Abstract
Sparsely populated transient states of proteins and their complexes play an important role in many biological processes including protein–protein and protein–DNA recognition, allostery, conformational selection, induced fit and self-assembly. These states are difficult to study as their low population and transient nature makes them effectively invisible to conventional structural and biophysical techniques. In the present article, I summarize recent NMR developments in our laboratory, including the use of paramagnetic relaxation enhancement, lifetime line broadening and dark-state exchange saturation transfer spectroscopy, that have permitted such sparsely populated states to be detected, characterized and, in some instances, visualized. I illustrate the application of these methods to the elucidation of mechanisms whereby transcription factors locate their specific target sites within an overwhelming sea of non-specific DNA, to the characterization of encounter complexes in protein–protein recognition, to largescale interdomain motions involved in ligand binding, and to the interaction of monomeric amyloid β-peptide with the surface of amyloid protofibrils and the internal cavity surface of the chaperonin GroEL.
Keywords: conformational selection, encounter complex, induced fit, macromolecular assembly, protein-DNA interaction, sparsely populated state
Introduction
Proteins and their complexes are not static but dynamic and are best described as an ensemble of states. The major species is located at the minimum of the free energy landscape and represents the species that has been studied with so much success by conventional structural and biophysical techniques, including crystallography and NMR. But the static picture revealed by these studies does not describe the complete picture. In addition to the major species, there exist highly transient sparsely populated states that arise from rare excursions between the minimum free energy configuration and other local minima of the free energy landscape [1]. Given that the populations of such sparsely populated states are so low that they are effectively invisible, one might well ask why they should even be studied. It turns out that many biological processes, including molecular recognition and binding, allostery, conformational selection, induced fit and self-assembly, proceed via such sparsely populated states. But little is known about the structural properties of sparsely populated states, since they are difficult to trap and therefore inaccessible (i.e. invisible) to conventional methods. Recent advances in solution NMR spectroscopy have opened the door to directly studying sparsely populated states of proteins and their complexes.
There are three NMR techniques that can be used to investigate sparsely populated states with occupancies ranging from 0.5 to 5 % and lifetimes spanning nanoseconds to milliseconds. PRE (paramagnetic relaxation enhancement) probes states with lifetimes less than 250–500 µs and is dependent on distances between a paramagnetic label and protons being shorter in the minor species than the major one [2,3]. Relaxation dispersion spectroscopy probes states with lifetimes typically ranging from 0.5 to 10 ms (although in particularly fortunate cases, lifetimes as short as 50 µs can be reached) and is dependent on significant chemical shift differences between the resonances of the major and minor species [4–6]. Lastly, DEST (dark-state exchange saturation transfer) spectroscopy and lifetime line broadening can probe events on timescales ranging from 500 µs to 100 ms and are dependent on very large differences in transverse relaxation (R2) rates (i.e. linewidths) between the major and minor species [7,8].
In the present article, I first describe the underlying basis for the use of the PRE to detect sparsely populated states and illustrate its application to protein-DNA and protein-protein interactions, as well as to the investigation of the interplay between conformational selection and induced fit. I then describe the basis for lifetime line broadening and DEST to study interactions between NMR visible molecules and large macromolecular assemblies, and illustrate this by studies on the interaction of soluble monomeric Aβ (amyloid β-peptide) with amyloid protofibrils and the prototypical chaperonin GroEL.
Fundamentals of the PRE
The PRE is caused by magnetic dipolar interactions between a nucleus and the unpaired electron of a paramagnetic centre [9,10]. In the absence of an intrinsic paramagnetic centre (e.g. paramagnetic metalloproteins), a paramagnetic label has to be covalently attached to the macromolecule of interest. In the case of proteins, this generally involves covalent linkage to an engineered surface cysteine residue [3,11]. The PRE results in an increase in the relaxation rate of 1H nuclear magnetization, the magnitude of which is directly proportional to <r−6>, where r is the distance between the unpaired electron and the proton of interest. Because the magnetic moment of an unpaired electron is very large, the PRE effect is detectable for sizeable separations, for example up to ~34 Å (1 Å = 0.1 nm) in the case of Mn2+. In the case of paramagnetic centres with an isotropic g tensor, the only paramagnetic effect is the PRE [3]. (For anisotropic g tensors, pseudo-contact shifts are also observed.) The most reliable method to make use of the PRE is to measure the transverse PRE rate, Γ2, by taking the difference in transverse relaxation rates (R2) between the paramagnetic sample and a diamagnetic control (e.g. Mn2+ compared with Ca2+) [12,13]. Because the paramagnetic centre is generally attached to macromolecules via a flexible linker, the original PRE formalism of Solomon [9] and Bloembergen and Morgan [10] has to be extended to a model-free formalism to take into account the influence of internal motions of the paramagnetic label on the PRE [12]. The availability of long-range distance information from PRE measurements can obviously be very useful in NMR structure determination, especially as one can refine directly against the PRE data [12].
In a system in which there is rapid exchange between various species, the large magnitude of the PRE at short distances can be taken advantage of to detect sparsely populated states [2,3]. For example, let us say that we have a system comprising a major species with an occupancy of 99 % and a paramagnetic centre-proton distance of 30 Å, and a minor species at an occupancy of 1 % with a corresponding distance to the same proton of 8 Å. For a ~20 kDa system, the PRE rate will have a value of ~2 s −1 for the major species and ~5600 s−1 for the minor species. The dependence of the observed PRE rate, , measured on the spectrum of the major species, can be calculated exactly using the McConnell [14] equations. In the fast exchange regime, where the exchange rate kex between the two species is significantly (> 10-fold) larger than the difference in Γ2 values, (~51 s−1) will simply be given by the population weighted average of the Γ2 rates for the major and minor species. Under these conditions, the footprint of the spectroscopically invisible minor species will be manifested on the PRE profiles measured on the NMR resonances of the major species, providing there are paramagnetic centre-proton distances that are shorter in the minor species than the major one. Even if exchange between the species is in the intermediate regime where the difference in Γ2 values is approximately equal to kex, the fast exchange approximation can still be used even though may be a factor of two smaller than the population weighted average. This is due to the <r−6> dependence of the PRE such that assuming that is a population-weighted average would only result in a 10–15 % overestimation in distance.
Detecting intermediates in specific protein-DNA interactions by PRE
Transcription factors have to be able to rapidly locate their specific target sites on DNA in an overwhelming sea of nonspecific DNA. The search process involves three possible mechanisms: one-dimensional diffusion along the DNA (intramolecular translocation; Figure 1B), direct transfer from one segment of DNA to another without dissociation into free solution (intermolecular translocation), and dissociation followed by reassociation at another site [15].
