Abstract
Multiple myeloma is a hematological cancer of plasma B-cells and remains incurable. Two major subtypes of myeloma, hyperdiploid (HMM) and non-hyperdiploid myeloma (NHMM), have distinct chromosomal alterations and different survival outcomes. Transcription factors (TrFs) have been implicated in myeloma oncogenesis but their dysregulation in myeloma subtypes are less studied. Here we develop a TrF-pathway co-expression analysis to identify altered co-expression between two sample types. We apply the method to the two myeloma subtypes and the cell cycle arrest pathway, which is significantly differentially expressed between the two subtypes. We find that TrFs MYC, NF-κB and HOXA9 have significantly lower co-expression with cell cycle arrest in HMM, co-occurring with their over-activation in HMM. In contrast, TrFs ESR1, SP1 and E2F1 have significantly lower co-expression with cell cycle arrest in NHMM. SP1 ChIP targets are enriched by cell cycle arrest genes. These results motivate a cooperation model of ESR1 and SP1 in regulating cell cycle arrest, and a hypothesis that their over-activation in NHMM disrupts proper regulation of cell cycle arrest. Co-targeting ESR1 and SP1 shows a synergistic effect on inhibiting myeloma proliferation in NHMM cell lines. Therefore, studying TrF-pathway co-expression dysregulation in human cancers facilitates forming novel hypotheses towards clinical utility.
Keywords: co-expression, cell cycle arrest, multiple myeloma, hyperdiploid
Introduction
Multiple myeloma (myeloma or MM) accounts for 10% of all hematological cancers1. It is a cancer of plasma B cells that undergoes monoclonal expansion in bone marrow, leading to symptoms such as kidney failures, frequent infections, anemia and bone fractures. The past decade has seen effective new treatment of MM such as proteasome inhibitors, but MM is still incurable and the median survival time of myeloma patients is 7–8 years2. Chromosomal alterations revealed by cytogenetics and genomics techniques divide MMs into two major subtypes: hyperdiploid multiple myeloma (HMM, 55–60% of MM) and non-hyperdiploid multiple myeloma (NHMM, the rest of MM). HMM usually contains trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, and 21 and lacks translocation events; while NHMM is often associated with one-copy deletion of chromosome 13 and translocations involving the immunoglobulin heavy chain (IgH) locus at 14q32, such as t(4;14), t(11;14), and t(14;16)2, 3. The survival outcome of HMM patients is better than that of NHMM2, 4. Therefore, it’s of biological and clinical interest to ask whether and how the different genomic alteration profiles in the two subtypes contribute to MM pathogenesis and response to treatments.
Genome-wide gene expression profiling has contributed to the understanding of transcriptional alterations in myeloma and its subtypes compared to normal plasma B cells. Dysregulation of cell cycle-related genes Cyclin D1, D2 or D3 by translocation or hyperdiploidy, has been found to be a unifying and early pathogenic event in MM5, 6. Chng and colleagues have compared the gene expression between HMM and NHMM and found that HMM is associated with the activation of NF-κB, MYC and MAPK pathways, whereas the NHMM is associated with oncogene-activating translocations7, 8. Agnelli and colleagues have reported that differentially expressed genes and pathways between HMM and NHMM are mainly involved in protein biosynthesis, transcriptional machinery and oxidative phosphorylation9. Most of these genes locate in the trisomy chromosomes of HMM and therefore could represent direct dosage effect of copy number gains. However, genomic alterations could also have indirect effect in gene expression through dysregulating transcription factors (TrFs) and members in signaling pathways10–12. In this project we study how dysregulated TrFs in MM contribute to such indirect effect.
TrFs are the downstream effectors of signaling pathways within cells that receive growth and other signals from microenvironment. They also survey internal cellular homeostasis and make decisions on cell survival and proliferation13. Consequently, mutations in TrF genes or dysregulation of TrFs could play significantly roles in cancer pathogenesis and drug resistance. In myeloma, over-activation of TrFs such as NF-κB and MYC is frequently observed and targeted as potential therapy14, 15. Several approaches have been developed to infer TrF dysregulation from global gene expression profiles. Gene set enrichment analysis identifies the enrichment of the target genes of a TrF in differentially expressed genes between two biological conditions as indication for the TrF’s own dysregulation16. Co-expression patterns between genes can also be analyzed to infer network changes between biological conditions17–20.
In this study, we hypothesize that between the HMM and NHMM myeloma subtypes, distinct TrF pathways dysregulate converging cancer pathways. This hypothesis motivates us to develop a TrF-pathway co-expression analysis method to identify TrFs that correlate with the cell cycle arrest pathway differently between the two subtypes. From this analysis, we find robust, subtype-specific co-expression patterns between TrFs and the cell cycle arrest pathway. These results support a novel cooperation model of ESR1, SP1 and E2F1 on dysregulating cell cycle arrest in NHMM and their potential therapeutic implication. We then validate this prediction by drug combination treatments using four NHMM cell lines.
Materials and Methods
Gene expression profiling data sets
We used the following public GEO data sets of myeloma gene expression profiling. GSE64777 contains 70 expression samples of HMM and 70 samples of NHMM. GSE1978421 contains 201 samples with HMM status out of 320 samples. HMM and NHMM status are measured by FISH in these two studies. Samples with ambiguous HMM status such as those with both FISH-measured trisomies and translocations were excluded from our analysis. GSE636522 has 90 samples without information of HMM status. We applied a KNN-based method to classify samples into HMM and NHMM by their expression profiles23. The accuracy of this method is more than 85% when compared to FISH-based and copy number microarray-based HMM status. All the data sets were normalized and processed by the RMA method of Bioconductor to obtain expression values.
We used a transcription factor list containing nearly 200 common TrFs24, which is downloaded from the UCSC Genome Browser database.
The TrF-pathway co-expression algorithm
1. Paired t-test and correlation calculation
Paired t-test was used to compare the overall expression change of cancer related pathways between the two subtypes of myeloma (Figure 1A). Each pathway gene has a pair of average expression value across the samples in the two subtypes, and the test assesses the overall expression difference of pathway genes between the two subtypes of myeloma.
Figure 1.
