Abstract
Motivation: Exome sequencing (exome-seq) data, which are typically used for calling exonic mutations, have also been utilized in detecting DNA copy number variations (CNVs). Despite the existence of several CNV detection tools, there is still a great need for a sensitive and an accurate CNV-calling algorithm with built-in QC steps, and does not require a paired reference for each sample.
Results: We developed a novel method named PatternCNV, which (i) accounts for the read coverage variations between exons while leveraging the consistencies of this variability across different samples; (ii) reduces alignment BAM files to WIG format and therefore greatly accelerates computation; (iii) incorporates multiple QC measures designed to identify outlier samples and batch effects; and (iv) provides a variety of visualization options including chromosome, gene and exon-level views of CNVs, along with a tabular summarization of the exon-level CNVs. Compared with other CNV-calling algorithms using data from a lymphoma exome-seq study, PatternCNV has higher sensitivity and specificity.
Availability and implementation: The software for PatternCNV is implemented using Perl and R, and can be used in Mac or Linux environments. Software and user manual are available at http://bioinformaticstools.mayo.edu/research/patterncnv/, and R package at https://github.com/topsoil/patternCNV/.
Contact: Asmann.Yan@mayo.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
DNA copy number variations (CNVs) are genomic structural changes that result in regional or chromosomal loss or gain of DNA copies (Hastings et al., 2009). Owing to the significant roles in human diseases, various laboratory techniques have been developed to detect CNVs, including recently advanced massive parallel sequencing of whole genomes and coding exomes. For exome-seq, it is commonly observed that coverage depths of short reads across regions vary, caused by different target capture efficiencies (Parla et al., 2011), as well as the differences in mappability of exons. Such coverage variations impose substantial challenges for reliable CNV detection. Most existing methods use a paired-sample approach, based on the intuitive assumption that somatic sample and its paired reference share similar coverage bias that can be cancelled out through pairing (Koboldt et al., 2012; Sathirapongsasuti et al., 2011). Although this assumption approximately holds, it oversimplifies the problem with two limitations unaddressed: (i) The region-specific noise (coverage variability) of a local region is not accounted for, leading to amplified noise in log-ratio values of coverage between sample and the paired reference. (ii) In the case of a missing or low-quality reference sample, CNV detection based on paired reference will be infeasible or have degraded accuracy/sensitivity. A recent published method, FishingCNV, tried to address the second limitation by using the average of multiple reference samples as the denominators in log-ratio calculation, but did not address the regional noises in individual samples (the numerator), which led to false CNV calls (details in Supplementary Section S2.3). Considering these issues, we proposed a novel method called PatternCNV, which summarizes overall consistent patterns of both depths and variability of exonic region coverage across samples, where ‘patterns’ of coverage and variability are summarized using multiple ‘normal’ or reference samples. We observed that the same patterns only exist between samples prepared using the same version of exome capture kit. During CNV detection, we compute the differences of observed coverage versus the common pattern, while penalizing regions associated with larger variability using a weighting scheme. Further, whole-genome CNV can be interpolated from exon-level CNV using any third-party segmentation method, e.g. circular binary segmentation (Olshen et al., 2004).
The PatternCNV was implemented in two different versions: a Mac and Linux/Unix version, and an R package version. We also developed a conversion tool to transform Binary version of sequence Alignment/Map (BAM) format files to much smaller wiggle (WIG) format files (<1% of BAM file size), which greatly speeds up pattern learning and CNV calculation. When compared with other state-of-the-art CNV algorithms in a lymphoma case study, PatternCNV displayed higher resolution and greater sensitivity/specificity.