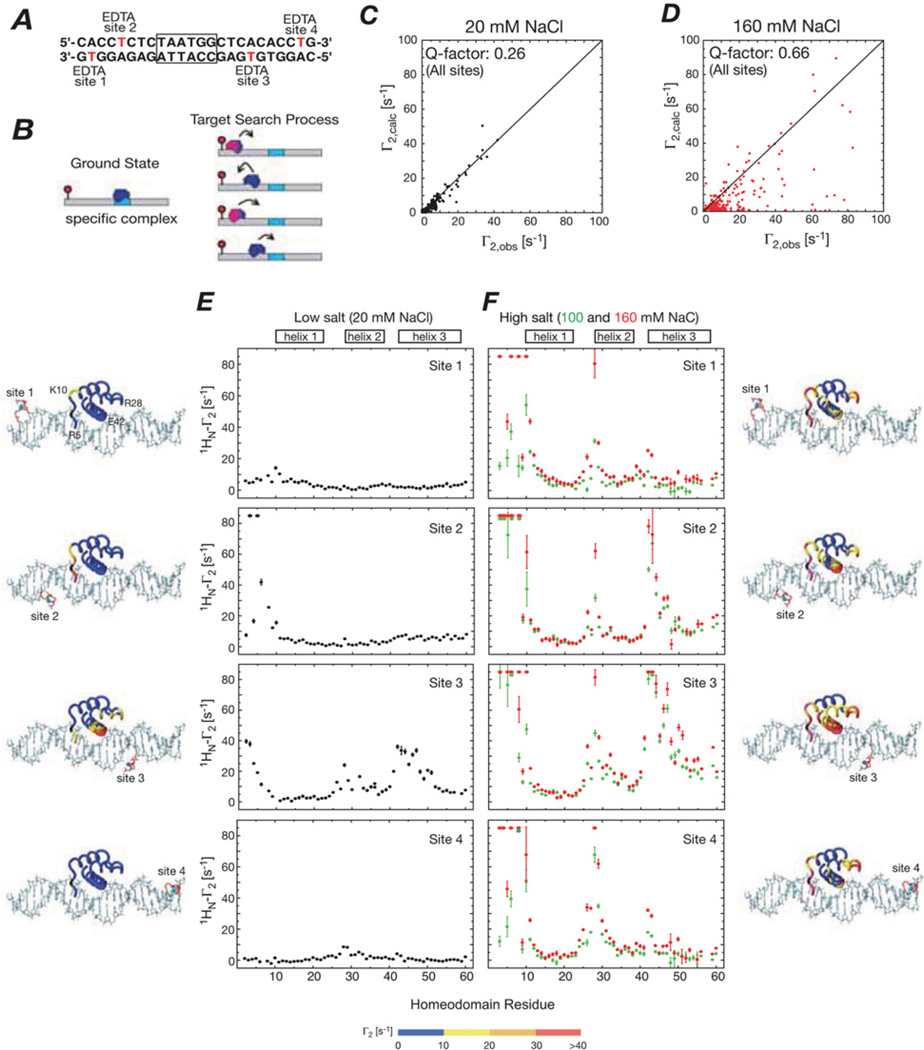
Figure 1. Intermolecular PREs observed for the specific HoxD9-DNA complex in low and high salt corresponding to slow and fast exchange regimes on the PRE relaxation time scale respectively for the interconversion between the specific complex and sparsely populated (<1 %) non-specific complexes.
(A) A 24-bp DNA duplex with the specific site centrally located (boxed) and the location of the four paramagnetic labels (one at a time) indicated. (B) Diagrammatic representation of the specific complex (left) and the target search process whereby the specific complex is located (right). (C and D) Correlation between observed and calculated PREs for all four sites at low (20 mM) and high (160 mM) NaCl concentrations respectively. The calculated Γ2 values are obtained from a model [2] derived from the crystal structure of the Antennapedia homeodomain/DNA complex [40]. (E and F) Intermolecular PRE profiles observed at low (20 mM) and high (100 and 160 mM) NaCl respectively. The PRE data are mapped on the structural model of the specific complex, with the colour-coding depicting the observed PRE values. Adapted from Iwahara, J. and Clore, G.M. (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 with permission.
We initially examined the binding of the homeodomain transcription factor HoxD9 to a 24-bp duplex DNA with the specific site centrally located [2] (Figure 1). The equilibrium dissociation constant at 100 mM NaCl is 1.5 nM with a dissociation rate constant koff of less than 0.01 s−1. Intermolecular PRE data for 15N-labelled HoxD9 were recorded using an EDTA–Mn2+ paramagnetic tag located at four different sites (individually) on the DNA (Figure 1A). At low salt, the PRE profiles from the four sites are in excellent agreement with the structure of the specific complex (Figures 1C and 1E). As the salt is raised to 100 or 160 mM NaCl, the PRE profiles bear no resemblance to those expected for the specific complex (Figures 1D and 1F); furthermore, the PRE profiles for sites 1 and 4 (paramagnetic labels located at bp 2 and 23) and for sites 2 and 3 (paramagnetic labels located at bp 5 and 18) are basically the same. Large PRE effects are observed for all residues close to the DNA-binding surface, even though many of the residues are far away from the paramagnetic label in the specific complex. This must reflect the presence of short-lived binding intermediates located at non-specific DNA-binding sites in close proximity to the paramagnetic labels. The occupancy of these minor states is ~0.5%, as judged from the ratio of specific to non-specific equilibrium binding constants.
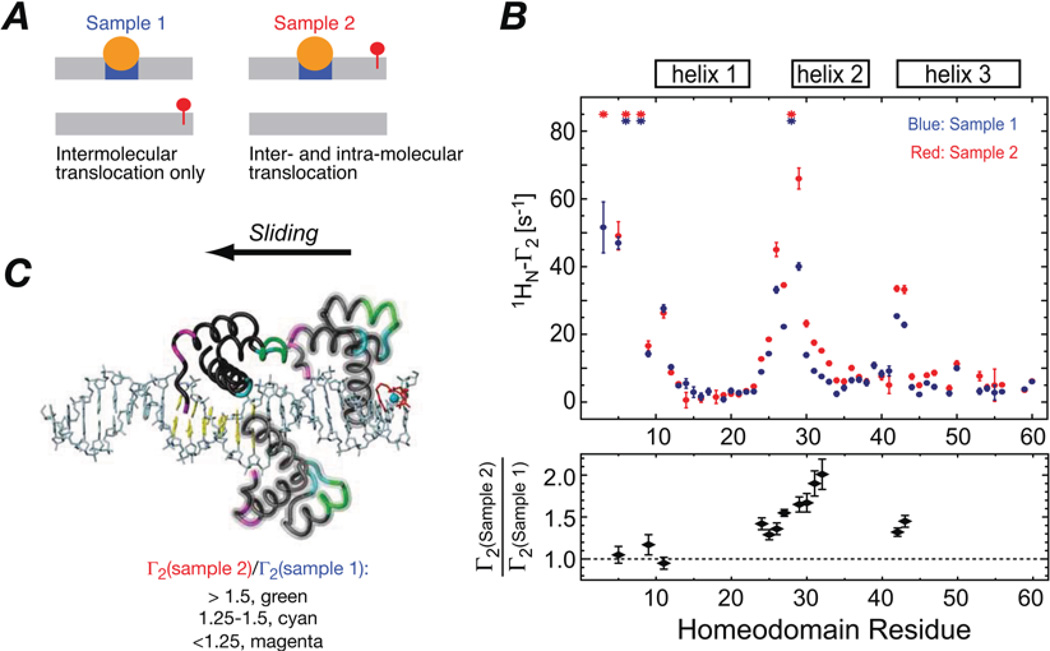
To separate the contributions to the intermolecular PREs from sliding and intermolecular translocation, PRE measurements were carried out on two samples containing an equal mixture of specific and non-specific DNA duplexes (Figure 2A). In the first sample, the paramagnetic label was attached to the end of the non-specific DNA duplex so that any intermolecular PREs must arise exclusively from intermolecular translocation involving transfer of HoxD9 from the specific duplex to the non-specific one and back again. (Dissociation from the DNA into free solution, followed by reassociation is excluded as the dissociation rate constant from the specific site is many orders of magnitude too low to permit a PRE to be observed.) In the second, the specific DNA duplex is paramagnetically labelled so that PREs can arise from both intra- and inter-molecular translocation. The PRE profiles observed for the two samples are very similar, but the PREs are larger for sample 2 in the region of the DNA recognition helix (helix 2), whereas those for the N-terminal tail are the same for both samples (Figure 2B). The larger PREs for helix 2 reflect sliding along the DNA as the DNA recognition helix can come into close proximity to the paramagnetic label while the N-terminal tail remains far away (Figure 2C). The N-terminal tail, on the other hand, can only come into close proximity to the paramagnetic label following an intermolecular translocation event coupled with a 180° change in binding orientation of the specific DNA.