(A) Differential expression of cancer-related pathways between HMM and NHMM. The bars show the gene counts (y-axis) vs. pathways ordered by the number of genes (x-axis). The gene lists are downloaded from the Gene Ontology database. P-values on the top of the bars are computed from paired t-test using each pathway gene’s average expression in HMM and in NHMM as paired data points. Data set used: GSE6477.
(B) Co-expression between TrF SP1 and a cell cycle gene CDK2 in HMM and NHMM. The gene expression values of SP1 (y-axis) is plotted against that of CDK2 (x-axis) for HMM samples (left) and for NHMM samples (right). The correlation coefficient is stronger in HMM. Both genes are not significantly differentially expressed between HMM and NHMM (CDK2, p-value=0.053, SP1, p-value=0.11). Data set used: GSE6477.
(C) Overview of the TrF-pathway co-expression analysis. Using gene expression profiling data, we identify differentially expressed cancer pathways between two biological conditions. Next, we use a formula to score TrFs for their differential co-expression patterns with pathways of interest between the two conditions. Permutation of sample group labels was used to assess the statistical significance of top-ranked TrFs.
Next, we apply Spearman’s rank correlation to calculate the correlation coefficient between TrF and each pathway’s genes in the HMM and NHMM subtypes respectively (Figure 1B). Defined as Pearson’s correlation between rank-transformed values, Spearman’s correlation can detect non-linear correlation and reduce bias caused by outlier samples.
2. Define Pattern Score to quantify TrF-pathway co-expression changes
In the next step, we have developed a formula to measure the change of TrF-pathway correlation pattern between the two myeloma subtypes. In the co-expression plot between a TrF and pathway genes (Figure 2A and 2B), the X value and Y value of each point represents the co-expression coefficient of a TrF and a gene in HMM and NHMM, respectively, rather than expression levels. Our analysis compares the co-expression alteration between the two subtypes.
Figure 2. Scoring significant TrF-pathway co-expression changes between two myeloma subtypes.
Key ideas are illustrated using GSE6477, a myeloma expression data set that compares global gene expression differences between HMM and NHMM. (A) The co-expression scatterplot between a TrF (COMP) and genes. Each point stands for the correlation coefficients between the TrF and a gene in HMM samples (X-axis) and in NHMM samples (Y-axis). Blue: genes in the pathway of interest (cell cycle arrest); black: all other genes. (B) A cartoon version of (A). Blue points stand for cell cycle arrest genes while black ones stand for other genes. The point (X1, Y1) indicates a gene that has respective correlation coefficients with the TrF in HMM (X1) and NHMM (Y1). The blue arrow stands for the distance between (X1, Y1) and the diagonal line, Y=X. (C) Two diagonal lines divide the correlation space into four regions. For the points (genes) in regions A and B, the absolute Y-axis value is larger than the absolute X-axis value. The opposite is true for the points in regions C and D. Panel C–E: Three typical TrF-pathway co-expression patterns. (D) The TrF COMP has no significant co-expression changes with cell cycle arrest genes between HMM and NHMM. (E) SP1 has a lower overall correlation with the pathway genes in NHMM (Y-axis) than in HMM (X-axis). (F) MYC has a lower overall correlation in HMM (X-axis) than in NHMM (Y-axis).
We then define a score for the scatterplot pattern based on individual pathway genes (x, y) as:
Pattern Score = Sum of all blue points {Distance between (x, y) and the diagonal line * sign(|y|-|x|) } * −log(p-value of Hotelling test)
The Hotelling p-value tests whether the correlation coefficients between the TrF and the pathway genes (blue points in Figure 2A) are differently distributed compared to the correlation coefficients between the TrF and all the other genes (black points). Hotelling's T-squared test is a multivariate version of the t-test25 and tests for the difference between the multivariate means of two populations. A significant p-value suggests that the TrF has a different functional relationship with this pathway compared to all other genes. The distance to the diagonal line of a point measures its degree of correlation coefficient change between the two subtypes, while the sign indicates in which subtype the point has higher absolute correlation with the TrF. For example, a gene point (x, y) in region A or B of Figure 2C has |y|>|x|, so this gene has higher absolute correlation with the TrF in NHMM (the Y-axis subtype) than in HMM (the X-axis subtype).
Pattern Score has larger absolute value when the scatterplot deviates from y=x (Figure 2E and 2F) than when it is close to y=x (Figure 2D). The significance of the Pattern Score is determined by permuting the subtype labels of the samples and comparing the observed Pattern Score to the distribution of the maximal score obtained from 1000 permutations (in the Bayesian model below we performed 100 permutations).
3. Linear regression to test the co-expression scatterplot’s deviation from y=x
A typical co-expression plot between a TrF and pathway genes is shown in Figure 2A, where blue points (pathway genes) and black points (the rest genes) represent the correlation coefficient between a specific TrF and a gene in HMM subtype (X-axis) and NHMM subtype (Y-axis). To detect the overall change of TrF-pathway co-expression between HMM and NHMM, we also checked whether or not the slope is 1 for the blue points (pathway genes) in Figure 2A. If the slope is significant different from 1, it indicates that the co-expression of TrF and pathway genes has a global change between the two subtypes of myeloma. We rotated the coordinates of all points by 45 degrees clockwise and then tested whether the regression slope in the new scatterplot is 0. A significant p-value indicates the change of TrF-pathway co-expression between the two subtypes.
4. Bayesian meta-analysis of multiple data sets
In the meta-analysis step, we used a Bayesian method to pool information across multiple data sets and down-weight the results specific to a single-data set26. Briefly, in each data set we first computed the correlation coefficient r between a transcription factor and a pathway gene, and then converted r into a standard normal statistic using Fisher’s r-to-z transformation, with associated variance determined by sample size. Next, a hierarchical Bayesian model was used to combine the z values from different data sets to arrive at the TrF-gene correlation coefficient pooled across multiple data sets. We followed the formulas in Choi et al. 200526. This meta-analysis was also performed on the null data generated from 100 permutations of sample group labels to assess the statistical significance of identified top TrFs.
Drug treatment, cell proliferation assay and isobologram analysis
U266, MM1S and RPMI-8226 myeloma cells were obtained from the American Type Culture Collection (ATCC). OPM2 were obtained from the German collection of microorganisms and cell cultures (DSMZ). The INA6 cell line was provided by Renate Burger. All the cell lines are NHMM-characterized subtype: INA-6 is a IL-6-dependent human myeloma cell line27, while U266, OPM-2, RPMI-8226 and MM1S contain Ig locus translocation28, 29.
