Abstract
We report herein the glycation sites in a vaccine candidate for cholera formed by conjugation of the synthetic hexasaccharide fragment of the O-specific polysaccharide of Vibrio cholerae, serotype Ogawa, to the recombinant tetanus toxin C-fragment (rTT–Hc) carrier. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of the vaccine revealed that it is composed of a mixture of neoglycoconjugates with carbohydrate:protein ratios of 1.9:1,3.0:1,4.0:1,4.9:1, 5.9:1, 6.9:1, 7.9:1 and 9.1:1. Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis of the tryptic and GluC V8 digests allowed identification of 12 glycation sites in the carbohydrate–protein neoglycoconjugate vaccine. The glycation sites are located exclusively on lysine (Lys) residues and are listed as follows: Lys 22, Lys 61, Lys 145, Lys 239, Lys 278, Lys 318, Lys 331, Lys 353, Lys 378, Lys 389, Lys 396 and Lys 437. Based on the 3-D representation of the rTT–Hc protein, all the glycation sites correspond to lysines located at the outer surface of the protein.
Keywords: nano-LC-ESI-QqTOF-MS/MS, neoglycoconjugate vaccine, recombinant tetanus toxin C-fragment (rTT–Hc), Trypsin digestion, GluC V8 digestion, MALDI-TOF-MS
Introduction
Carbohydrate-based vaccines are widely used to prevent, control and eliminate an array of diseases.[1] They consist of a carbohydrate antigen attached to a medically acceptable protein. Different methods have been used for the conjugation of carbohydrates to proteins.[2–4] The squaric acid chemistry that has been used extensively by the group of Kováč[4] is arguably the most efficient conjugation method currently used for single-point attachment of synthetic carbohydrate antigens to proteins.[5] This approach offers two main advantages. First, cross-linked products are not formed, and second, although the squaric acid derivative is typically used in excess to force the reaction to completion, any unconjugated antigen can be recovered following conjugation.[6,7]
Kováč’s group also established a method for a one-pot preparation of a series of neoglycoconjugates with predetermined carbohydrate–protein ratios.[4,8,9] Using this method, they synthesized different carbohydrate-protein neoglycoconjugates by attachment of fragments of the O-specific polysaccharide of Vibrio cholerae O1 serotype Ogawa[9,10] and lnaba[11,12] to bovine serum albumin (BSA), and also the Ogawa hexasaccharide to the medically acceptable carrier, rTT–Hc.[13] These hapten–protein glycoconjugates are not only immunogenic but also offer protective capacity.[14–16]
Tetanus toxoid (TT) is a protein that has been used as a protective vaccine against tetanus, since the 1930s. Tetanus toxoid is prepared from tetanus toxin by treatment with formaldehyde to inactivate its neurotoxicity. Tetanus toxin has an average molecular weight of ~159 kDa and comprised three ~50 kDa functional domains, namely, the translocational HN chain, the catalytic N-terminal L chain and the receptor-binding Hc chain.[17,18] The toxoid form of this protein has also been used as a carrier protein for antigens in experimental carbohydrate-protein conjugate vaccines.[13] However, because the structure of TT is not fully characterized, the reproducibility of the synthesis of a well-defined polysaccharide–TT neoglycoconjugate vaccine was found to be poor. Consequently, the use of a fully characterized Hc fragment for the preparation of such vaccines is generally preferred.
In view of the potential of the medically acceptable rTT–Hc in vaccine preparation, the purpose of this study was to determine the average carbohydrate: protein ratio and the glycation sites in the title neoglycoconjugate using LC-MS/MS.
Matrix-assisted laser desorption/ionization (MALDI) and surface-enhanced laser desorption/ionization coupled to a time-of-flight (TOF) mass analyzer are the methods of choice for the determination of the molecular weight of glycoproteins[16,19] and carbohydrate hapten-to-carrier protein ratios.[12,20–22] Recently, we reported on the determination of the glycation sites of several different vaccine models prepared by dialkyl squarate chemistry: a synthetic tetrasaccharide hapten of Bacillus anthracis–BSA conjugate vaccine, a synthetic lactose-BSA conjugate and neoglycoconjugates from the terminal monosaccharide hapten of the O-specific polysaccharide of Vibrio cholerae O1, serotype Ogawa and BSA.[20–23] Our strategy then, as well as during the present work, was based on the enzymatic digestion of the hapten–BSA glycoconjugates, followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis of the digests.
Material and methods
Preparation of the hapten–rTT–Hc neoglycoconjugate
Conjugation of the hexasaccharide of Vibrio choleras serotype Ogawa (targeted hexasaccharide–carrier ratio 5:1) to rTT–Hc was carried out as described in the literature.[13] Briefly, rTT–Hc (26.8 mg, 1 eq) was dissolved in pH 9 borate buffer (775 µ1) to form a clear solution. Ogawa hexasaccharide squarate[4] (5.5 mg, 6 eq) was added, and the clear solution was stirred at room temperature for 24 h. The mixture was dialyzed (eight times) against 10 mM ammonium carbonate using Millipore Amicon Ultra centrifugal filter device (30 kDa cut-off, 4°C). The retentate was lyophilized to give a white fluffy solid conjugate, which was then analyzed by MALDI-TOF-MS.