Figure 2. Assessing the contributions of intramolecular sliding and intermolecular translocation for the HoxD9-DNA complex.
(A) PRE data were collected on two samples containing an equimolar concentration of specific and non-specific DNA duplexes with the paramagnetic label attached to the end of the non-specific duplex in sample 1 and to the end of the specific duplex in sample 2. The location of the specific site and the paramagnetic label are indicated in blue and red respectively. (B) PRE profiles observed for samples 1 (blue) and 2 (red) are shown in the upper panel, and the ratio of the observed PRE rates for the two samples is shown in the lower panel. (C) Schematic representation of sliding along the DNA starting from the specific site with HoxD9 coloured according to the Γ2 (sample 2)/Γ2(sample 1) ratio. Adapted from Iwahara, J. and Clore, G.M. (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 with permission.
We have since conducted similar experiments on Oct1 [16], a transcription factor containing two DNA-binding domains known as POUS and POUHD; the minor-groove DNA binding HMG (high-mobility group)-box protein Sox2 [17]; and finally a ternary Oct1–Sox2–DNA complex [17]. In the case of Oct1, the POUHD domain predominantly samples the DNA by sliding, whereas the POUS domain functions as an antenna to promote intermolecular translocation. The latter involves the formation of a bridged intermediate in which the POUHD domainislocated onthe first DNA duplex and the POUS domain is on the second, thereby promoting the completion of intermolecular translocation by a first-order process involving dissociation of POUHD from the first DNA duplex followed by association on to the second duplex. The cross-talk between the POUS and POUHD domains, each fulfilling different roles in the search process ensures efficient sampling of DNA, thereby facilitating the location of Oct1 target sites. Sox2, on the other hand, searches the DNA primarily via intermolecular translocation, and sliding only plays a minor role. In the ternary Oct1– Sox2–DNA complex, Sox2 interacts directly with the POUS domain of Oct1, thereby effectively locking POUS on to its specific DNA site, leaving the POUHD domain to search for other DNA sites predominantly via intermolecular translo-cation.
Characterizing encounter complexes in protein–protein association by PRE
Generally, protein–protein recognition proceeds via a two-step process involving the initial formation of an ensemble of short-lived encounter complexes generated by diffusion-controlled intermolecular collisions, followed by translations and rotations of the two partners down a two-dimensional funnel-like energy landscape to reach a well-defined specific complex stabilized by a set of complementary van der Waals and electrostatic interactions [18]. In essence, encounter complexes in protein–protein recognition are the two-dimensional equivalent of one-dimensional sliding in specific protein–DNA recognition.
We investigated this phenomenon using a complex between the N-terminal domain of Enzyme I (EIN) and the histidine phosphocarrier protein HPr of the bacterial PTS (phosphotransferase system) [19–21]. Paramagnetic labels in the form of EDTA–Mn2+ were conjugated one at a time to three surface-engineered cysteine residues of HPr. When PREs are recorded on a sample comprising EIN at natural isotopic abundance and HPr both uniformly 15N-labelled and paramagnetically tagged, there is excellent agreement between the observed PREs which, under these conditions, are exclusively intramolecular, and those calculated from the structure of HPr. However, when EIN is 15N-labelled and HPr is at natural isotopic abundance, but paramagnetically tagged so that the observed PREs are exclusively intermolecular, the agreement between experimental PREs and those calculated from the structure of the specific complex reveal significant discrepancies (Figure 2A). Although there are regions of the experimental PRE profiles that reflect the specific complex, it is clear that there are stretches of observed PREs that are much larger than expected. Thus there are regions on EIN that spend a small proportion of the time much closer to the paramagnetic tags on HPr than in the specific complex. Similar findings were obtained for complexes of HPr with other proteins of the PTS. The discrepancies between the observed PRE profiles and those predicted from the structure of the specific complex are indicative of the existence of encounter complexes populated at the 5–10% level. The distribution of HPr on the surface of EIN in the encounter complex ensemble can then be calculated by PRE-driven ensemble simulated annealing.