2 FEATURES
2.1 Input, output and major functions
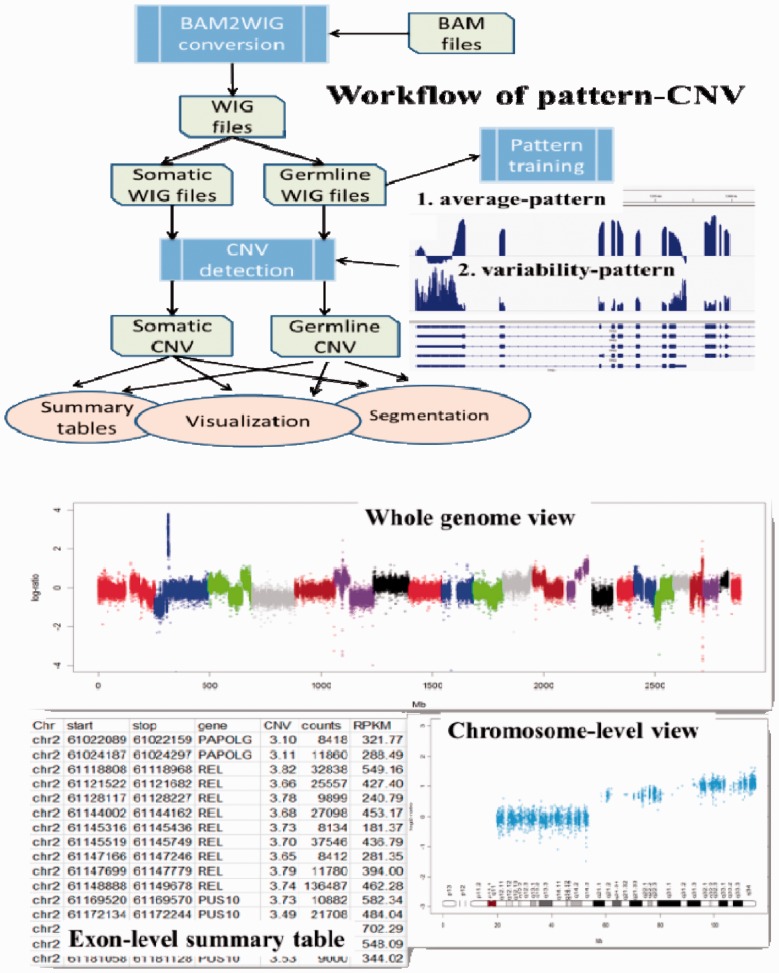
PatternCNV is divided into three major functional components: (i) BAM-to-WIG conversion for improved computational performance: a BAM2WIG converter using SAMtools (Li et al., 2009) and BEDtools (Quinlan and Hall 2010), which takes as input a BAM file, a file of Browser Extensible Data (BED) format defining exon regions and a second BED file for capture targets defined by the exome capture kit. The outputs are WIG files with greatly reduced file sizes compared with BAM files; (ii) CNV detection: starting with WIG files, PatternCNV estimates the coverage and variability patterns from multiple reference samples and calculates CNVs relative to the pattern for all samples including the references; and (iii) CNV summary and visualization: this module outputs a detailed exon-level CNV summary file per sample, and provides several visualization options for viewing CNVs at the whole-genome level or chromosome level. In addition, there are built-in QA/QC steps to detect sample outliers and batch effects. Figure 1 displays the overall workflow of PatternCNV along with illustrative examples of program output.
Fig. 1.
PatternCNV workflow is demonstrated in the upper panel. Examples of whole-genome and chromosome-level visulization are displayed in the bottom panel, along with Exon-level CNV summary table
2.2 Description of the PatternCNV algorithm
Each exon is first divided into consecutive bins of user-defined size (e.g. 10 base pairs). To make the exon coverage of different samples comparable, log2-transformed RPKM (reads per kilo-base per million total reads) is used to standardize the bin coverage. Denoting as log2-transformed RPKM coverage of l-th bin in a given exon, the standard coverage of a bin without CNV is assumed to approximately follow a normal distribution . The and are estimated from a pool of reference samples as the coverage and variability patterns. For a bin with a copy number of C, the bin signal is calculated as , ∼ . Hence, a bin-level CNV can be estimated as . Considering variability of bin coverage depending on its relative position in an exon or with respect to capture probe, we further smooth multiple bins within k-th exon (we denote related bin indices as ), leading to a maximum likelihood estimation: , where is designed to take variability of each bin into consideration (details of the statistical formulation are described in Supplementary Section S1).