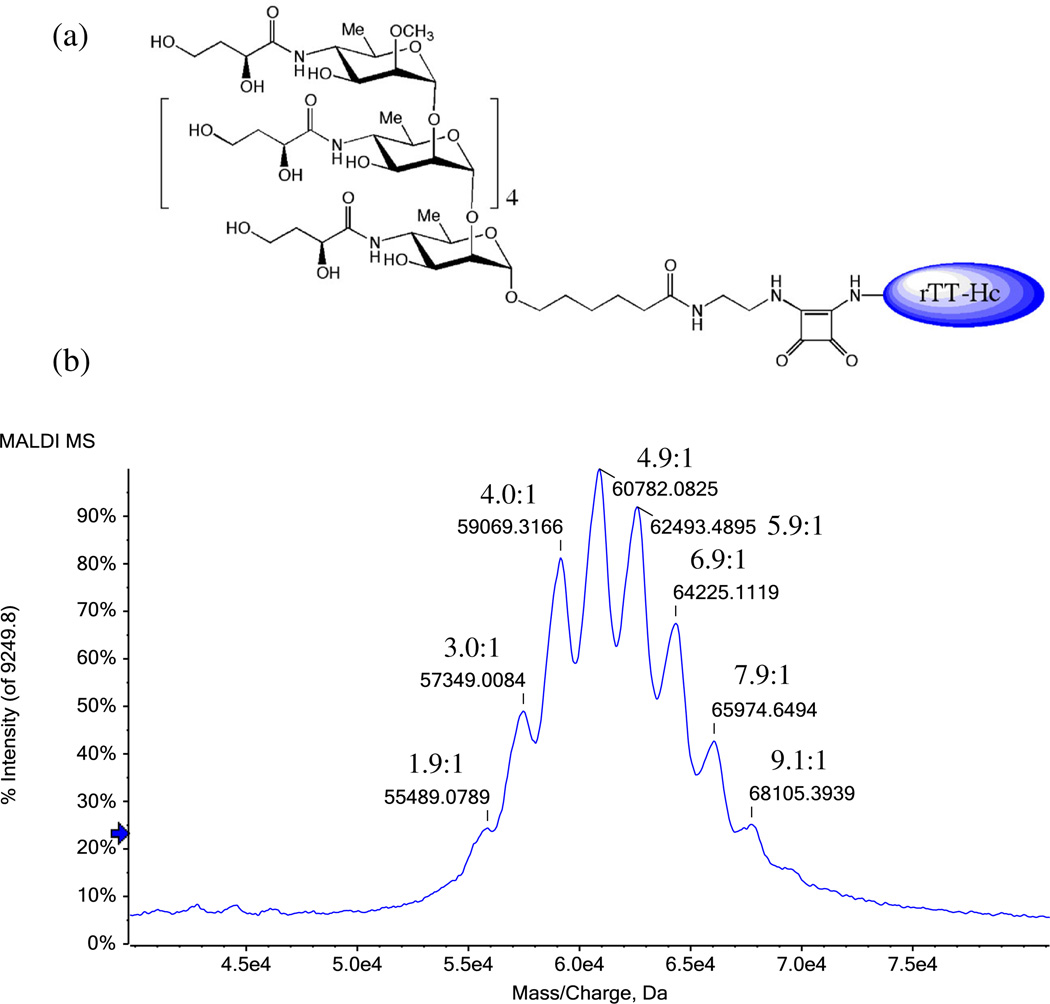
Figure 1(a) shows the structure of the hexasaccharide of Vibrio cholerae serotype Ogawa conjugated through a squarate spacer to the rTT–Hc carrier.
Figure 1.
(a) Schematic representation of the hexasaccharide antigen of the Vibrio cholera serotype Ogawa conjugated to the rTT–Hc protein, using the squaric acid chemistry, (b) MALDI-TOF-MS analysis of the the hexasaccharide antigen of the Vibrio cholera serotype Ogawa conjugated to the rTT–Hc protein vaccine. The carbohydrate: protein ratios are displayed with the identified molecularions.
Digestion
The digestion of the hapten–rTT–Hc glycoconjugate was carried out with trypsin and GluC V8 protease (Sigma Aldrich, Saint Louis, MO, USA). Thus, 100 µg of the glycoconjugate (4.3 nmol) was dissolved in a mixture of 0.1% RapiGest SF Surfactant (1 µg, Waters, Milford, MA, USA) in 50 mM of NH4HCO3 (100 µl) at a pH of 8.0 and reduced by treatment with 2 µl of 10 mM dithiothreitol (Sigma Aldrich, Saint Louis, MO, USA) for 30 min at room temperature, followed by alkylation with 2 µl of 50 mM iodoacetamide (Sigma Aldrich, Saint Louis, MO, USA) for 1 h at room temperature. A portion (50 µg, 4.3 nmol) of the glycoconjugate was digested with trypsin using a 20 ng/ml solution of trypsin dissolved in NH4HCO3 (50 mM, 1 ml) at a trypsin-glycoprotein ratio of 1:25 (w/w) and incubated at 37°C overnight with shaking. The other 50 µg (4.3 nmol) of the tetanus toxin glycoconjugate was digested using the GluC V8 endoprotease dissolved in NH4HCO3 (50 mM, 1 ml) at a protease–glycoprotein ratio of 1:25 (w/w) and incubated at 37°C overnight with shaking. The sample was then dried under vacuum, and the residue was dissolved in 20µl of 1% acetic acid (Sigma Aldrich, Saint Louis, MO, USA). An aliquot of each sample (10 µl) was then cleaned up using ZipTip C18 (Millipore, Bedford, MA, USA) before mass spectral analysis.
MALDI-TOF-MS analysis
The MALDI-TOF-MS analysis was carried out on a 4800 Proteomics analyzer with TOFATOF optics (Applied Biosystems Foster City, CA) and a 200-Hz frequency-tripled Nd:YAG laser. α-Cyano-4-hydroxycinnamic acid (α-CHCA) was used as matrix for the analysis of rTT–Hc and hapten–rTT–Hc conjugates with an average of 5000–8000 laser shots per spectra. Briefly, 1 µl of a 20 mg/ml solution of α-CHCA [dissolved in acetone, 0.1% trifluoroacetic acid (TFA)] was spotted on the MALDI plate and dried at room temperature (the use of acetone allows a good homogeneity of the matrix in the spot). Then, an aliquot of 1 µl of sample was spotted on the top of the dried matrix and allowed to dry before the MALDI-MS experiments. The analysis was achieved in the linear mode (positive ion mode), and the MALDI-TOF-MS was calibrated using BSA.
LC-ESI-QqTOF-MS/MS analysis
The peptides were separated on a DIONEX UltiMate3000 Nano LC System (Germering, Germany). Digested glycoprotein of 250 fmol was dissolved in 0.1% TFA and loaded onto a precolumn [300 µm i.d. × 5 mm, C18 PepMap100, 5 µm (LC Packing, Sunnyvale, CA)] in order to desalt and concentrate the sample. After their elution from the precolumn, the peptides and glycopeptides mixtures were separated on a nanoflow analytical column [75 µm i.d.×15cm, C18 PepMap 100,3 µm, 100 A, (LC Packing, Sunnyvale, CA)] at a flow rate of 180 nl/min. The elution of the peptides and glycopeptides was achieved using the following mobile phases: 0.1% formic acid (FA)/0.01% TFA/2% ACN (A) and 0.08% FA/0.008% TFA/98% ACN (B). The elution started with 0% B for 10 min, followed by a gradient of 0–60% B in 55 min and 60–90% B in 3 min and was kept at 90% B for 3 min. The tandem mass spectrometry analysis of the eluted peptides and glycopeptides was accomplished using an Applied Biosystems API-QSTAR XL quadrupole orthogonal time-of-flight (QqTOF)-MS/MS hybrid tandem mass spectrometer (Applied Biosystems Intemational-MDS Sciex, Foster City, CA, USA) equipped with a nanoelectrospray source (Protana XYZ manipulator), which produces the electrospray through a PicoTip needle (10 µm i.d., New Objectives, Wobum, MA, USA) carrying a voltage of 2400V. The TOF analyzer was calibrated based on the ions at m/z 586.9815 and m/z 879.9723, derived from renin (1 pmol/µl solution). The collision energies used during the CID-MS/MS analyses were determined automatically using the Information Dependent Acquisition method integrated in the Analyst software.
Results and discussion
Determination of the carbohydrate: protein ratio
Before the conjugation, the rTT–Hc protein was analyzed by surface-enhanced laser desorption/ionization-TOF-MS. The spectrum was characterized by the formation of a protonated molecular ion at m/z 52,149.09 (data not shown).