The MM cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. In the combination treatment experiments, cells were cultured with or without Tamoxifen (TMX) in the absence or presence of Terameprocol (TMP) for 48 hours. MM cell proliferation was measured by DNA synthesis through [3H]thymidine (Perkin-Elmer) incorporation assay, as previously described30. All experiments were carried out in triplicates.
The interaction between TMP and TMX was analyzed using the CalcuSyn software program (Biosoft, Ferguson, MO), which is based on the Chou-Talalay method31. When combination index (CI) = 1, it represents the conservation isobologram and indicates additive effects. CI < 1 indicates synergism and CI > 1 indicates antagonism.
Results
Motivations for studying the cell cycle arrest pathway and its co-expression with TrFs
Using the Chng data set and major cancer pathways32, 33, we found that the cell cycle arrest pathway is significantly up-regulated in HMM compared to NHMM (Figure 1A), and is up-regulated in both subtypes compared to normal plasma cells. We also observed co-expression changes between TrFs and their target genes. For example, the co-expression pattern between a TrF SP1 and cyclin-dependent kinase CDK2, a transcriptional target of SP134, is much stronger in HMM than in NHMM (Figure 1B). The co-expression analysis can detect such changes that are not obvious from gene-wise differential expression analysis of either SP1 or CDK2 (p-value is 0.11 and 0.053, respectively). These results and existing knowledge of cell cycle’s dysregulation in myeloma motivated us to develop a TrF-pathway co-expression analysis method to identify TrFs that differently correlate with the cell cycle arrest pathway between the two subtypes.
TrF-pathway differential co-expression analysis
We developed a statistical analysis method to detect co-expression changes between transcription factors and genes in cancer-related pathways. Figure 1C outlines the analysis pipeline. Differentially expressed, cancer-related pathways between two biological conditions were first identified. We next scored TrFs that have significant co-expression changes with pathways of interest between two sample conditions. This method ranks TrFs by the degree of TrF-pathway co-expression changes between the two conditions. Finally, permutation of sample group labels was used to generate null data sets and call significantly TrFs.
We used this pipeline to ask what TrF’s co-expression with the cell cycle arrest pathway has significant difference between the two myeloma subtypes, HMM and NHMM. The Pattern Score (defined in Methods) measures whether a TrF-pathway pair has an overall different correlation pattern between the two subtypes. If the pathway genes (blue points in Figure 2) and a TrF have largely unchanged co-expression between the two subtypes, the scatterplot of blue points centers along the diagonal line (y=x) and the Pattern Score is close to zero (Figure 2D). If the scatterplot has a slope far from 1, it indicates an overall change of TrF-pathway co-expression between the two subtypes and the Pattern Score will be much larger or smaller than 0 (Figure 2E and 2F). We also used a linear regression to test whether the pattern has significant deviation from the diagonal line (see Methods). The p-value of this test is 0.896, < 2.2e-16 and 3.7e-08 for the patterns in Figure 2D, E, F, respectively.
TrFs involved in IL-6 and MAPK signaling have significantly lower co-expression with cell cycle arrest genes in HMM
We first applied the TrF-pathway co-expression analysis on the data set GSE64777 and compared our findings to those in the original study for evaluation of the co-expression analysis. Table 1 lists top-ranked TrFs that have significantly lower co-expression with cell cycle arrest genes in HMM compared to NHMM, including HOXA9, MYC and NF-κB family member c-Rel (REL). Also from Table 1, CEBPB and STAT3 are involved in the IL-6 pathway, and NF-κB, MYC and ATF2 are involved in the MAPK signaling pathway35, 36. Both pathways promote myeloma initiation and progression2. Furthermore, RELA (p65, an NF-κB subunit, score=−138) and FOS (an IL-6 pathway member; score=−141) also have high scores but their permutation p-values are not as significant. These two TrFs also associate with the MAPK signaling pathway and the IL-6 pathway. Our co-expression analysis arrived at findings consistent with the original study of GSE6477, which has reported that MYC, NF-κB, MAPK and IL-6 pathways are over-expressed in HMM than in NHMM7.
Table 1.
Top-ranked TrFs that have significantly lower co-expression with cell cycle arrest genes in HMM compared to NHMM from the GSE6477 analysis. The TrFs in Tables 1–3 are selected by the permutation p-value threshold of 0.06 and linear regression p-value threshold of 1E−5. For the column of “Known roles in cancer”, see Supplementary Tables for literature references.
| Symbol | Gene | Pattern Score | Chromosome | Regression p-value |
Permutation p-value |
Known roles in cancer |
|---|---|---|---|---|---|---|
| HOXA9 | NM_152739 | −311.0993844 | Chr7 | 7.76E-29 | 0.018 | myeloma |
| FOXD1 | NM_004472 | −284.6487066 | Chr5 | 8.72E-21 | 0.016 | associated with placental growth factor (PLGF) |
| CEBPB | NM_005194 | −258.7742785 | Chr20 | 3.68E-12 | 0.015 | myeloma |
| BPTF | NM_004459 | −237.042492 | Chr17 | 6.31E-21 | 0.03 | |
| PAX3 | NM_181460 | −257.9273348 | Chr2 | 5.4E-09 | 0.039 | PAX3-FKHR fusion; chromosomal rearrangements |
| IRF1 | NM_002198 | −231.9772177 | Chr5 | 7.76E-11 | 0.012 | myeloma |
| TAL1 | NM_003189 | −228.7284582 | Chr1 | 3.4E-10 | 0.051 | TAL1/SCL; leukemia |
| MZF1 | NM_003422 | −226.4785758 | Chr19 | 2.44E-15 | 0.052 | Myeloid malignancies |
| MYC | NM_002467 | −206.9462599 | Chr8 | 3.71E-08 | 0.06 | myeloma; hyperdiploid |
| NFE2L1 | NM_003204 | −185.2599543 | Chr17 | 0.000000467 | 0.018 | |
| ATF2 | NM_001880 | −160.7918875 | Chr2 | 0.000000271 | 0.028 | associated with AP-1 complex |
| STAT3 | NM_003150 | −159.6504808 | Chr17 | 0.00000699 | 0.057 | myeloma |
| ATF6 | NM_007348 | −140.8047495 | Chr1 | 0.00000868 | 0.018 | myeloma |
| REL | NM_002908 | −94.94920668 | Chr2 | 4.78E-09 | 0.047 | myeloma |
ESR1, SP1 and E2F1 have significantly lower co-expression with cell cycle arrest genes in NHMM
In contrast to HMM, the transcriptional dysregulation in NHMM has been less studied, although the translocation events are more associated with NHMM. Table 2 lists top-ranked TrFs that have significantly lower co-expression with cell cycle arrest genes in NHMM compared to HMM. Among them, ESR1 (Estrogen receptor alpha), SP1 and E2F1 are three TrFs known to influence cellular processes that include proliferation and apoptosis37–39. These results have not been reported by the original study of GSE64777.
