Abstract
Summary: With the ubiquitous generation of complete genome assemblies for a variety of species, efficient tools for whole-genome alignment along with user-friendly visualization are critically important. Our VISTA family of tools for comparative genomics, based on algorithms for pairwise and multiple alignments of genomic sequences and whole-genome assemblies, has become one of the standard techniques for comparative analysis. Most of the VISTA programs have been implemented as Web-accessible servers and are extensively used by the biomedical community. In this manuscript, we introduce GenomeVISTA: a novel implementation that incorporates most features of the VISTA family—fast and accurate alignment, visualization capabilities, GUI and analytical tools within a stand-alone software package. GenomeVISTA thus provides flexibility and security for users who need to conduct whole-genome comparisons on their own computers.
Availability and implementation: Implemented in Perl, C/C++ and Java, the source code is freely available for download at the VISTA Web site: http://genome.lbl.gov/vista/
Contact: avpoliakov@lbl.gov or ildubchak@lbl.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Comparing genomic sequences across related species has become a source of invaluable data on the functional elements in various genomes (Ponting and Hardison, 2011). There are a number of individual programs developed separately for genome alignment (Chen and Tompa, 2010) and visualization of comparative information (Chan et al., 2012), but few programs and Web servers integrate the two, giving researchers an opportunity to analyze results interactively. VISTA (Frazer et al., 2004), Dcode.org (Loots and Ovcharenko, 2005), the PipMaker suite of tools (Schwartz et al., 2000) and Mauve (Darling et al., 2010) are some examples of such integration.
VISTA online servers provide a wide range of services, which allow a user to align and compare sequences from multiple species up to 10 Mb long using different algorithms (Bray et al., 2003; Brudno et al., 2003a, b; Dubchak et al., 2009), locate regulatory sequences using comparative sequence analysis and transcription factor binding site search (Loots et al., 2002), compare user’s sequences against whole-genome assemblies and browse pre-computed alignments of hundreds of microbial, fungal, plant, vertebrate, and other genomes. Comparative results can be examined through a highly interactive graphic user interface (GUI) featuring the visualization of the level of conservation in the format of a continuous VISTA curve based on the conservation in a sliding window. This concept proved to be extremely successful owing to the easy interpretation of the resulting plots.
A novel stand-alone software GenomeVISTA integrates all well-established popular features of the VISTA family of tools (Dubchak et al., 2009; Frazer et al., 2004) in one package, and provides users the opportunity to carry out comparative analysis of whole genomes on their own computers, allowing for more flexibility and security of computations. It runs an extensively tested and recently improved alignment algorithm (Dubchak et al., 2009; Earl et al., 2014). Simultaneously, the built-in interactive GUI allows real-time examination of results of the comparative analysis. VISTA Point, a novel visualization program, is provided as a part of the GenomeVISTA package.
2 IMPLEMENTATION
Architecture. The whole-genome alignment pipeline is a combination of Perl and C/C++ programs and MySQL relational database to store both input genomic sequences and generated alignments. The pipeline uses the open-source BLAT program (Kent, 2002) to obtain local hits. The interactive GUI for data input and the examination of results was written in Java. GenomeVISTA can be run on any major platform—Windows, Mac OS X and Linux. In its minimal setup, it requires only a single machine, but it can also be configured to use a computer cluster using SGE/UGE, Condor or Torque batch systems.