The MALDI-TOF-MS analysis of the hapten–rTT–Hc glycoconjugate vaccine showed the presence of different protonated molecular ions at m/z 55489.08, m/z 57349.01, m/z 59069.32, m/z 60782.08, m/z 62493.49, m/z 64225.11, m/z 65974.65 and m/z 68105.39 [Fig. 1(b)]. These correspond to vaccine glycoconjugates at different hapten–protein ratios. Thus, knowing that the molecular weight of the antigen hexasaccharide of Vibrio cholerae serotype Ogawa coupled to the squaric acid linker is 1750.78 Da and by comparing the m/z values of the hapten–rTT–Hc glycoconjugates to that of the rTT–Hc protein, we determined that the vaccine is composed of a mixture of eight hapten–rTT–Hc glycoconjugates with the following carbohydrate:protein ratios (Table 1): 1.9:1, 3.0:1, 4.0:1, 4.9:1, 5.9:1,6.9:1, 7.9:1 and 9.1 :1. In addition, the average carbohydrate: protein ratio was found to be 5.3:1.
Table 1.
Protonated molecular ions identified on the Matrix-assisted laser desorption/ionization mass spectrometry spectrum of the hexasaccharide of Vibrio Cholerae serotype Ogawa conjugated to the recombinant tetanus toxin C-fragment and the corresponding carbohydrate-to-protein ratio
| [M + H]+ | Carbohydrate-to-protein ratio |
|---|---|
| 55489.08 | 1.9 |
| 57349.01 | 3.0 |
| 59069.32 | 4.0 |
| 60782.08 | 4.9 |
| 62493.49 | 5.9 |
| 64225.11 | 6.9 |
| 65974.65 | 7.9 |
| 68105.39 | 9.1 |
Determination of the glycation sites of the hexasaccharide– rTT–Hc glycoconjugate
The glycoconjugate was digested with Trypsin and GluC V8, and the digests were analyzed using LC-MS/MS, to ensure the highest possible protein coverage as well as the identification of the highest possible number of glycation sites.
The data acquired during LC-MS/MS analysis of the trypsin digests were submitted to the Mascot database, which allowed identification of peptides from the tetanus toxin (Clostridium tetani) protein with sequence coverage of 58%.
We showed in our previous studies that conjugation by squaric acid chemistry occurs exclusively through lysine residues.[20–23] Based on this, the m/z value of the peptides that did not match to any protein were compared with a table model containing hypothetical molecular weight values of all possible hapten hexasaccharide conjugated to lysine residues of tryptic peptide of the tetanus toxin protein. Subsequently, the mass spectra of each possible glycated peptide were manually processed, looking for the carbohydrate-linker signature product ions, as well as the corresponding peptide sequence. Eleven tryptic glycated peptides were identified (Table 2): FIIK*R (Lys 318) at m/z 1213.1000 (+2), ALNPK*EIEK (Lys 239) at m/z 930.8000 (+3), VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3), LRDLK*TYSVQLK (Lys 389) at m/z 1071.5000 (+3), TYSVQLK*LYDDK (Lys 396) at m/z 1074.5200 (+3), GNNLIWTLK*DSAGEVR (Lys 145) at m/z 1174.5500 (+3), YTPNNEIDSFVK*SGDFIK (Lys 331) at m/z 1274.9400 (+3), NLDCWVDNEEDIDVILK*K (Lys 22) at m/z 1322.9500 (+3), YDTEYYLIPVASSSK*DVQLK (Lys 278) at m/z 1356.6600 (+3), DK*ILGCDWYFVPTDEGWTND (Lys 437) at m/z 1393.9400 (+3) and LYVSYNNNEHIVGYPK*DGNAFNNLDR (Lys 353) at m/z 1194.5800 (+4).
Table 2.
Glycated peptides identified from the tryptic digestion of the hexasaccharide of Vibrio Cholerae serotype Ogawa conjugated to the recombinant tetanus toxin C-fragment carrier by LC-ESI-QqTOF-MS/MS
| Sequence of the glycated peptides (* = glycation site) | ||||
|---|---|---|---|---|
| Molecular weight |
Calculated m/z (charge) |
Observed m/z (charge) |
Mass difference (Da) |
|
| FIIK*R(Lys 318) | 2424.2029 | 1213.1093 (+2) | 1213.1000 (+2) | 0.0093 |
| ALNPK*EIEK(Lys 239) | 2789.3463 | 930.7899 (+3) | 930.8000 (+3) | −0.0101 |
| VGYNAPGIPLYK*K (Lys 378) | 3167.5519 | 1056.8584 (+3) | 1056.8600 (+3) | −0.0016 |
| LRDLK*TYSVQLK (Lys 389) | 3211.6105 | 1071.5446 (+3) | 1071.5000 (+3) | 0.0446 |
| TYSVQLK*LYDDK (Lys 396) | 3220.5156 | 1074.5130 (+3) | 1074.5200 (+3) | −0.0070 |
| GNNLIWTLK*DSAGEVR (Lys 145) | 3520.6814 | 1174.5683 (+3) | 1174.5500 (+3) | 0.0183 |
| YTPNNEIDSFVK*SGDFIK (Lys 331) | 3821.7652 | 1274.9295 (+3) | 1274.9400 (+3) | −0.0105 |
| NLDCWVDNEEDIDVILK*K (Lys 22) | 3965.8220 | 1322.9485 (+3) | 1322.9500 (+3) | −0.0015 |
| YDTEYYLIPVASSSK*DVQLK (Lys 278) | 4066.9279 | 1356.6504 (+3) | 1356.6600 (+3) | −0.0096 |
| DK*ILGCDWYFVPTDEGWTND (Lys 437) | 4178.8071 | 1393.9435 (+3) | 1393.9400 (+3) | 0.0035 |
| LYVSYNNNEHIVGYPK*DGNAFNNLDR (Lys 353) | 4774.1915 | 1194.5557 (+4) | 1194.5800 (+4) | −0.0243 |
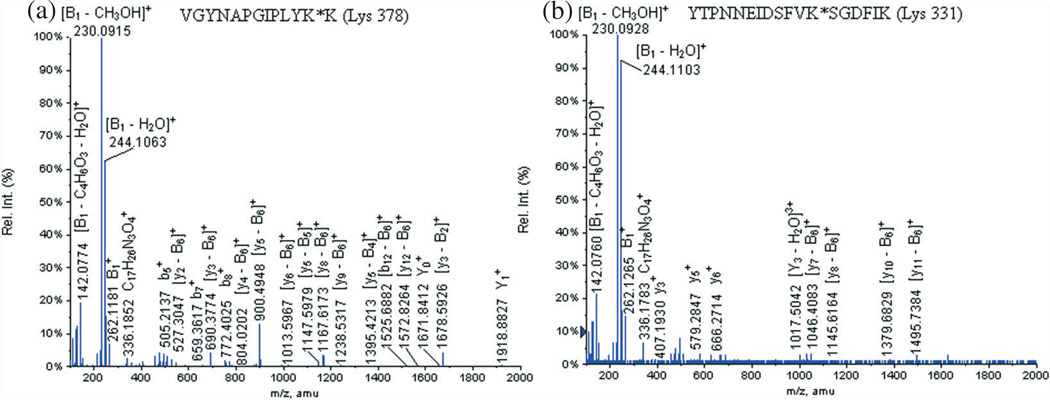
Figure 2(a) shows the MS/MS of the glycated peptide VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3), Table 3 displays the identified product ions and Fig. 3 describes the signature product ions of the hexasaccharide carbohydrate-linker identified on the glycated peptide VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3). The nomenclature established by Domon and Costello[24] was used for the assignment of the carbohydrate product ions. Basically, the fragment ions containing the aglycone are labeled X, Y and Z, and their complementary fragment ions are named A, B and C.