Table 2.
Top-ranked TrFs that have significantly lower co-expression with cell cycle arrest genes in NHMM compared to HMM from the GSE6477 analysis. See Table 1 legend for the selection criteria.
| Symbol | Gene | Pattern Score |
Chromosome | Regression p-value |
Permutation p-value |
Known roles in cancer |
|---|---|---|---|---|---|---|
| SP1 | NM_003109 | 407.6157627 | Chr7 | 1.23E-40 | 0.011 | myeloma |
| EGR3 | NM_004430 | 383.5931729 | Chr8 | 1.67E-21 | 0.006 | breast cancer |
| NFIC | NM_205843 | 366.5650056 | Chr19 | 3.88E-15 | 0.013 | |
| SREBF1 | NM_001005291 | 352.3150398 | Chr17 | 1.26E-20 | 0.032 | breast cancer |
| ESR1 | NM_001122741 | 312.6539295 | Chr6 | 4.75E-25 | 0.036 | myeloma |
| RFX1 | NM_002918 | 309.4206141 | Chr19 | 8.29E-23 | 0.028 | |
| FOXL1 | NM_005250 | 292.2963268 | Chr16 | 7.02E-10 | 0.026 | colon cancer; gastrointestinal |
| FOXC1 | NM_001453 | 289.9529049 | Chr6 | 1.54E-16 | 0.055 | associated with VEGF expression; cancer |
| YY1 | NM_003403 | 254.8226128 | Chr14 | 0.00000111 | 0.029 | cancer biology |
| E2F1 | NM_005225 | 252.5363106 | Chr20 | 9.85E-15 | 0.025 | myeloma; Myc |
| TFAP2C | NM_003222 | 245.9572093 | Chr20 | 1E-23 | 0.036 | invasive breast cancer |
Bayesian meta-analysis of multiple data sets improves the robustness of analysis
We next asked whether these findings are specific to one data set (GSE6477) or general in myeloma. We added two other myeloma expression data sets that have both HMM and NHMM samples, GSE636522 and GSE1978421. We used a Bayesian hierarchical model to compute correlation coefficients between TrFs and genes by pooling across data sets and weighting their respective sample sizes26. The meta-analysis down-weights TrF-pathway co-expression patterns specific to one data set, even if this pattern may be significant in the data set (Figure 3A); and enhances the patterns consistent across data sets but may not be significant in any one data set (Figure 3B). Table 3 lists the TrFs with significantly different co-expression with cell cycle arrest genes between HMM and NHMM after pooling the three data sets. Table 3B and Figure 3B show that both SP1 and ESR1 are still top-ranked as having significantly lower co-expression with cell cycle arrest genes in NHMM compared to HMM across different patient cohorts.
Figure 3. The co-expression patterns of TrFs AHR and ESR1 in individual data sets and in the meta-analysis.
Meta-analysis averages out conflicting patterns in (A) and strengthens consistent patterns in (B). (A) The TrF AHR does not have a robust co-expression alteration between HMM and NHMM across the three individual data sets. The red and green dots represent two pathway genes and their correlation coefficients with the TrF. Because the coefficients of the same genes are quite different in the three data sets, the meta-analysis does not lead to a significant Pattern Score. (B) ESR1 has a robust co-expression alteration between HMM and NHMM across the three data sets. Most pathway genes such as those highlighted by the red and green dots remain at similar correlation coefficient positions in the three data sets, so the meta-analysis leads to a significant Pattern Score.
Table 3.
Top-ranked TrFs that have significantly lower co-expression with cell cycle arrest genes in HMM (a) and in NHMM (b) from Bayesian meta-analysis of three data sets. See Table 1 legend for the selection criteria.
| (a) | |||||
|---|---|---|---|---|---|
| Symbol | Gene | Pattern Score | Chromosome | Regression p-value |
Permutation p-value |
| HOX9 | NM_152739_at | −267.4627552 | Chr12 | 6.8E-31 | 0.03 |
| PAX3 | NM_181460_at | −197.9994386 | Chr2 | 2.31E-15 | 0.05 |
| CREB1 | NM_004379_at | −187.2967718 | Chr2 | 7.74E-16 | 0.03 |
| TAL1 | NM_003189_at | −176.9745554 | Chr1 | 1.27E-14 | 0.05 |
| TP53 | NM_000546_at | −155.3202053 | Chr17 | 1.83E-09 | 0.06 |
| ATF2 | NM_001880_at | −147.651126 | Chr2 | 2.51E-12 | 0.04 |
| HLF | NM_002126_at | −147.014587 | Chr17 | 5.86E-11 | 0.04 |
| HOXA5 | NM_019102_at | −119.8556556 | Chr7 | 1.07E-10 | 0.02 |
| ZNF238 | NM_006352_at | −93.4493948 | Chr1 | 1.13E-07 | 0.03 |
| (b) | |||||
| ESR1 | NM_000125_at | 260.5433561 | Chr6 | 1.13E-25 | 0.04 |
| SOX5 | NM_006940_at | 238.5539368 | chr12 | 3.90E-26 | 0.02 |
| MYC | NM_002467_at | 148.8921877 | Chr8 | 1.32E-07 | 0.03 |
| POU6F1 | NM_002702_at | 156.3538726 | Chr12 | 1.08E-19 | 0.02 |
| TCF3 | NM_003200_at | 170.4257588 | Chr19 | 7.76E-10 | 0.03 |
| SP1 | NM_003109_at | 51.73820256 | Chr7 | 3.25E-07 | 0.02 |
Cell cycle arrest genes are enriched by SP1 ChIP targets
Although co-expression analysis suggests regulatory role of SP1 on cell cycle arrest genes, it cannot determine whether such regulation is direct or indirect. We next analyzed SP1 ChIP-seq (chromatin immunoprecipitation followed by sequencing) data of GM12878, a lymphoblastoid cell line from the ENCODE project, to check if SP1 targets are enriched by cell cycle arrest pathway genes (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeHaibTfbs). An enrichment supports a direct regulatory role of SP1 on the cell cycle arrest pathway. There are 2150 genes whose transcript starting site (TSS) is within 3 kb upstream or downstream of the top 5000 SP1 peaks. Among the 310 cell cycle arrest genes we used in the co-expression analysis (Suppl. Table 1), 85 have strong SP1 binding peaks near their TSS regions (27%). Therefore, cell cycle arrest genes are enriched by SP1 targets (p-value = 3.86e-08) in this B lymphocyte cell line. Since myeloma cells come from the B cell lineage, SP1 dysregulation is likely to directly affect the gene expression of cell cycle arrest pathway in myeloma.
