowtie algorithm (Langmead et al., 2009) to identify all matches to these words anywhere in the selected genome. Genomic locations with up to three mismatches to any of the words in the list are reported by Bowtie. Finally, GT-Scan creates its interactive output report after determining which off-target locations correspond to each of the candidate targets. GT-Scan’s report, which can be downloaded as an HTML file, is retained on the server for at least 7 days.
GT-Scan fills a gap in the existing toolset for identifying optimal genomic targets. CRISPR Design (Hsu et al., 2013) is specifically engineered for target selection for the CRISPR/Cas9 system, does not allow for user-defined rules and currently limits the candidate region to 250 bp (versus 4000 bp for GT-Scan). Cas-OFFinder (Bae et al., 2014) is also designed specifically for CRISPR/Cas systems, does not allow user-specified rules and requires the user to specify the candidate targets. Cas-OT (Xiao et al., 2014) cannot currently be run via the web.
ACKNOWLEDGEMENT
We thank Fabian A. Buske for allowing us to use components from Triplex-Inspector (Buske et al., 2013).
Funding: T.L.B. and A.O. are funded by a National Institutes of Health grant (RO-1 RR021692-01).
Conflict of interest: none declared.
REFERENCES
- Bae S, et al. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, et al. Triplex-Inspector: an analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics. 2013;29:1895–1897. doi: 10.1093/bioinformatics/btt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, et al. RNA-guided editing of bacterial genomes using crispr-cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Xiao A, et al. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]