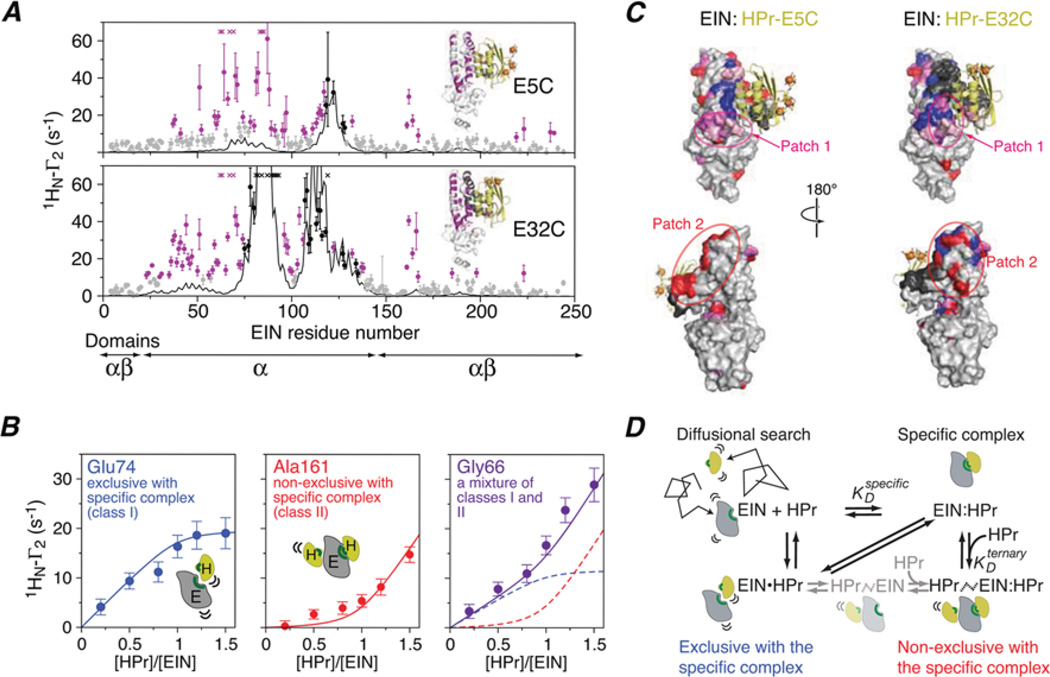
Further analysis of the intermolecular PRE intensities as a function of added paramagnetically labelled HPr yield further insights into the nature of the encounter complexes [21] (Figure 3B). The PREs attributable to the specific complex titrate as a simple one-site binding isotherm with an equilibrium dissociation constant Kd of ~7 µM, in agreement with the Kd determined by isothermal titration calorimetry. The PREs arising from the encounter complexes, however, titrate as three classes (Figure 3B). Class I PREs display the same titration behaviour as the intermolecular PREs directly attributable to the specific complex. Thus these intermolecular PREs arise from encounter complexes that are exclusive with the specific complex (i.e. the binding sites must overlap such that the specific complex and the class I encounter complexes cannot coexist simultaneously). The magnitude of the class II PREs follows the concentration of free HPr, and therefore class II PREs arise from encounter complexes that are non-exclusive with the specific complex (i.e. class II encounter complexes coexist with the specific complex). Finally, class III PREs exhibit a mixture of class I and II behaviour. When the three classes of encounter complex PREs are mapped on to the surface of EIN (Figure 3C), it is clear that class I PREs report on encounter complexes near the active site of EIN that are occluded by the specific complex. The class II PREs report on ternary HPrnon-specific–EIN–HPr complexes that predominate when the active site is occupied by the specific complex. These findings can be summarized by the equilibrium binding model shown in Figure 3(D).
Figure 3. Characterization of transient sparsely populated encounter complexes for the interaction of EIN and HPr.
(A) Comparison of experimental backbone amide intermolecular PREs (1HN-Γ2) (circles) observed on 15N-labelled EIN and arising from covalently attached paramagnetic tags (EDTA-Mn2+) located at two positions on HPr (E5C and E32C) with the PRE profiles calculated from the structure of the specific complex (black line). Black and purple circles indicate PREs attributable to the specific complex and to an ensemble of encounter complexes respectively. (B) Intermolecular PREs as a function of added paramagnetically labelled HPr(E5C) illustrating three types of titration behaviour. (C) Mapping of intermolecular PREs attributable to the specific complex (black) and to the encounter complexes (class I, blue; class II, red; mixture of classes I and II, purple; and encounter complex PREs that are too large to measure accurately, pink). (D) Equilibrium binding model for the EIN-HPr association pathway. Adapted from Fawzi, N.L., Doucleff, M., Suh, J.Y. and Clore, G.M. (2010) Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U.S.A. 107, 1379–1384 with permission.
The transient HPrnon-specific–EIN–HPr ternary complexes inferred from the PRE titration experiments may help Enzyme I compete for the cellular pool of HPr, even while phosphotransfer is occurring at the EIN active site, and may be important for efficiently reloading the Enzyme I active site, thereby facilitating higher rates of sugar uptake when substrate is transiently abundant [21]. Given the intracellular concentrations of Enzyme I and HPr (5 and 20–100 µM respectively), one can estimate that the population of ternary complex ensembles in vivo is ~1%. This estimate may be revised higher if intracellular crowding and compartmentalization further favours the formation of transient ternary complexes.
Looking at the interplay between conformational selection and induced fit by PRE
In its simplest form, ligand binding to a protein can proceed via one of two pathways involving either conformational selection or induced fit [22]. In the former, a small population with the same conformation as that of the ligand-bound state is selected by the ligand. In the latter, binding of the ligand to the protein results in a conformational change. For a broad class of proteins, ligand binding is associated with large-scale domain reorientation that effectively traps the ligand, thereby rendering it solvent-inaccessible. This is the case for both MBP (maltose-binding protein) and CaM (calmodulin).
MBP (Figure 4) and CaM (Figure 5) comprise N- and C-terminal domains connected by a linker which is rigid in the former [23] and highly flexible [24] in the latter. Binding of sugars to MBP results in a ~35° reorientation of the two domains of MBP, giving rise to a closed state in which the bound sugar is trapped akin to a Venus flytrap [25,26] (Figure 4D). Binding of peptides to CaM-4Ca2+ also results in apposition of the two domains which clamp the bound peptide in a helical conformation [27]. PRE measurements on MBP [28] and CaM-4Ca2+ [29] reveal the existence of a small (5–10%) population of configurations in the absence of ligand that resembles, but is not identical with, the holo conformation. For MBP, the PRE data can be quantitatively accounted for by a single minor partially closed state (Figure 4), whereas for CaM–4Ca2+ an ensemble of closed states is required (Figure 5).
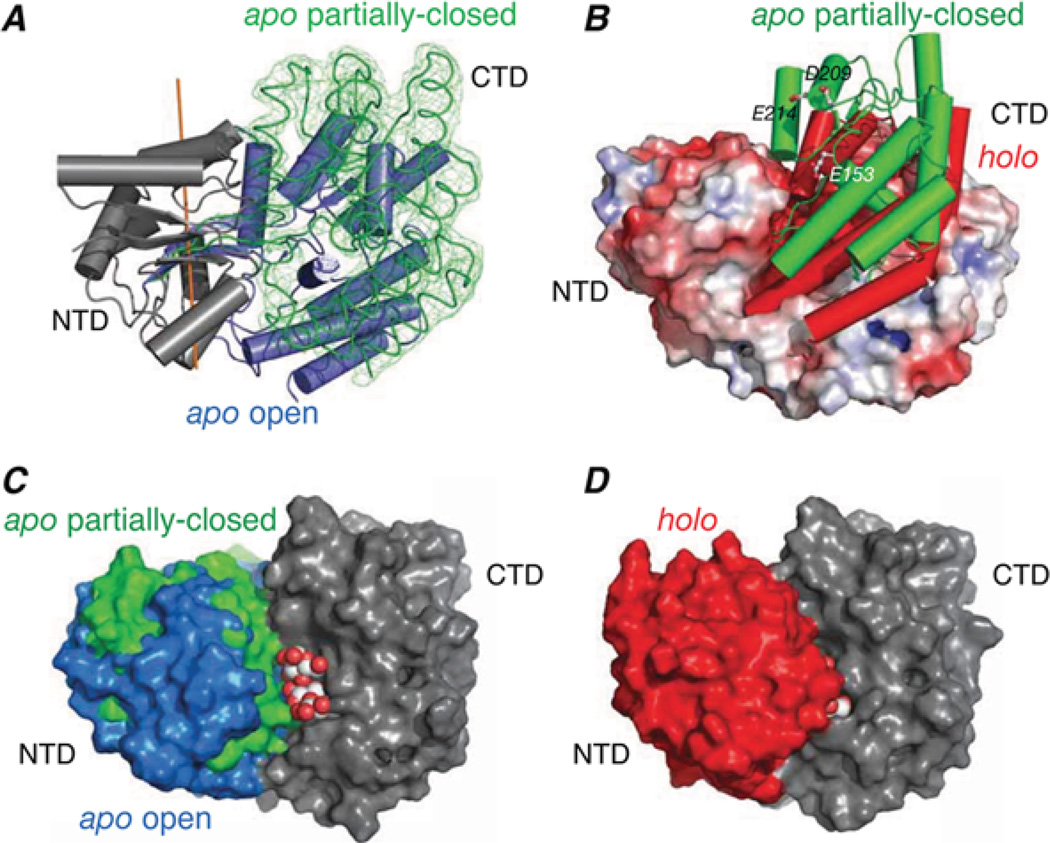
Figure 4. Structure of sparsely populated partially closed apo state of MBP derived from PRE measurements.