We then analyzed an ESR1 ChIP-seq data in a breast cancer cell line MCF-7 (GSE25021), since ESR1 ChIP data are not available for B cells. There is no enrichment of ESR1 targets in cell cycle arrest genes. However, ESR is known to regulate genes indirectly through interactions with Sp1 transcription factor40, supporting its possible indirect regulation of the cell cycle arrest pathway.
A model of cooperation between SP1 and ESR1 on regulating cell cycle arrest
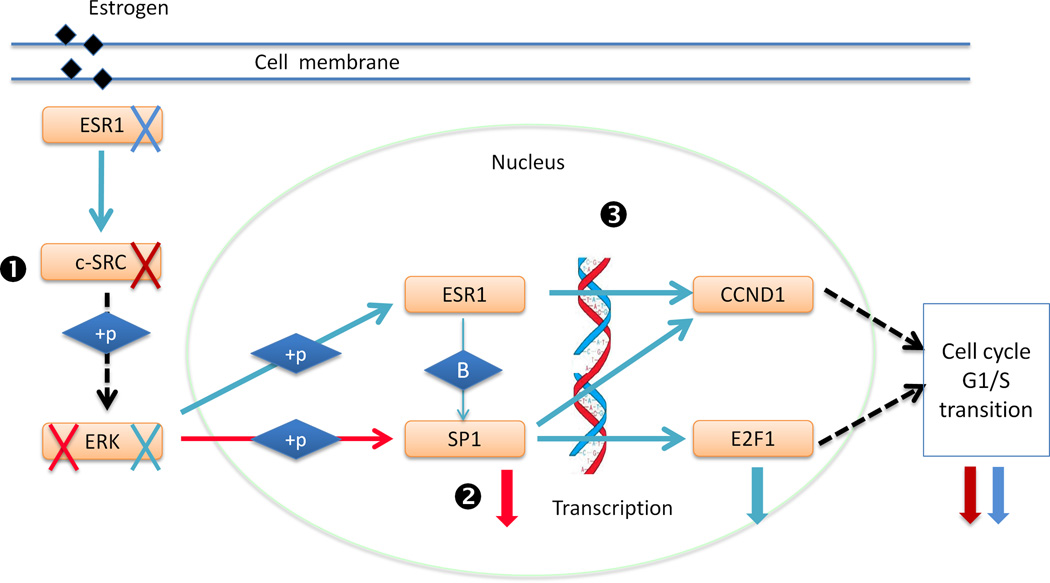
Based on these results and literature evidences, we proposed a model of cooperation between SP1 and ESR1 on regulating cell cycle arrest and their over-activation in NHMM (Figure 4). Over-activation of ESR1 in myeloma cells induces MAPK/ERK pathways through c-SRC41 (Step 1 in Figure 4), which in turn activate SP1, leading to increased expression of the downstream factor E2F1 (Step 2). Furthermore, both SP1 and ESR1 regulate Cyclin D1 (CCND1)42, 43, which is frequently abnormal in myeloma5, and lead to cell cycle progression (Step 3). This model suggests a combination therapy to inhibit both SP1 and ESR1 activities to overcome the limitations of single-drug treatments that inhibit either SP1 or ESR1 alone. For example, anti-estrogens therapy, especially the most commonly used tamoxifen, has major limitations including its partial agonist activity and constitutive or acquired resistance44. Tamoxifen resistance has been shown to be a result of increased ERα (product of ESR1) and Sp1 interaction that enhances the expression of E2F145, 46.
Figure 4. A model of cooperation between SP1 and ESR1 on regulating cell cycle arrest and their over-activation in NHMM.
ESR1, SP1 and their target E2F1 all have significantly lower co-expression with the cell cycle arrest pathway in NHMM compared to HMM. See Discussion for details. Blue arrows refer to literature-supported regulation in breast cancer, and red arrows in myeloma. “+p” stands for phosphorylation. Black dotted arrows indicate indirect regulation. Red and blue crosses and corresponding downward thick arrows indicate literature-supported knock-down of upstream genes or TrFs and the corresponding effects in reducing downstream genes’ activity or expression in breast cancer (blue) and in myeloma (red).
Synergistic growth inhibition effect of co-targeting SP1 and ESR1 in myeloma cells
The cooperation model predicts an approach to preventing cell proliferation by intervening SP1 and ESR1’s dysregulation in myeloma cells. We investigated the effect of co-targeting SP1 and ESR1 proteins on MM cell growth. Our recent work showed that Sp1 plays a regulatory role in multiple myeloma cell growth and survival, and it can be inhibited by the anti-MM activity of TMP, a small molecule that specifically competes with Sp1-DNA binding in vitro and in vivo30. In addition, tamoxifen (TMX), an antagonist of estrogen receptor, has been shown to induce growth arrest and apoptosis in multiple myeloma cell lines37, 44. In our combination treatment experiments, NHMM cells were cultured for 48 hours with TMX (0.5 and 20 µM), TMP (5 and 10 µM) or their combinations, and cell proliferation was measured by [3H]thymidine uptake at 48 hours. In four of the five cell lines treated, all combinations of TMX and TMP resulted in significant (p-value < 0.05) and synergistic anti-proliferative effect (Figure 5), except the treatment TMP 5 uM and TMX 20 uM for the OPM2 cell line. For example, treatment of TMX at 0.5 and 20 µM alone triggers 10% and 40% growth inhibition in U266 cells, respectively (Figure 5A), which was further enhanced to 40% (combination index = 0.68) and 70% (combination index = 0.5) by combined treatment with TMP at 5 µM (Figure 5C, table rows in blue). In MM1S cells, a synergistic effect was achieved only at 72 hours (data not shown).