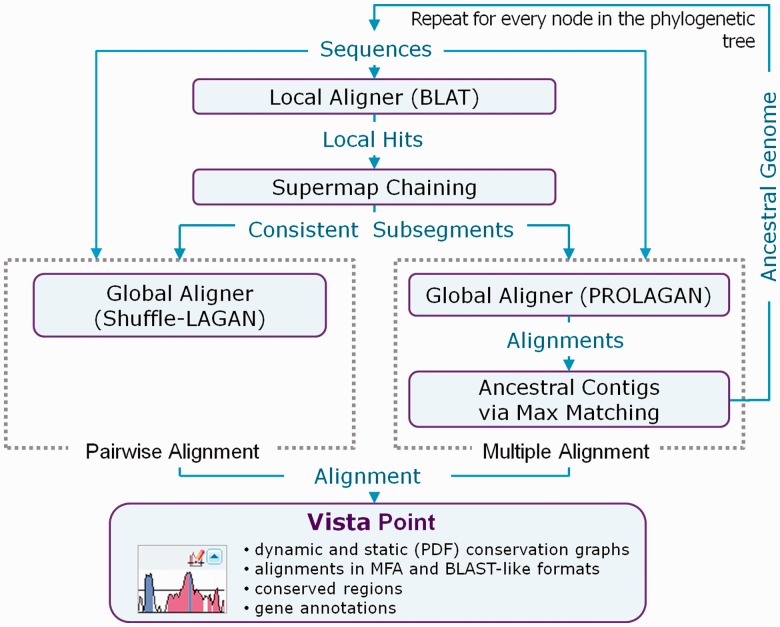
Design. Figure 1 shows the workflow of pairwise and multiple whole-genome alignment computations performed by GenomeVISTA. In the pairwise alignment, the local anchors between all sequences are computed using BLAT, which is run in a translated DNA mode, indexing all five-amino acid words. Then, Supermap (Dubchak et al., 2009), the fully symmetric whole-genome extension to the original Shuffle-LAGAN chaining algorithm (Brudno et al., 2003a, b), is used to obtain a map of large blocks of conserved synteny between the two species. Finally, regions of conserved synteny are aligned using Shuffle-LAGAN. The major difference in the multiple alignment pipeline is the use of PROLAGAN, a variation of the original Multi-LAGAN program (Brudno et al., 2003a, b) that allows the alignment of two alignments (profiles) and includes an additional step of predicting ancestral contigs using a maximum matching algorithm. The four stages (local hits, chaining, global alignment and ancestral reconstruction) are repeated for every node in the phylogenetic tree.
Fig. 1.
Schematic view of the comparative sequence analysis by the GenomeVISTA software
Runtime. Estimated runtimes for GenomeVISTA depend on the length and the number of genomic regions submitted to the program. It varies from several minutes to several hours for genomes from 1 to 50 Mb long (Supplementary Table S1). We recommend using a computer cluster for improved run times.
Output. GenomeVISTA provides users with an interactive GUI similar to VISTA Point used for analysis and visualization of alignments in all online VISTA applications. It displays a level of conservation in the format of a conventional VISTA plot and allows an interactive change of parameters, such as level of conservation and resolution of a plot. It also gives convenient access to all data used and produced in the alignment (Fig. 1).
3 DISCUSSION
GenomeVISTA unifies in one package multiple capabilities necessary to carry out various types of comparative analysis of genomic sequences and whole-genome assemblies. It aligns sequences both in finished and draft format, thus allowing to use it for multiple applications such as genome assembly, mapping newly sequence reads on the reference genome and calculating syntenic regions on complete genome assemblies. Importantly, it also gives access to the results of the alignment through a highly interactive interface that makes comparative analysis of genomic data fast and efficient.
Supplementary Material
Acknowledgements
The authors are grateful to all VISTA developers, collaborators and users for support and suggestions for its ongoing development.
Funding: National Heart, Lung and Blood Institute, National Institute of Health, Grant R01GM081080A. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract No. (DE-AC02-05CH11231).
Conflict of Interest: none declared.
REFERENCES
- Bray N, et al. AVID: a global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003a;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, et al. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003b;19(Suppl. 1):i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- Chan PP, et al. The UCSC archaeal genome browser: 2012 update. Nucleic Acids Res. 2012;40:D646–D652. doi: 10.1093/nar/gkr990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tompa M. Comparative assessment of methods for aligning multiple genome sequences. Nat. Biotechnol. 2010;28:567–572. doi: 10.1038/nbt.1637. [DOI] [PubMed] [Google Scholar]
- Darling AE, et al. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, et al. Multiple whole-genome alignments without a reference organism. Genome Res. 2009;19:682–689. doi: 10.1101/gr.081778.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D, et al. 2014 Alignathon: a competitive assessment of whole genome alignment methods. bioRxiv—the preprint server for biology. [Google Scholar]
- Frazer KA, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. Dcode.org anthology of comparative genomic tools. Nucleic Acids Res. 2005;33:W56–W64. doi: 10.1093/nar/gki355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, et al. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Hardison RC. What fraction of the human genome is functional? Genome Res. 2011;21:1769–1776. doi: 10.1101/gr.116814.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, et al. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.