(A) Superimposition of the major open (blue cylinders [25]) and minor partially closed (green trace [28]) states of apo-MBP with the N-terminal domains (grey) of the two species superimposed. The reweighted atomic probability map for the backbone heavy atoms of the C-terminal domain in the partially closed state is displayed as a green mesh plotted at a threshold of 20 %. (B) Comparison of the C-terminal domain orientation in the partially closed form of apo-MBP (green cylinders) and holo-MBP (red cylinders [26]) with the apo open state shown as a molecular surface colour-coded according to electrostatic potential. (C) Molecular surface representation of the major open and minor partially closed states of apo-MBP best-fitted to the C-terminal domain (CTD; grey) with the N-terminal domain (NTD) displayed in blue and green respectively. A space-filling representation of maltotriose is modelled bound to the C-terminal domain. (E) Holo-MBP shown in the same view as in (C) with the N-terminal domain in red and the C-terminal domain in grey; the substrate is buried in holo-MBP and is barely visible. Adapted from Tang, C., Schwieters, C.D. and Clore, G.M. (2007) Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 449, 1078–1082 with permission.
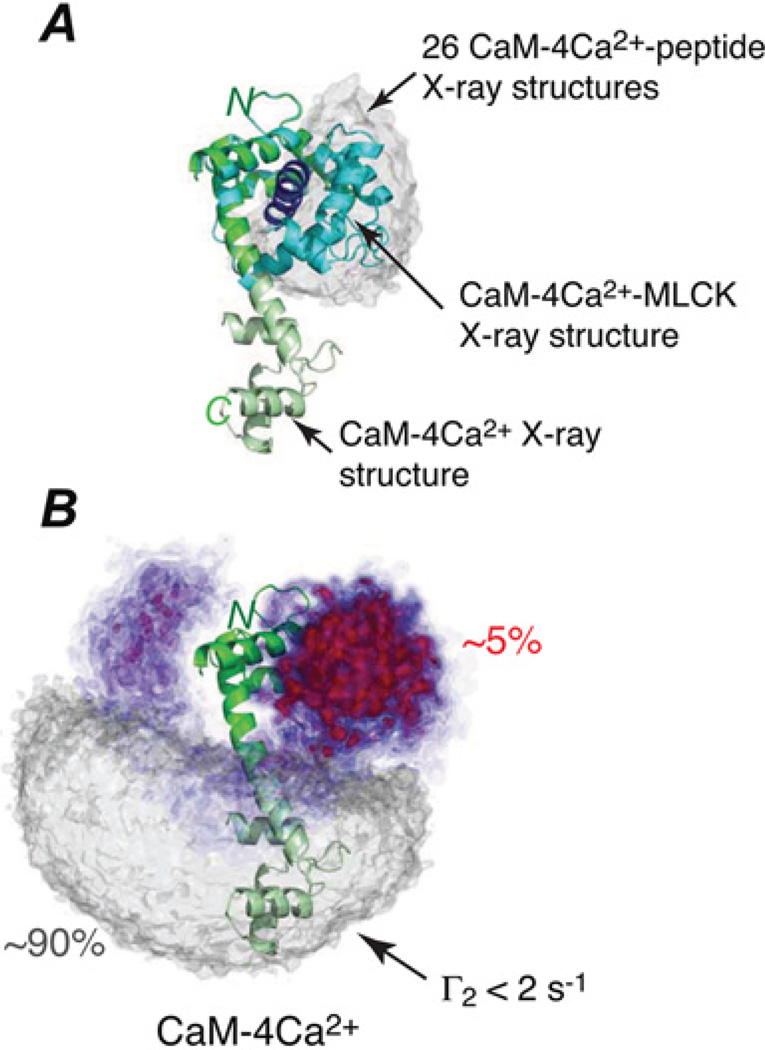
Figure 5. Visualization of the minor closed-state ensemble of CaM–4Ca2+.
(A) Crystal structure of the CaM–4Ca2+ –MLCK (myosin light chain kinase) complex (CaM, cyan; MLCK, blue [41]) overlaid on the CaM–4Ca2+ dumbbell crystal structure (green [42]), best-fitted to the N-terminal domain. An additional 26 peptide-bound crystal structures were overlaid in the same manner, and the grey atomic probability map represents their distributions for the C-terminal domain. (B) Atomic probability density maps showing the conformational space sampled by the minor species ensemble derived from PRE measurements with ensemble members best-fitted to the N-terminal domain. The minor-state atomic probability maps, calculated from 100 independent simulated annealing calculations (with an ensemble size of eight and a population of 10 %), are plotted at multiple contour levels ranging from 0.1 (transparent blue) to 0.5 (opaque red) of maximum. The grey atomic probability density maps, plotted at a single contour level of 0.1 of maximum, show the conformational space sampled by the major species ensemble (90 % occupancy), characterized by no interdomain contacts (i.e. interdomain PRE values restrained to values less than 2 s−1). The extended dumbbell structure is displayed as a ribbon diagram for reference. Approximately half of the minor species ensemble occupies a region of conformational space that is in the vicinity of and overlaps with that of the peptide-bound structures (red probability map contours. Adapted with permission from Anthis, N.J., Doucleff, M. and Clore, G.M. (2011) Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: interplay of conformational selection and induced fit. J. Am. Chem. Soc. 133, 18966–18974.
The partially closed state of apo-MBP involves both reorientation and translation of the two domains that leaves the sugar-binding site on the C-terminal domain accessible and separates the negatively charged residues on the sugar-binding surfaces of the N- and C-terminal domains sufficiently to circumvent the highly unfavourable interdomain electrostatic interactions that would be present in the holo conformation in the absence of bound ligand [28] (Figures 4B and 4C). The closed state ensemble of CaM– 4Ca2+ samples a region of conformational space that overlaps with that sampled by crystal structures of various CaM– 4Ca2+ –peptide complexes [29] (Figure 5).