Figure 5. Synergistic growth inhibitory effect of Sp1 inhibitor TMP and ESR1 antagonist tamoxifen (TMX) on MM cells.
Cell proliferation is measured as percentage of untreated cells at 48 hours for four different MM cell lines: (A) U266, and (B) INA6, RPMI-8226 and OPM2. Data are means ± SD (n = 3). (C) Combination indices (CI) and the fraction of cells affected by drug concentration (Fa) for the combinations of TMX and TMP are shown in the tables. CI < 1, CI = 1, CI values >1 indicate synergism, additive effect and antagonism, respectively. For analyses of combinations, different doses of each drug were used, but only the most representative doses of each combination were included in the figure.
Discussion
This study starts from the two myeloma subtypes that have distinct and characteristic genomic alterations, which may have been selected during cancer cell evolution and lead to different paths to oncogenesis and differential survival outcomes. We hypothesize that the dysregulation of TrFs can induce co-expression alteration between a TrF and the genes it regulates directly or indirectly. Because the cell cycle arrest pathway has significant expression level changes between the HMM and NHMM subtypes among cancer-related pathways, and that cell cycle dysregulation could be a common pathogenic mechanism in myeloma5, we have focused on the cell cycle arrest pathway. We have developed a co-expression analysis method to identify TrF-pathway co-expression dysregulation in two major myeloma subtypes. Our analysis has revealed specific TrFs that could be dysregulated in each myeloma subtype which in turn may lead to their reduced regulation of cell cycle arrest. Based on these results and literature evidences, we have proposed a cooperation model between TrFs SP1 and ESR1 in contributing to cell cycle dysregulation in myeloma. Our experiments show that combination treatment targeting both TrFs has synergistic effect on growth inhibition of myeloma cell lines, and therefore support the proposed model.
Compared to the traditional method of differential expression analysis followed by gene set enrichment of pathways or TrF target sets, our co-expression method may identify TrFs whose targets are not differentially expressed but are differentially regulated by the TrFs, which are biologically relevant and complementary results to existing methods. In addition, the co-expression analysis can identify TrFs that dysregulate downstream genes in indirect ways, while TrF target enrichment analysis cannot. For example, using predicted TrF target information based on TRANSFAC24, we identified only a few TrFs, whose target genes are enriched in cell cycles arrest genes and which are also differentially expressed between HMM and NHMM in the GSE6477 dataset, including MYC, MYB, CREB1, E2F3 and USF2 (Suppl. Table 4). This is in contrast to many TFs that we identified to differentially regulate cell cycle arrest genes between the two myeloma subtypes (Table 1 and 2). In particular, our analysis identified ESR1’s dysregulation in NHMM. ESR1 had been reported to regulate genes through indirect mechanisms such as by interacting with cofactors AP-1 and Sp140, and our ChIP-seq analysis of ESR1 binding peaks showed no enrichment of cell cycle arrest genes. Finally, although TrF target enrichment analysis may suggest direct regulation, it is restricted to available target prediction or ChIP-seq data; while the co-expression does not require such information.
The TrF-pathway co-expression analysis has revealed dysregulated pathways that are consistent with previous analysis of HMM. Both our analysis and the original study of GSE6477 have found MYC and NF-κB pathways as dysregulated in HMM. In addition, we have identified HOXA9 as the top TrF dysregulated in HMM due to its reduced co-expression with cell cycle arrest genes both in the GSE6477 data and in the Bayesian meta-analysis (Tables 1 and 3A). Although HOXA9 was not identified in Chng et al.7, it has recently been suggested as a candidate oncogene in myeloma by a genome-wide sequencing study47. HOXA9 is a transcription factor regulating gene expression, morphogenesis and differentiation48. A translocation event causing a fusion between HOXA9 and NUP98 has been associated with myeloid leukemogenesis49, 50. In MM, it has high expression level in patients lacking IgH translocations47, agreeing with that HOXA9 has a higher expression in HMM than in NHMM in the three data sets we analyzed (p-values of 0.076, 0.019, 0.008). HOXA9-depleted cells show a competitive growth disadvantage47, suggesting its expression-related carcinogenesis role in myeloma. Therefore, HOXA9 with higher expression level in HMM is associated with oncogenesis, co-occurring with our analysis results of HOXA9’s reduced co-expression with cell cycle arrest pathway in HMM.
We have also identified TrFs SP1, ESR1 and E2F1 as having significantly lower co-expression with the cell cycle arrest pathway in NHMM compared to HMM (Tables 2 and 3B). SP1 regulates a variety of processes such as cell growth, apoptosis, differentiation and immune responses38. It also mediates the transcription of cyclins and cyclin-dependent kinases43, 51. It has been recently shown to play an important regulatory role in MM cell growth and survival30. MM cells display increased SP1 protein (Sp1) nuclear levels and DNA binding activity compared to normal cells, and their interaction with bone marrow stromal cells (BMSC) lead to Sp1 activation via induction of the ERK signaling pathway30. Moreover, genetic inhibition of Sp1 with siRNA and shRNA technologies and pharmacological inhibition with a specific Sp1 inhibitor significantly reduced MM cell growth and survival in vitro and in vivo30. Estrogen receptors (ESR1 and ESR2) are nuclear receptors that are activated by the hormone estrogen and then bind to DNA to regulate downstream genes52. They are over-expressed in 70% of breast cancer samples, and these ER-positive cancers respond to estrogen receptor antagonist such as tamoxifen46, 53. Studies show that MM cells often express and over-activate estrogen receptors (ER), and selective estrogen receptor modulators induce G1-S cell cycle arrest and apoptosis in MM cells44, 54, 55. Moreover, ESR1 interacts with Sp1 to regulate transcription of downstream genes, including E2F1, a key regulator of the G1-S cell cycle checkpoint 46, 56.