Figure 2.
LC-ESI-QqTOF-MS/MS spectra of the glycated peptides (a) VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3) and (b) YTPNNEIDSFVK*SGDFIK (Lys 331) at m/z 1356.6600 (+3).
Table 3.
Identified product ions LC-ESI-QqTOF-MS/MS analysis of the glycated peptide VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3)
| Production | Calculated m/z |
Observed m/z |
Deviation (Da) |
|
|---|---|---|---|---|
|
|
1919.0155 | 1918.8827 | −0.1328 | |
| [y3−B2]+ | 1678.8036 | 1678.5926 | −0.2110 | |
|
|
1671.9099 | 1671.8412 | −0.0687 | |
| [y12−B6]+ | 1572.8410 | 1572.8264 | −0.0146 | |
| [b12−B6]+ | 1525.8049 | 1525.6882 | −0.1167 | |
| [y2-B2]+ | 1515.7400 | 1515.7789 | 0.0389 | |
| [y7−B5]+ | 1414.7817 | 1414.6484 | −0.1333 | |
| [y5−B4]+ | 1394.7290 | 1395.4213 | 0.6923 | |
| [y10−B6]+ | 1352.7562 | 1352.6128 | −0.1434 | |
| [y9 − b6]+ | 1238.7132 | 1238.5317 | −0.1815 | |
| [ys−B6]+ | 1167.6761 | 1167.6173 | −0.0588 | |
| [y5−B5]+ | 1147.6234 | 1147.5979 | −0.0255 | |
| [y7 − B6]+ | 1070.6234 | 1070.6128 | −0.0106 | |
| [y2 − B4]+ | 1021.5289 | 1021.5341 | 0.0052 | |
| [y6 − B6]+ | 1013.6019 | 1013.5967 | −0.0052 | |
| [y5 − b6]+ | 900.5178 | 900.4948 | −0.0230 | |
|
|
836.4589 | 836.4098 | −0.0491 | |
| [y4−B6]+ | 803.4651 | 804.0202 | 0.5551 | |
|
|
772.3994 | 772.4025 | 0.0031 | |
| [y3−B6]+ | 690.3810 | 690.3774 | −0.0036 | |
|
|
659.3153 | 659.3617 | 0.0464 | |
| [y2 − B6]+ | 527.3177 | 527.3047 | −0.0130 | |
|
|
509.2347 | 509.2240 | −0.0107 | |
|
|
505.2411 | 505.2137 | −0.0274 | |
| [B3−B1]+ | 495.2184 | 495.2060 | −0.0124 | |
| [B3 − B1 − H2O]+ | 477.2079 | 477.1967 | −0.0112 | |
|
|
336.1918 | 336.1852 | −0.0066 | |
|
|
262.1285 | 262.1181 | −0.0104 | |
| [B2−B1]+ | 248.1129 | 248.1006 | −0.0123 | |
| [B, − H2O]+ | 244.1179 | 244.1063 | −0.0116 | |
| [B1−CH3OH]+ | 230.1023 | 230.0915 | −0.0108 | |
|
|
157.0977 | 156.0629 | −1.0348 | |
| [B1−C4H6O3−H2O]+ | 142.0863 | 142.0774 | −0.0089 |
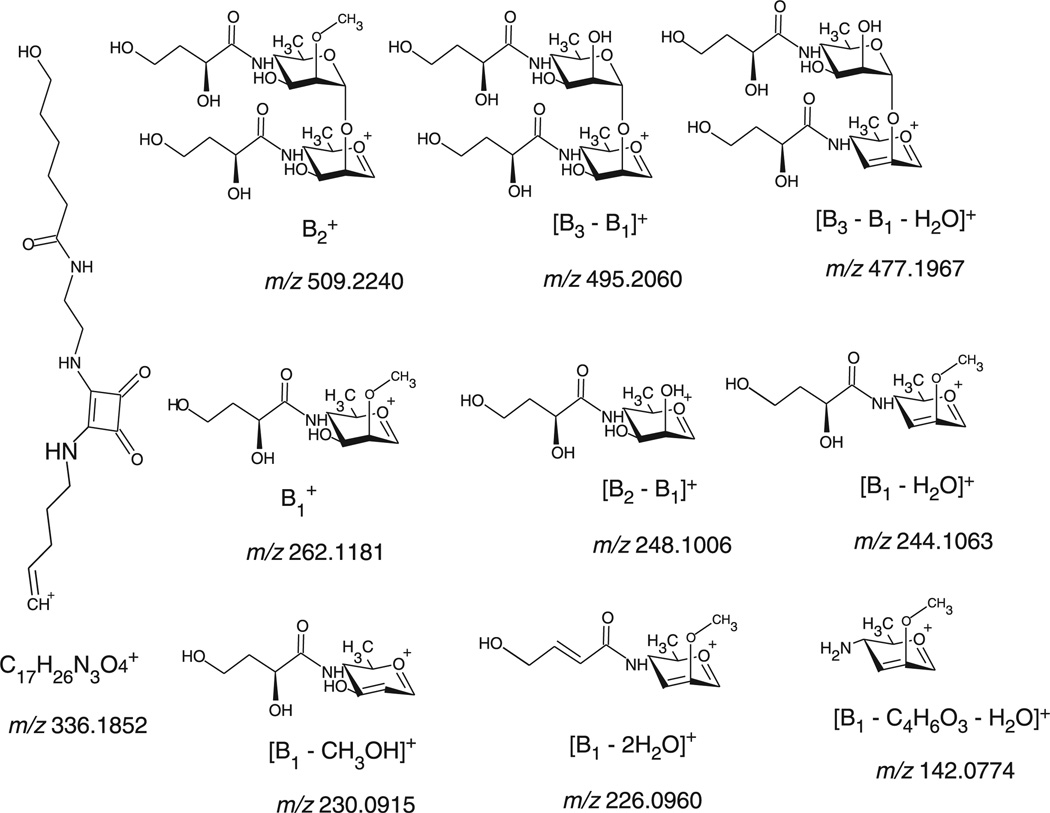
Figure 3.