The existence of sparsely populated states that are partially closed, occupy a region of conformational space in close proximity to that of the holo state and are available for ligand binding may facilitate the transition to the closed ligand-bound complexes. As such, both MBP and CaM highlight the complementarity and interplay of conformational selection and induced fit, with the former characterized by the existence of partially closed states and the latter by the transition to the final holo state, subsequent to initial ligand binding to either the open or partially closed apo states. Which pathway predominates will depend on the timescale of the open to partially closed state transition in the absence of ligand, the characteristic ligand diffusion time, the concentration of protein and ligand, and the relative affinities of the open and partially closed apo states for the ligand.
A visibility cloak for molecules bound to large macromolecular assemblies: lifetime line broadening and DEST spectroscopy
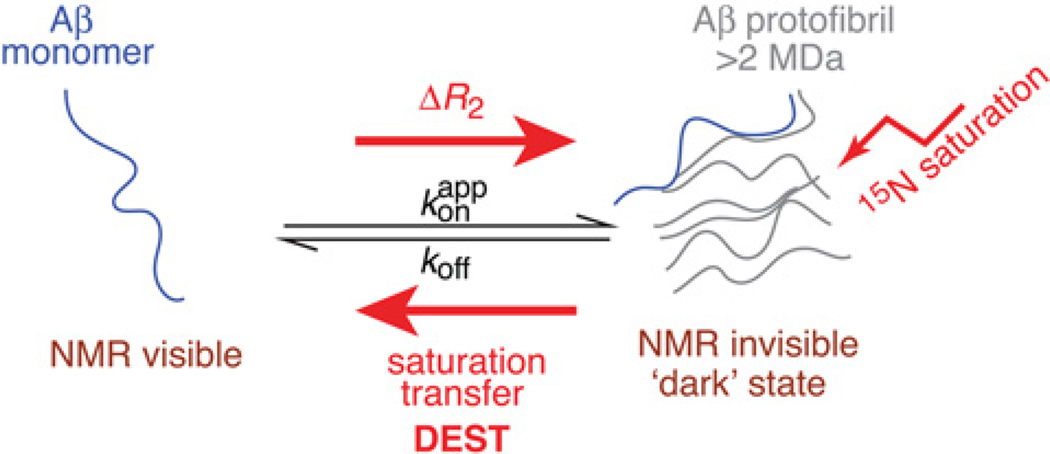
Many biological processes involve exchange phenomena in which the free state of a molecule or macromolecule is NMR-visible, whereas the bound state is in an NMR-invisible ‘dark’ state owing to interaction with a very-high-molecular-mass particle such as an aggregate, a fibril or a supramolecular assembly. Such systems are amenable to study by solution NMR using lifetime line broadening (ΔR2) [30] and DEST spectroscopy [7].
Exchange line broadening can arise from either differences in chemical shifts (chemical exchange line broadening) or transverse relaxation rates (lifetime line broadening) between the free and bound states. When a molecule binds to a high (>700 kDa)-molecular mass entity, the reduced rate of molecular tumbling leads to a marked increase in transverse (R2) relaxation rates (i.e. severe line broadening) which precludes direct observation of the bound state by standard NMR techniques. If the dissociation rate constant koff is considerably smaller (by two orders of magnitude or more) than the R2 in the bound state , the difference in R2 values (ΔR2) for the NMR-visible species in the presence and absence of the high-molecular-mass species will be equal to the pseudo-first-order association rate constant [kon (app)] [30]. If koff is comparable with or larger than , ΔR2 will be dependent on koff, kon (app) and [8].
The essence of the DEST experiment (Figure 6) is that the large R2 values in the bound state that preclude direct observation by NMR allow for efficient partial saturation of longitudinal magnetization of bound state resonances by a weak radiofrequency field, even at offsets where the magnetization of the free species is completely unaffected [7,31] (Figure 7B). In other words, even though the bound resonances are broadened beyond detection, they can be perturbed and partially saturated. Saturation of the bound resonances is then transferred back to the corresponding resonances of the free species by chemical exchange and subsequently measured as attenuation of the easily observed resonances of the NMR visible species. In our applications, we have largely made use of 15N-DEST since cross-relaxation with neighbouring protons is removed. Operationally, the DEST experiment involves the creation of an action profile by applying weak saturation at set intervals from, say, + 35 kHz to — 35 kHz, and measuring the cross-peak intensities in a 2D correlation spectrum as a function of the frequency offset of the saturation pulse. The resulting profiles are dependent on the association and dissociation rate constants, and the transverse relaxation rates in the bound state.
Figure 6. Schematic illustration of lifetime line broadening (Δ/R2) and DEST effects for the exchange of Aβ monomer on the surface of large (>2 MDa) amyloid protofibrils.
Adapted from Fawzi, N.L., Ying, J., Ghirlando, R., Torchia, D.A. and Clore, G.M. (2011) Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature 480, 268–272 with permission.
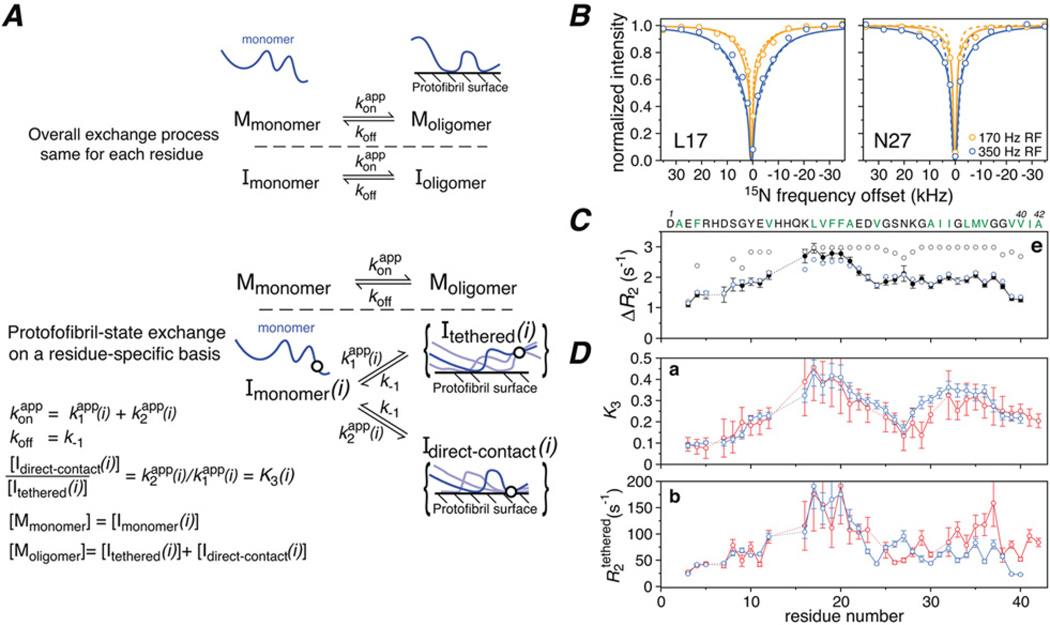
Figure 7. Exchange of Aβ monomer on the surface of large polydisperse protofibrils studied by 15N lifetime line broadening (Δ/R2) and 15N-DEST.