Drug combination is widely used in treating cancers, helping achieve synergistic therapeutic effect and minimizing drug resistance31. Based on the TrF-pathway results and literature reviews, we have proposed a cooperation model of SP1 and ESR1’s roles in regulating cell cycle arrest and their dysregulation in NHMM. A combination treatment targeting both SP1 and ESR1 has synergistic growth inhibition effect on NHMM cell lines. These result support the SP1-ESR1 cooperation model and point to further experimental follow-up.
There are areas for further improvement on this study. First, since myeloma is a cancer of bone marrow, no normal bone marrows are available in large numbers for comparative expression profiling. Due to small sample size or lack of normal samples in the data sets used, we have not included normal samples in the analysis. In this study, we have used the evidence that TrFs such as MYC and HOXA9 are both over-activated in HMM and have lower co-expression with the cell cycle arrest pathway to associate a TrF’s lower co-expression in a subtype with its dysregulation in the subtype. Comparing MM subtypes with normal samples in future studies using the co-expression analysis could help confirm this association. Comparing to normal samples can also reveal TrFs that are commonly dysregulated in both subtypes. The second limitation is the use of TrFs that are based on TRANSFAC TrFs, which are well studied58. Although this helps interpreting these TrFs’ roles in cell cycle pathways and designing experimental validation, we may miss novel mechanisms involving TrFs not in the list. Expanding the analysis to predicted TrFs based on their DNA-binding domains will alleviate this limitation59. The third limitation is that when a TrF has significant co-expression with a gene in a pathway, relationship between the TrF and the gene may not be causal or direct. Other genetic regulators may regulate both the TrF and the gene, or the TrF regulates the gene via intermediate regulators. We have checked the ChIP-seq based gene targets of SP1 and found that they are enriched by cell cycle arrest genes. This lends support that SP1 directly regulates many cell cycle arrest genes, but wet experiments are needed to validate these predictions. Bioinformatic algorithms such as ARACNE may be used to better distinguish direct and indirect co-expression relationship60. These limitations point to areas to improve the TrF-pathway co-expression analysis for better understanding of regulatory alterations in cancer. Another interesting direction is to use co-expression signatures as biomarker for patient prognosis57. The TrF-pathway co-expression analysis code is available at http://chenglilab.weebly.com/.
Supplementary Material
Acknowledgements
This work was supported by National Basic Research Program of China (973 Program; No. 2010CB944904) (X Wang, Y Li, Y Zhang, C Li) and NIH R01 GM077122 (C Li, Z Yan). This work was also supported in part by NIH grants RO1-124929, PO1-155258, P50-100007 and PO1-78378 to NC Munshi and by the Dept. of Veterans Affairs Merit Review Awards to NC Munshi. PK Shah was supported by Claudia Adams Barr Program in Innovative Basic Cancer Research and the Multiple Myeloma Career Development award. We thank the reviewers and Cheng Li group members for constructive comments and discussion.
Abbreviations
- TrF
Transcription Factor
- MM
Multiple Myeloma
- CI
Combination Index
Footnotes
Authorship Contributions
X Wang, Z Yan and C Li designed the study. X Wang implemented the methodology, analyzed the data, and interpreted the results in the context of biological literature. Y Li predicted the HMM status of one data set. PK Shah, SB Amin and Y Zhang contributed to study design and data analysis. M Fulciniti, M Gkotzamanidou and NC Munshi provided biological interpretation of the results and carried out drug treatment experiments. X Wang and C Li wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict of Interest
The authors declare no conflict of interest.
Supplementary information is available at the journal's website.
Contributor Information
Nikhil C Munshi, Email: Nikhil_Munshi@dfci.harvard.edu.
Cheng Li, Email: lch3000@gmail.com.
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111(6):2962–2972. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuilleme S, Robillard N, Lode L, Magrangeas F, Beris H, Harousseau JL, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19(2):275–278. doi: 10.1038/sj.leu.2403586. [DOI] [PubMed] [Google Scholar]
- 5.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106(1):296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raje N, Hideshima T, Mukherjee S, Raab M, Vallet S, Chhetri S, et al. Preclinical activity of P276-00, a novel small-molecule cyclin-dependent kinase inhibitor in the therapy of multiple myeloma. Leukemia. 2009;23(5):961–970. doi: 10.1038/leu.2008.378. [DOI] [PubMed] [Google Scholar]
- 7.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67(7):2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 8.Chng WJ, Santana-Davila R, Van Wier SA, Ahmann GJ, Jalal SM, Bergsagel PL, et al. Prognostic factors for hyperdiploid-myeloma: effects of chromosome 13 deletions and IgH translocations. Leukemia. 2006;20(5):807–813. doi: 10.1038/sj.leu.2404172. [DOI] [PubMed] [Google Scholar]
- 9.Agnelli L, Fabris S, Bicciato S, Basso D, Baldini L, Morabito F, et al. Upregulation of translational machinery and distinct genetic subgroups characterise hyperdiploidy in multiple myeloma. Br J Haematol. 2007;136(4):565–573. doi: 10.1111/j.1365-2141.2006.06467.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455(7215):971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26(8):1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23(1):10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitada M, Rowitch DH. Transcription factor co-expression patterns indicate heterogeneity of oligodendroglial subpopulations in adult spinal cord. Glia. 2006;54(1):35–46. doi: 10.1002/glia.20354. [DOI] [PubMed] [Google Scholar]