Signature product ions of the hexasaccharide-linker portion identified during the LC-MS/MS analysis of the glycated peptide VGYNAPGIPLYK*K (Lys 378) at m/z 1056.8600 (+3).
The diagnostic product ions of the carbohydrate-linker portion were observed in all the identified glycated peptides [Fig. 2(a) and Fig. 3, Table 3]: at m/z 336.1852, at m/z 509.2240, [B3 − B1]+ at m/z 495.2060, [B3 − B1 − H2O]+ at m/z 477.1967, at m/z 262.1181, [B2−B1]+ at m/z 248.1006, [B1, − H2O]+ at m/z 244.1063, [B1 − CH3OH]+ at m/z 230.0915 and [B1 − C4H6O3 − H2O]+ at m/z 142.0774. Other distinguishing fragment ions of these glycopeptides are the product ions formed by the glycated peptide that lost part or the full carbohydrate portion [Fig. 2(a)]: at m/z 1918.8827, at m/z 1671.8412 and at m/z 836.4098.
In addition, the MS/MS spectra also revealed fragmented glycated peptide product ions, which lost the carbohydrate portion (B6 =−1496.6Da): [y3−B2] at m/z 1678.5926, [y12−B6] at m/z 1572.8264, [b12−B6] at m/z 1525.6882, [y2−B2] at m/z 1515.7789, [y7−B5]+ at m/z 1414.6484, [y5−B4] at m/z 1395.4213, [y10−B6]+ at m/z 1352.6128, [y9−B6]+ at m/z 1238.5317, [y8 − B6]+ at m/z 1167.6173, [y5 - B5]+ at m/z 1147.5979, [y7 − B6]+ at m/z 1070.6128, [y2 − B4]+ at m/z 1021.5341, [y6 −B6]+ at m/z 1013.5967, [y5 − B6]+ at m/z 900.4948, [y4−B6]+ at m/z 804.0202, [y3 − B6]+ at m/z 690.3774 and [y2 − B6]+ at m/z 527.3047.
An additional example of identified glycated peptide during the LC-MS/MS analysis worth discussing is YTPNNEIDSFVK*SGDFIK (Lys 331) at m/z 1356.6600 (+3) [Fig. 2(b), Table 4]. This glycated peptide followed the same fragmentation pathway as the previously described one. The following signature product ions of the carbohydrate-linker moiety were detected [Fig. 2(b) and Fig. 3, Table 4]: at m/z 336.1783, at m/z 509.2237, [B3 − B1]+ at m/z 495.3028, [B3 − B1 − H2O]+ at m/z 477.1977, at m/z 262.1265, [B2 − B1]+ at m/z 248.1046, [B1 − H2O]+ at m/z 244.1103, [B1−CH3OH]+ at m/z 230.0928 and [B1 − C4H6O3 − H2O]+ at m/z 142.0760. The following product ion composed of the entire glycated peptide that lost a carbohydrate portion was also detected: [Y3−H2O]3+ at m/z 1017.5042. The loss of the entire carbohydrate portion (B6 = − 1496.6 Da) was also detected on the following peptide product ions: [y11 − B6]+ at m/z 1495.7384, [y10− B6]+ at m/z 1379.6829, [y8 − B6]+ at m/z 1145.6164 and [y7 − B6]+ at m/z 1046.4083.
Table 4.
Identified product ions LC-ESI-QqTOF-MS/MS analysis of the glycated peptide YTPNNEIDSFVK*SGDFIK (Lys 331) at m/z 1356.6600 (+3)
| Production | Calculated m/z |
Observed m/z |
Deviation (Da) |
|
|---|---|---|---|---|
| [y11−B6]+ | 1494.7464 | 1495.7384 | 0.9920 | |
| [y10−B6]+ | 1379.7194 | 1379.6829 | −0.0365 | |
| [y8−B6]+ | 1145.6190 | 1145.6164 | −0.0026 | |
| [y7−B6]+ | 1046.5506 | 1046.4083 | −0.1423 | |
| [Y3 − H2O]3+ | 1017.1480 | 1017.5042 | 0.3562 | |
|
|
666.3452 | 666.2714 | −0.0738 | |
|
|
579.3131 | 579.2847 | −0.0284 | |
|
|
509.2347 | 509.2237 | −0.0110 | |
| [B3−B1]+ | 495.2184 | 495.3028 | 0.0844 | |
| [B3 − B1 − H2O]+ | 477.2079 | 477.1977 | −0.0102 | |
|
|
407.2647 | 407.1930 | −0.0717 | |
|
|
336.1918 | 336.1783 | −0.0135 | |
|
|
262.1285 | 262.1265 | −0.0020 | |
|
|
260.1963 | 260.1838 | −0.0125 | |
| [B2−B1]+ | 248.1129 | 248.1046 | −0.0083 | |
| [B, − H2O]+ | 244.1179 | 244.1103 | −0.0076 | |
| [B1−CH3OH]+ | 230.1023 | 230.0928 | −0.0095 | |
|
|
147.1123 | 147.1096 | −0.0027 | |
| [B1−C4H6O3−H2O]+ | 142.0863 | 142.0760 | −0.0103 |
Thus, our analytical strategy allowed us to discover in the tryptic digests of the hexasaccharide–rTT–Hc vaccine a total of 11 glycations sites, exclusively on lysine residues: Lys 22, Lys 145, Lys 239, Lys 278, Lys 318, Lys 331, Lys 353, Lys 378, Lys 389, Lys 396 and Lys 437.
The submission of the LC-MS/MS data of the GluC V8 digests to the Mascot library allowed the identification of the tetanus toxin (Clostridium tetani) protein, with sequence coverage of 39%. The molecular accurate weight of the precursor ions of the peptides that did not match any known peptides in the Mascot library were also compared to a table containing molecular weight values of all possible glycated peptides of the tetanus toxin protein GluC V8 digests. Two potential glycated peptides were identified, each one presenting only one glycation site (Table 5): KFRIFCKALNPKE at m/z 1133.9000 (+3) and AQLVPGINGKAIHLVNNE at m/z 1212.5994 (+3).
Table 5.