(A) Kinetic schemes for Aβ monomer exchange on the surface of amyloid protofibrils in which the protofibril-bound peptide (Moligomer) exists in only a single state (top), or a large ensemble of states such that each residue can be either tethered or in direct contact with the surface of the oligomer with K3(i)=k2 (app) (i)/ki (app) (i) (bottom). The circle in the diagrammatic representation of the states represents a single residue that is either tethered or in direct contact and for which three possible chain configurations are shown. (B) 15N-DEST profiles for Leu17 and Asn27 at two radiofrequency (RF) fields (170 Hz, orange; 350 Hz, blue) with the experimental points shown as circles, and the best-fit curves obtained with the single state or ensemble of states models shown as dashed and continuous lines respectively. (C) Comparison of the experimental 15N-ΔR2 profile (black closed circles) with the calculated profiles obtained with the single state (grey open circles) and ensemble of states (blue open circles) models. (D) Profiles for the residue-specific partition coefficient K3 (given by the ratio of direct contact to tethered states; see A, bottom panel) and 15N-R2 values for the tethered states derived from the fits to the experimental ΔR2 and DEST data. Adapted from Fawzi, N.L., Ying, J., Ghirlando, R., Torchia, D.A. and Clore, G.M. (2011) Atomic-resolution dynamics on the surface of amyloid-^ protofibrils probed by solution NMR. Nature 480, 268–272 with permission.
Exchange of monomeric Aβ on the surface of protofibrils studied by DEST
Aβ is associated with Alzheimer’s disease, and aggregated forms of Aβ are thought to be highly toxic. At low concentration (50 µM), both Aβ40 and Aβ42 are monomeric. At higher concentrations (> 100 µM), however, the intensities of the cross-peaks in a 1H-15N correlation spectrum decrease significantly over a period of approximately a week, after which a pseudo-equilibrium is established between monomeric Aβ and NMR invisible polydisperse protofibrils ranging from ~2 to ~80 MDa in size [7]. The measured ΔR2 values between the concentrated and dilute Aβ samples are independent of field (600 or 900 MHz 1H frequency) and nucleus (1H or 15N), and display very small (albeit significant) variations across the protein sequence [30]. The increase in R2 in the presence of protofibrils is a lifetime effect owing to the very rapid decay of transverse magnetization in the protofibril-bound state. In this particular instance, the maximum ΔR2 value corresponds to the pseudo-first-order association rate constant [kon (app)] which in the case of the 270 µM Aβ40 sample has a value of 3 s −1.
The 15N-ΔR2 and 15N-DEST data for Aβ could not be fitted simultaneously by a simple two-state exchange model [7] (Figure 7A). Rather, the simplest model required to account for the experimental data comprises an overall two-state exchange process in which the exchanging species on the surface of the protofibrils consists of an ensemble of sparsely populated states for which each residue is either in direct contact with the surface or tethered to the surface through other residues (Figure 7B). The ratio of direct contact to tethered states is given by a residue-specific partition coefficient K3(i) (Figure 7B). The results are summarized in Figure 7. koff has a value of 50–60 s−1, the R2 value for residues in direct contact with the protofibril surface is ~ 19 000 s−1, and the R2 values for residues that are tethered to the surface range from 25 to 200 s−1 (Figure 7D, bottom). The population of Aβ transiently bound to the protofibril surface is ~5 % with a lifetime of ~20 ms. In the ‘dark’ protofibril-bound state, the first eight residues of Aβ exist predominantly in a mobile tethered state, whereas the largely hydrophobic central region and part of the C-terminal hydrophobic region are in direct contact with the protofibril surface for a significant proportion of the time (Figure 7D, top). The C-terminal residues of both Aβ40 and Aβ42 display lower affinity for the protofibril surface indicating that they are likely to be surface-exposed rather than buried as in structures of Aβ fibrils, and may therefore comprise the critical nucleus for fibril formation. Interestingly, the 15N-R2 values in the tethered state are significantly larger for the C-terminal residues of Aβ42 than Aβ40 (Figure 7D, bottom) which might explain the higher propensity of Aβ42 for rapid aggregation and fibril formation.
Probing the transient dark state of substrate binding to GroEL
GroEL is a prototypical example of a chaperonin, a class of molecular machine involved in facilitating protein folding, transport, oligomeric assembly and conformational switching. GroEL has a molecular mass of 800 kDa, considerably smaller than the amyloid protofibrils discussed above. The mechanism whereby GroEL performs work has not been fully elucidated owing to a lack of detailed structural and dynamic information on the state of GroEL-bound polypeptides as the complexes are dynamic and transient. This has resulted in conflicting reports on the state of GroEL-bound polypeptides. Well-ordered β-hairpin and extended conformations have been observed crystal-lographically [32,33], but in all likelihood represent states that were preferentially selected during crystallization. NMR transferred nuclear Overhauser enhancement studies have suggested the existence of well-defined helical and hairpin conformations [34,35], but interpretation is complicated because of extensive spin-diffusion within GroEL as a result of slow tumbling [36]. Hydrogen-deuterium exchange experiments using MS [37] and NMR [38], on the other hand, indicate that the secondary structure of GroEL-bound proteins is sufficiently destabilized or disrupted to allow substantial, if not complete, backbone hydrogen-deuterium exchange to occur. Lastly, direct observation of 1H-15N correlations for GroEL-bound proteins using an NMR technique known as CRINEPT (cross-correlated relaxation enhanced polarization transfer), specifically designed for directly looking at species with molecular masses in excess of 100 kDa, suggests that the bound protein substrates are dynamic and largely unfolded as the few cross-peaks that were observed are located at characteristic random coil positions [39]. This conclusion, however, is one of exclusion since very few cross-peaks could actually be observed.
To address the conformational preferences and dynamics of a GroEL-bound substrate, we chose to probe directly and quantitatively the ‘dark’ NMR-invisible state of a model intrinsically disordered polypeptide, Aβ40, bound to GroEL [8]. Because the lifetime of GroEL-bound Aβ40 (<1 ms) is much shorter than in the example above of the Aβ40-protofibril interaction, and the rotational correlation time of GroEL is much shorter than that of the protofibrils on account of the smaller size of GroEL, the interaction of Aβ40 with GroEL could not be probed using 15N-DEST and 15N-ΔR2 measurements alone. Rather, it required a combination of four different relaxation experiments, all of which are influenced in different ways on the interplay of the global exchange rates and populations of free and GroEL-bound Aβ40, and on the residue-specific dynamics and structure within the bound state, as reported by transverse relaxation rates and backbone chemical shifts. Since in this instance , lifetime line broadening (15N- ΔR2) is dependent on kon (app), koff and 15, as are the 15N-DEST profiles. CPMG (Carr-Purcell-Meinboom-Gill) 15N-relaxation dispersion and exchange-induced chemical shifts are also dependent on the exchange rates and values, but in addition are influenced by the differences in 15N chemical shifts between the free and bound species. Simultaneous fitting of these four experiments at different magnetic fields and GroEL concentrations is consistent with a simple two-state exchange model with values of 2×107 M−1·s−1 and 1400 s−1 for kon and koff respectively, and an occupancy of 2 % for the transiently bound Aβ40 (in a sample containing 50 µM Aβ40 and 20 µM GroEL subunits). Addition of Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase), a tight binding GroEL substrate, displaces bound Aβ40 and abolishes all lifetime broadening and exchange-induced shifts, indicating that the interactions of Aβ40 with GroEL are specific and involve the same binding sites as Rubisco.