2.3 Lymphoma case study
We applied PatternCNV to a set of 15 germ line–tumor pairs of diffuse large B-cell lymphoma exome-seq data (Lohr et al., 2012). When comparing CNV results derived from exome-seq using PatternCNV with those calculated from SNP microarray data profiled on the same samples, the two sets of results largely correlate for large CNVs. As expected, PatternCNV identified many small CNV regions at the single exon and/or multiple exon level (Supplementary Section S2.3) that the SNP array failed to detect owing to lack of probe coverage/density at the region. In addtion, thanks to the digitalized dynamic range of read coverages, PatternCNV can differentiate high versus low amplifications, while microarrays are limited by the saturation of probe hybridization signal. We compared PatternCNV with three other exome-seq-based CNV detection methods, ExomeCNV (Sathirapongsasuti et al., 2011), Varscan2 (Koboldt et al., 2012) and FishingCNV (Shi and Majewski 2013) using CNV detected by SNP microarrays as the ground truth. PatternCNV displayed superior visual resolution and achieved better specifity and sensitivity when compared with the paired approaches used by ExomeCNV and Varscan2 (Supplementary Section S2.2), and had much less false positives compared with FishingCNV (Supplementary Section S2.3). In several focused comparisons, we also saw an increased resolution of PatternCNV-based estimations compared with these two methods (Supplementary Section S2.1). In situations where a reference sample had less reliable quality than its paired counterpart, we often observed dramatically reduced performance of both Varscan2 and ExomeCNV for CNV detection, but not PatternCNV (Supplementary Section S2.1). This highlights the robustness of the pattern-based approach over conventional paried approaches. FishingCNV uses a method of taking the average across normal samples, which is more similar to PatternCNV than the paired methods used by the other two tools. However, a detailed comparison shows that FishingCNV has different data processing and CNV detection methods (Supplementary Section S2.3). FishingCNV’s principle component analysis (PCA) step over corrects batch effects and consequently removes CNV signals, resulting in false negative calls. We recommend that the users do not perform the default PCA step of FishingCNV. Moreover, it also oversimplifies average read-depth approach, producing an alarmingly high number of false-positive CNV calls (Supplementary Section S2.3). In contrast, PatternCNV’s novel use of both the weighted average read depth and coverage variability produces results that are superior and simpler to use by improving true positives and greatly reducing false-positive CNV calls.
3 DISCUSIONS AND CONCLUSIONS
We introduce PatternCNV, a software package designed to focus on exon-level CNV detection from exome-seq data. CNV estimate is based on coverage and variability patterns summarized from multiple reference samples. The implemented algorithm uses WIG file format, which improves the runtime and space efficiency. Several post-processing functions are included to facilitate interpretation, through visualization, segmentation and tabular summarization. As demonstrated by the case study, we believe it is a useful utility for exome-seq studies where robust detection of germ line and/or somatic CNVs is of interest.
Funding: Support for this work was provided by Center for Individualized Medicine at Mayo Clinic and the NIH (P50 CA97274). We thank Dr Todd R. Golub and colleagues at the Broad Institute, where the genomic data were generated.
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Hastings PJ, et al. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment-map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Parla JS, et al. A comparative analysis of exome capture. Genome Biol. 2011;12:R97. doi: 10.1186/gb-2011-12-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: exomeCNV. Bioinformatics. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Majewski J. FishingCNV: a graphical software package for detecting rare copy number variations in exome-sequencing data. Bioinformatics. 2013;29:1461–1462. doi: 10.1093/bioinformatics/btt151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.