- 18.Watson M. CoXpress: differential co-expression in gene expression data. BMC Bioinformatics. 2006;7:509. doi: 10.1186/1471-2105-7-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 20.de la Fuente A. From 'differential expression' to 'differential networking' - identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010;26(7):326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116(14):2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 22.Agnelli L, Bicciato S, Fabris S, Baldini L, Morabito F, Intini D, et al. Integrative genomic analysis reveals distinct transcriptional and genetic features associated with chromosome 13 deletion in multiple myeloma. Haematologica. 2007;92(1):56–65. doi: 10.3324/haematol.10414. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Wang X, Zheng H, Wang C, Minvielle S, Magrangeas F, et al. Classify hyperdiploidy status of multiple myeloma patients using gene expression profiles. PLoS One. 2013;8(3):e58809. doi: 10.1371/journal.pone.0058809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Z, Shah PK, Amin SB, Samur MK, Huang N, Wang X, et al. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic Acids Res. 2012;40(17):e135. doi: 10.1093/nar/gks395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson TW. An introduction to multivariate statistical analysis. 3rd edn. Hoboken, N.J.: Wiley-Interscience; 2003. [Google Scholar]
- 26.Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21(24):4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- 27.Brocke-Heidrich K, Kretzschmar AK, Pfeifer G, Henze C, Loffler D, Koczan D, et al. Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood. 2004;103(1):242–251. doi: 10.1182/blood-2003-04-1048. [DOI] [PubMed] [Google Scholar]
- 28.Largo C, Alvarez S, Saez B, Blesa D, Martin-Subero JI, Gonzalez-Garcia I, et al. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica. 2006;91(2):184–191. [PubMed] [Google Scholar]
- 29.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulciniti M, Amin S, Nanjappa P, Rodig S, Prabhala R, Li C, et al. Significant biological role of sp1 transactivation in multiple myeloma. Clin Cancer Res. 2011;17(20):6500–6509. doi: 10.1158/1078-0432.CCR-11-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 33.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 34.Xie RL, Gupta S, Miele A, Shiffman D, Stein JL, Stein GS, et al. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J Biol Chem. 2003;278(29):26589–26596. doi: 10.1074/jbc.M301491200. [DOI] [PubMed] [Google Scholar]
- 35.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 36.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 37.Thomas C, Gustafsson JA. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11(8):597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 38.Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol. 2009;29(10):2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3(6):e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai Y, Chen S, Shah R, Pei XY, Wang L, Almenara JA, et al. Disruption of Src function potentiates Chk1-inhibitor-induced apoptosis in human multiple myeloma cells in vitro and in vivo. Blood. 2011;117(6):1947–1957. doi: 10.1182/blood-2010-06-291146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castro-Rivera E, Samudio I, Safe S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J Biol Chem. 2001;276(33):30853–30861. doi: 10.1074/jbc.M103339200. [DOI] [PubMed] [Google Scholar]
- 43.Bartusel T, Schubert S, Klempnauer KH. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene. 2005;351:171–180. doi: 10.1016/j.gene.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 44.Sola B, Renoir JM. Antiestrogenic therapies in solid cancers and multiple myeloma. Curr Mol Med. 2006;6(4):359–368. doi: 10.2174/156652406777435408. [DOI] [PubMed] [Google Scholar]
- 45.Kim K, Thu N, Saville B, Safe S. Domains of estrogen receptor alpha (ERalpha) required for ERalpha/Sp1-mediated activation of GC-rich promoters by estrogens and antiestrogens in breast cancer cells. Mol Endocrinol. 2003;17(5):804–817. doi: 10.1210/me.2002-0406. [DOI] [PubMed] [Google Scholar]
- 46.Louie MC, McClellan A, Siewit C, Kawabata L. Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol Cancer Res. 2010;8(3):343–352. doi: 10.1158/1541-7786.MCR-09-0395. [DOI] [PubMed] [Google Scholar]
- 47.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan JT, Ludwig DL, Bertrand FE. HoxA9 induces insulin-like growth factor-1 receptor expression in B-lineage acute lymphoblastic leukemia. Leukemia. 2008;22(6):1161–1169. doi: 10.1038/leu.2008.57. [DOI] [PubMed] [Google Scholar]
- 49.Lam DH, Aplan PD. NUP98 gene fusions in hematologic malignancies. Leukemia. 2001;15(11):1689–1695. doi: 10.1038/sj.leu.2402269. [DOI] [PubMed] [Google Scholar]
- 50.Romana SP, Radford-Weiss I, Ben Abdelali R, Schluth C, Petit A, Dastugue N, et al. NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogenetique Hematologique. Leukemia. 2006;20(4):696–706. doi: 10.1038/sj.leu.2404130. [DOI] [PubMed] [Google Scholar]
- 51.Willoughby JA, Sr, Sundar SN, Cheung M, Tin AS, Modiano J, Firestone GL. Artemisinin blocks prostate cancer growth and cell cycle progression by disrupting Sp1 interactions with the cyclin-dependent kinase-4 (CDK4) promoter and inhibiting CDK4 gene expression. J Biol Chem. 2009;284(4):2203–2213. doi: 10.1074/jbc.M804491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levin ER. Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol. 2005;19(8):1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clemons M, Danson S, Howell A. Tamoxifen ("Nolvadex"): a review. Cancer Treat Rev. 2002;28(4):165–180. doi: 10.1016/s0305-7372(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 54.Otsuki T, Yamada O, Kurebayashi J, Moriya T, Sakaguchi H, Kunisue H, et al. Estrogen receptors in human myeloma cells. Cancer Res. 2000;60(5):1434–1441. [PubMed] [Google Scholar]
- 55.Gauduchon J, Gouilleux F, Maillard S, Marsaud V, Renoir JM, Sola B. 4-Hydroxytamoxifen inhibits proliferation of multiple myeloma cells in vitro through down-regulation of c-Myc, up-regulation of p27Kip1, and modulation of Bcl-2 family members. Clin Cancer Res. 2005;11(6):2345–2354. doi: 10.1158/1078-0432.CCR-04-1668. [DOI] [PubMed] [Google Scholar]
- 56.Wang W, Dong L, Saville B, Safe S. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol Endocrinol. 1999;13(8):1373–1387. doi: 10.1210/mend.13.8.0323. [DOI] [PubMed] [Google Scholar]
- 57.Fujiwara T, Hiramatsu M, Isagawa T, Ninomiya H, Inamura K, Ishikawa S, et al. ASCL1-coexpression profiling but not single gene expression profiling defines lung adenocarcinomas of neuroendocrine nature with poor prognosis. Lung Cancer. 2012;75(1):119–125. doi: 10.1016/j.lungcan.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 58.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 60.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.