Glycated peptides identified on the GluC V8 digests of the hexasaccharide of Vibrio Cholerae serotype Ogawa conjugated to the recombinant tetanus toxin C-fragment carrier the using LC-ESI-QqTOF-MS/MS
| Sequence of the glycated peptides (* = glycation site) | Molecular weight |
Calculated m/z (charge) |
Observed m/z (charge) |
Mass difference (Da) |
|---|---|---|---|---|
| KFRIFCKALNPK*E (Lys 239) | 3399.6751 | 1133.8969 (+3) | 1133.9000 (+3) | 0.0031 |
| AQLVPGINGK*AIHLVNNE (Lys 61) | 3635.8049 | 1212.6068 (+3) | 1212.5994 (+3) | −0.0074 |
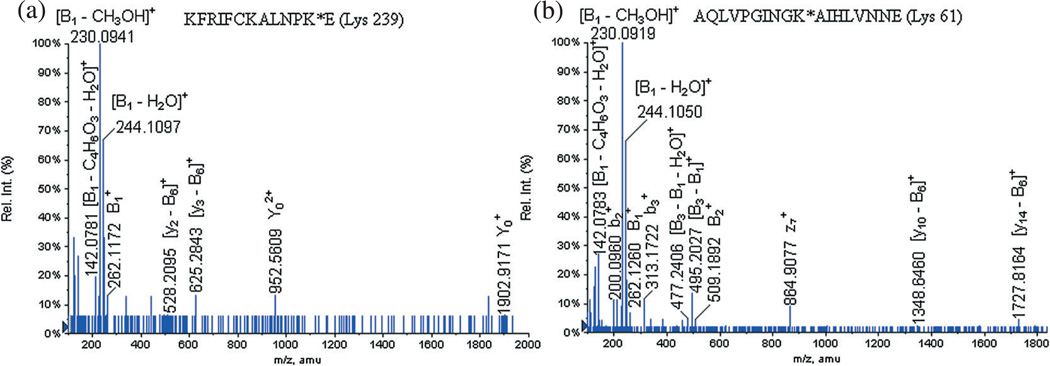
The LC-MS/MS analysis of the glycated peptide KFRIFCKALNPKE at m/z 1133.9000 (+3) allowed the observation of the signature product ions of the carbohydrate-linker moiety [Fig. 4(a), Table 6]: [B3 − B1]+ at m/z 495.2170, at m/z 262.1172, [B1 − H2O]+ at m/z 244.1097, [B1 − CH3OH]+ at m/z 230.0941, [B1 − 2H2O]+ at m/z 226.0956 and [B1 − C4H6O3 − H2O]+ at m/z 142.0781. Also, two formed product ions corresponding to the loss of the entire carbohydrate moiety were detected: at m/z 1902.9171 and at m/z 952.5609. Finally, the loss of the entire carbohydrate portion was also observed on two peptidic fragments: [y3 − B6]+ at m/z 625.2843 and [y2 − B6]+ at m/z 528.2095.
Figure 4.
LC-MS/MS spectra of the glycated peptides KFRIFCKALNPK*E (Lys 239) at m/z 1133.9000 (+3) and AQLVPGINGK*AIHLVNNE (Lys 61) at m/z 1212.5994 (+3) identified on the GluC V8 digests.
Table 6.
Identified product ions LC-ESI-QqTOF-MS/MS analysis of the glycated peptide KFRIFCKALNPK*E (Lys 239) at m/z 1133.9000 (+3)
| Production | Calculated m/z |
Observed m/z |
Deviation (Da) |
|
|---|---|---|---|---|
|
|
1903.0253 | 1902.9171 | 0.1082 | |
|
|
951.9625 | 952.5609 | −0.5984 | |
| [y3−B6]+ | 625.3181 | 625.2843 | 0.0338 | |
| [y2 − b6]+ | 528.2653 | 528.2095 | 0.0558 | |
| [B3−B1] | 495.2184 | 495.2170 | 0.0014 | |
|
|
262.1285 | 262.1172 | 0.0113 | |
| [B1 − H2O]+ | 244.1179 | 244.1097 | 0.0082 | |
| [B1−CH3OH]+ | 230.1023 | 230.0941 | 0.0082 | |
| [B1−2H2O]+ | 226.1074 | 226.0956 | 0.0118 | |
| [B1−C4H6O3−H2O]+ | 142.0863 | 142.0781 | 0.0082 |
It is interesting to remark that the LC-MS/MS of the identified glycated peptide at m/z 1133.9000 (+3) did not contain enough peptidic product ions (y- and b-) to perform full de novo sequencing. We propose that the characterization of the sole carbohydrate portion product ions was due to the privileged effectiveness of fragmentation of the carbohydrate ions when compared to peptide ions.
However, it is interesting to mention the presence of a unique glycopeptide KFRIFCKALNPKE specific to the hexasaccharide of Vibrio cholerae serotype Ogawa at m/z 1133.9000 (+3); this glycated peptide appears to contain only one glycation site on the peptide: [Fig. 4(a), Table 6]. The precursorion scan of this glycopeptide at m/z 1133.9000 afforded the productions: at m/z 262.1172, [B1 − H2O]+ at m/z 244.1097, [B1 −CH3OH]+ at m/z 230.0941, [B1 - 2H2O]+ at m/z 226.0956 and [B1 − C4H6O3 − H2O]+ at m/z 142.0781. It was found that this glycated peptide contains three lysine residues (Lys 228, Lys 234 and Lys 239), and one of them is conjugated to the hexasaccharide of Vibrio cholerae serotype Ogawa. We have also detected the two peptidic product ions formed by the loss of the entire carbohydrate portion, and these were assigned as [y3 − B6]+ at m/z 625.2843 and [y2 − B6]+ at m/z 528.2095. This permits us simply to conclude that the original glycation site was on the Lys 239 residue. These results seem to confirm our original finding that during the LC-MS/MS analysis of the tryptic digests, a glycation site was also identified on the Lys 239 residue.
The LC-MS/MS analysis of the glycated peptide AQLVPGINGKAIHLVNNE at m/z 1212.5994 (+3) also allowed us to identify product ions corresponding to the carbohydrate portion [Fig. 4(b), Table 7]: at m/z 509.1892, [B3−B1]+ at m/z 495.2027, [B3 − B1 − H2O]+ at m/z 477.2406, at m/z 262.1260, [B2−B1]+ at m/z 248.1016, [B1 − H2O]+ at m/z 244.1050, [B1−CH3OH]+ at m/z 230.0919 and [B1−C4H6O3− H2O]+ at m/z 142.0783. As for the previously discussed glycated peptides, we also observed product ions corresponding to the loss of the entire carbohydrate portion on peptidic fragment ions: [y14−B6]+ at m/z 1727.8164 and [y10−B6]+ at m/z 1348.6460. The glycated peptide only contains one lysine residue (Lys 61), and as we already reported that the squaric acid chemistry allows the conjugation on exclusively lysine residues[15,17–19] we can conclude that the glycation site corresponds to the Lys 61.
Table 7.
Identified product ions LC-ESI-QqTOF-MS/MS analysis of the glycated peptide AQLVPGINGK*AIHLVNNE (Lys 61) at m/z 1212.5994 (+3)
| Production | Calculated m/z |
Observed m/z |
Deviation (Da) |
|
|---|---|---|---|---|
| [y14−B6]+ | 1727.9064 | 1727.8164 | 0.0900 | |
| [y10−B6]+ | 1346.7052 | 1348.6460 | −1.9408 | |
|
|
864.4216 | 864.9077 | −0.4861 | |
|
|
509.2347 | 509.1892 | 0.0455 | |
| [B3−B1]+ | 495.2184 | 495.2027 | 0.0157 | |
| [B3 − B1 − H2O]+ | 477.2079 | 477.2406 | −0.0327 | |
|
|
313.1876 | 313.1722 | 0.0154 | |
|
|
262.1285 | 262.1260 | 0.0025 | |
| [B2−B1]+ | 248.1129 | 248.1016 | 0.0113 | |
| [B1 − H2O]+ | 244.1179 | 244.1050 | 0.0129 | |
| [B1−CH3OH]+ | 230.1023 | 230.0919 | 0.0104 | |
|
|
200.1035 | 200.0960 | 0.0075 | |
| [B1−C4H6O3−H2O]+ | 142.0863 | 142.0783 | 0.0080 |
As a result, the LC-MS/MS analysis of the GluC V8 digests of the hexasaccharide of Vibrio cholerae serotype Ogawa conjugated to the rTT–Hc carrier allowed us to identify two glycation sites on the following amino acids: Lys 61 and Lys 239. In addition, the total protein coverage calculated using the different peptides and glycated peptides identified on the trypsin and GluC V8 digests was estimated to be 83.6%.
Conclusion
We mapped the glycation sites on the neoglycoconjugate made from the hexasaccharide antigen of Vibrio cholerae, serotype Ogawa, conjugated to the rTT–Hc carrier, composed of glycoconjugates having hapten-to-protein ratios of 1.9:1, 3.0:1, 4.0:1, 4.9:1, 5.9:1, 6.9:1, 7.9:1 and 9.1:1. The LC-MS/MS analysis of the tryptic and GluC V8 digests allowed us to identify 12 glycation sites on the glycoconjugate (Fig. 5). As observed in our previous studies, all the identified glycation sites are located exclusively on lysine residues.[20–23] This suggests that while the squaric acid chemistry allows an amino acid specific conjugation, it does not allow a site specific conjugation.
Figure 5.
Amino acid sequence of the rTT–Hc protein. The identified glycated lysine residues are highlighted with a star.
The group of Vann also carried out the chemoenzymatic synthesis of immunogenic meningococcal group C polysialic acid-tetanus Hc fragment glycoconjugates. They conjugated an alkyne squarate to the rTT–Hc protein and analyzed the tryptic digests of this neoglycoconjugate using LC-ESI-MS, in order to reveal the modification sites on the carrier protein.[25] They were thus able to identify a maximum of 14 modifications on the rTT–Hc protein, most of them corresponding to the glycation sites we identified in this manuscript. This increases the confidence of the method we are using to reveal the modification sites on the neoglycoconjugates formed by the attachment of antigenic carbohydrates to a protein carrier, using the squaric acid chemistry.
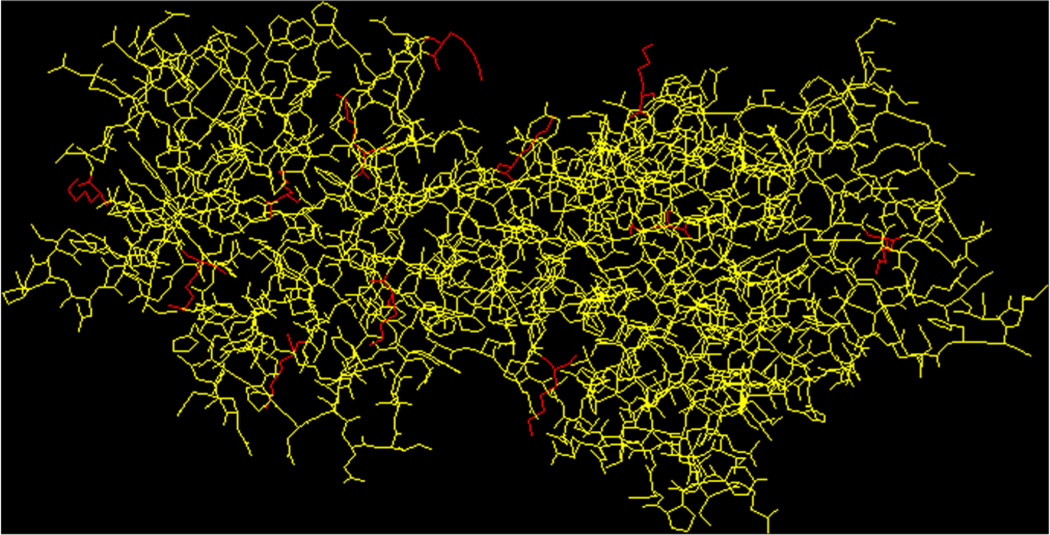
Moreover, the three-dimensional structure of the rTT–Hc protein, constructed using the Protein Database (Fig. 6),[26–28] showed that the lysine residues involved in conjugation (representation in red in Fig. 6) are located at the outer surface of the protein. Presumably, the presence of carbohydrate antigens at or near the outer surface of protein carriers makes them available to interact with elements of the immune system following vaccination.[29]
Figure 6.
Three-dimensional representation of the rTT–Hc protein. The glycated lysine residues are displayed in red.
In our previous studies,[21] we demonstrated the advantage of using LC-ESI-QqTOF-MS/MS, rather than MALDI-MS/MS, for establishing the glycation sites in glycoconjugates in which the carbohydrate portion is of low molecular weight (up to 950 Da).[21] Indeed, the LC-MS/MS analysis of the Bacillus anthracis neoglycoconjugate vaccine digests allowed us to identify 18 glycation sites, while only five glycation sites were observed during the MALDI-MS/MS analysis.[21]
References
- 1.Kuberan B, Linhardt RJ. Carbohydrate based vaccines. Curr. Org. Chem. 2000;4:653. [Google Scholar]
- 2.Lay L, Panza L, Poletti L, Prosperi D, Canevari S, Perico ME. Improvement of the synthesis of immunological carbohydrate vaccines containing the tumour associate antigen CaMBr1. Eur. J. Org. Chem. 2001;2001:4331. [Google Scholar]
- 3.Costantino P, Rappuoli R, Berti F. The design of semi-synthetic and synthetic glycoconjugate vaccines. Expert Opin. Drug Discov. 2011;6:1045. doi: 10.1517/17460441.2011.609554. [DOI] [PubMed] [Google Scholar]
- 4.Hou SJ, Saksena R, Kováč P. Preparation of glycoconjugates by dialkyl squarate chemistry revisited. Carbohydr. Res. 2008;343:196. doi: 10.1016/j.carres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bundle DR. New frontiers in the chemistry of glycoconjugate vaccines. In: Bagnoli F, Rappuoli R, editors. Vaccine Design: Innovative Approaches and Novel Strategies. Caister Academic Press; 2011. p. 69. [Google Scholar]
- 6.Saksena R, Chernyak A, Poirot E, Kovác P. Conjugating low molecular mass carbohydrates to proteins 2 Recovery of the excess ligand used in the conjugation reaction. Methods Enzymol. 2003;362:140. doi: 10.1016/S0076-6879(03)01011-5. [DOI] [PubMed] [Google Scholar]
- 7.I.Auzanneau F, Pinto M. Preparation of antigens and immunosirbents corresponding to the streptococcus group A cell-wall polysaccharide. Bioorg. Med. Chem. 2003;4 doi: 10.1016/s0968-0896(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Yergey A, Kowalak J, Kováč P. Studies towards neoglycoconjugates from the monosaccharide determinant of Vibrio chlerae 0:1 serotype Ogawa, using the diethyl squarate reagent. Carbohydr. Res. 1998;313:15. doi: 10.1016/s0008-6215(98)00261-4. [DOI] [PubMed] [Google Scholar]
- 9.Saksena R, Ma X, Kováč P. One-pot preparation of a series of glycoconjugates with predetermined antigen-carrier ratio from oligosaccharides that mimic the O–PS of Vibrio cholerae O:1, serotype Ogawa. Carbohydr. Res. 2003;338:2591. doi: 10.1016/s0008-6215(03)00273-8. [DOI] [PubMed] [Google Scholar]
- 10.Chernyak A, Karavanov A, Ogawa Y, Kováč P. Conjugating oligosaccharides to proteins by squaric acid diester chemistry: rapid monitoring of the progress of conjugation, and recovery of the unused ligand. Carbohydr. Res. 2001;330:479. doi: 10.1016/s0008-6215(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, Saksena R, Chernyak A, Karavanov A, Kováč P. Neoglycoconjugates from synthetic tetra- and hexasaccharides that mimic the terminus of the O–PS of Vibrio cholerae 0:1, serotype Inaba. Org. Biomol. Chem. 2003;7:775. doi: 10.1039/b211660j. [DOI] [PubMed] [Google Scholar]
- 12.Saksena R, Chernyak A, Karavanov A, Kováč P. Conjugating low molecular mass carbohydrates to proteins 1 Monitoring the progress of conjugation. Meth. Enzymol. 2003;362:125. doi: 10.1016/S0076-6879(03)01010-3. [DOI] [PubMed] [Google Scholar]
- 13.Bongat AFG, Saksena R, Adamo R, Fujimoto Y, Shiokawa Z, Peterson DC, Fukase K, Vann WF, Kováč P. Multimeric bivalent immunogens from recombinant tetanus toxin HC fragment, synthetic hexasaccharides and a glycopeptide adjuvant. Glycoconj. J. 2010;27:69. doi: 10.1007/s10719-009-9259-4. [DOI] [PubMed] [Google Scholar]
- 14.Rollenhagen JE, Kalsy A, Saksena R, Sheikh A, Alam MM, Qadri F, Calderwood SB, Wade WF, Kováč P, Ryan ET. Transcutaneous immunization with a synthetic hexasaccharide-protein conjugate induces anti-Vibrio cholerae lipopolysaccharide responses in mice. Vaccine. 2009;27:4917. doi: 10.1016/j.vaccine.2009.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarique AA, Kalsy A, Arifuzzaman M, Rollins SM, Charles RC, Leung DT, Harris JB, LaRocque RC, Sheikh A, Bhuiyan MS, Saksena R, Clements JD, Calderwood SB, Qadri F, Kováč P, Ryan ET. Transcutaneous immunization with a Vibrio cholerae O1 Ogawa synthetic hexasaccharide conjugate following oral whole-cell cholera vaccination boosts vibriocidal responses and induces protective immunity in mice. Clin. Vaccine Immunol. 2012;19:594. doi: 10.1128/CVI.05689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chernyak A, Kondo S, Wade TK, Meeks MD, Alzari PM, Fournier JM, Taylor RK, Kovac P, Wade WF. Induction of protective immunity by synthetic Vibrio Cholerae hexasaccharide derived from Vibrio cholerae O:1 Ogawa lipopolysaccharide bound to a protein carrier. J. Infect. Dis. 2002;185:950. doi: 10.1086/339583. [DOI] [PubMed] [Google Scholar]
- 17.Helting TB, Zwisler O, Wiegandt H. Structure of tetanus toxin. II. Toxin binding to ganglioside. J. Biol. Chem. 1977;252:194. [PubMed] [Google Scholar]
- 18.Umland TC, Wingert LM, Swaminathan S, Furey WF, Schmidt JJ, Sax M. Structure of the receptor binding fragment HC of tetanus neurotoxin. Nat. Struct. Biol. 1997;4:788. doi: 10.1038/nsb1097-788. [DOI] [PubMed] [Google Scholar]
- 19.Giménez E, Benavente F, Barbosa J, Sanz-Nebot V. Towards a reliable molecular mass determination of intact glycoproteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:2555. doi: 10.1002/rcm.3109. [DOI] [PubMed] [Google Scholar]
- 20.Jahouh F, Saksena R, Aiello D, Napoli A, Sindona G, Kováč P, Banoub JH. Glycation sites in neoglycoconjugates from the terminal monosaccharide antigen of the O-PS of Vibrio cholerae O1, serotype Ogawa, and BSA revealed by matrix-assisted laser desorption-ionization tandem mass spectrometry. J. Mass Spectrom. 2010;10:1148. doi: 10.1002/jms.1796. [DOI] [PubMed] [Google Scholar]
- 21.Jahouh F, Hou SJ, Kováč P, Banoub JH. Determination of the glycation sites of Bacillus anthracis neoglycoconjugate vaccine by MALDI-TOFATOF-CID-MS/MS and LC-ESI-QqTOF-tandem mass spectrometry. J. Mass Spectrom. 2011;46:993. doi: 10.1002/jms.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahouh F, Hou SJ, Kováč P, Banoub JH. Determination of glycation sites by tandem mass spectrometry in a synthetic lactose-bovine serum albumin conjugate, a vaccine model prepared by dialkyl squarate chemistry. Rapid Commun. Mass Spectrom. 2012;26:1. doi: 10.1002/rcm.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahouh F, Saksena R, Kováč P, Banoub JH. Revealing the glycation sites in synthetic neoglycoconjugates formed by conjugation of the antigenic monosaccharide hapten of Vibrio cholerae O1 serotype Ogawa with the BSA protein carrier using LC-ESI-QqTOF-MS/MS. J. Mass Spectrom. 2012;47:890. doi: 10.1002/jms.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domon B, Costello C. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 1988;5:397. [Google Scholar]
- 25.McCarthy PC, Saksena R, Peterson DC, Lee C-H, An Y, Cipollo JF, Vann WF. Chemoenzymatic synthesis of immunogenic meningococcal group C polysialic acid-tetanus Hc fragment Glycoconjugates. Glycoconj. J. doi: 10.1007/s10719-013-9490-x. submitted. [DOI] [PubMed] [Google Scholar]
- 26.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinf. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 27.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophor. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Rappuoli R, De Gregorio E. A sweet T cell response. Nat. Med. 2001;17:1551. doi: 10.1038/nm.2587. [DOI] [PubMed] [Google Scholar]