The profile indicates that binding to GroEL largely occurs via the central (residues 16–21) and C-terminal (residues 30–36) hydrophobic regions [8]. The separation between these two hydrophobic segments suggests that Aβ40 can easily span the apical domains of two adjacent subunits of GroEL. The maximum values of (~600s−1 at 900 MHz), however, are 60–70% of the expected value for a rigid molecule bound to GroEL, suggesting either that the two hydrophobic regions retain considerable internal mobility in the bound state and/or that the two hydrophobic regions bind to GroEL independently of one another. In the latter case, there would be two hemi-associated states as well as a fully associated state that interconvert on a timescale that is significantly shorter (<700 µs) than the overall lifetime of the bound state. The bound backbone (1HN, 15N, 13Cα and 13Cβ) chemical shifts derived from analysis of the exchange-induced shifts indicate that Aβ40 remains intrinsically disordered upon binding to GroEL.
The absence of structural ordering upon binding has important implications for the mechanism of substrate capture and release by GroEL. In particular, binding entails minimal entropy loss and extensive conformational flexibility which facilitates the binding of any solvent-exposed five to ten residue non-polar sequence, whereas binding of multiple segments to adjacent subunits increases avidity, but still allows for rapid dissociation [8].
Conclusion
The work summarized in the present article illustrates how modern solution NMR can be used to study spectroscopically invisible states of proteins and their complexes over a wide range of timescales, and provide fundamental new insights into a wide range of biological and biophysical processes. The power of NMR in this regard lies in the fact that many sites and interactions are probed simultaneously, and therefore interpretation of the data is not reliant on a single or very limited number of interactions. Furthermore, the availability of a large number of NMR observables (e.g. paramagnetic centre–proton distances, chemical shifts or relaxation rates) provides the key to obtaining detailed structural information on sparsely populated states.
Acknowledgements
I thank my postdoctoral fellows and collaborators, past and present, who participated in the work summarized in this review, specifically N. Anthis, M. Doucleff, N. Fawzi, J. Iwahara, D. Libich, C. Schwieters, J.-Y. Suh, Y. Takayama, C. Tang, D. Torchia and J. Ying.
Funding
This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health, and by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health.
Abbreviations used
- Aβ
amyloid β-peptide
- CaM
calmodulin
- DEST
dark-state exchange saturation transfer
- EIN
N-terminal domain of Enzyme I
- HPr
histidine phosphocarrier protein
- MBP
maltose-binding protein
- PRE
paramagnetic relaxation enhancement
- PTS
phosphotransferase system
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
Biography

Marius Clore
Footnotes
Centenary Award and Sir Frederick Gowland Hopkins Memorial Lecture
Delivered at the MRC Laboratory of Molecular Biology, Cambridge, on 13 December 2012
References
- 1.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 3.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 5.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc. Chem. Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 7.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libich DS, Fawzi NL, Ying J, Clore GM. Probing the transient dark state of substrate binding to GroEL by relaxation-based solution NMR. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11361–11366. doi: 10.1073/pnas.1305715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon I. Relaxation processes in a system of two spins. Phys. Rev. 1955;99:559–565. [Google Scholar]
- 10.Bloembergen N, Morgan LO. Proton relaxation times in paramagnetic solutions: effects of electron spin relaxation. J. Chem. Phys. 1961;34:842–850. [Google Scholar]
- 11.Kosen PA. Spin labeling of proteins. Methods Enzymol. 1989;177:86–121. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- 12.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 13.Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McConnell HM. Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 1958;28:430–431. [Google Scholar]
- 15.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 16.Takayama Y, Clore GM. Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E169–E176. doi: 10.1073/pnas.1100050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takayama Y, Clore GM. Interplay between minor and major groove-binding transcription factors Sox2 and Oct1 in translocation on DNA studied by paramagnetic and diamagnetic NMR. J. Biol. Chem. 2012;287:14349–14363. doi: 10.1074/jbc.M112.352864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 20.Suh JY, Tang C, Clore GM. Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2007;129:12954–12955. doi: 10.1021/ja0760978. [DOI] [PubMed] [Google Scholar]
- 21.Fawzi NL, Doucleff M, Suh JY, Clore GM. Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1379–1384. doi: 10.1073/pnas.0909370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evena¨ s J, Tugarinov V, Skrynnikov NR, Goto NK, Muhandiram R, Kay LE. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J. Mol. Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
- 24.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 25.Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 26.Quiocho FA, Spurlino JC, Rodseth LE. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 27.Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 28.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 29.Anthis NJ, Doucleff M, Clore GM. Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: interplay of conformational selection and induced fit. J. Am. Chem. Soc. 2011;133:18966–18974. doi: 10.1021/ja2082813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fawzi NL, Ying J, Torchia DA, Clore GM. Kinetics of amyloid β monomer-to-oligomer exchange by NMR relaxation. J. Am. Chem. Soc. 2010;132:9948–9951. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawzi NL, Ying J, Torchia DA, Clore GM. Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy. Nat. Protoc. 2012;7:1523–1533. doi: 10.1038/nprot.2012.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Sigler PB. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- 33.Buckle AM, Zahn R, Fersht AR. A structural model for GroEL-polypeptide recognition. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3571–3575. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry SJ, Jordan R, McMacken R, Gierasch LM. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992;355:455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Feng H, Landry SJ, Maxwell J, Gierasch LM. Basis of substrate binding by the chaperonin GroEL. Biochemistry. 1999;38:12537–12546. doi: 10.1021/bi991070p. [DOI] [PubMed] [Google Scholar]
- 36.Bodner CR, Dobson CM, Bax A. Multiple tight phospholipid-binding modes of α-synuclein revealed by solution NMR spectroscopy. J. Mol. Biol. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson CV, Gross M, Eyles SJ, Ewbank JJ, Mayhew M, Hartl FU, Dobson CM, Radford SE. Conformation of GroEL-bound α-lactalbumin probed by mass spectrometry. Nature. 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- 38.Zahn R, Spitzfaden C, Ottiger M, Wu¨ thrich K, Plu¨ ckthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994;368:261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]
- 39.Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wüthrich K. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 41.Frankael E, Pabo C. Comparison of X-ray and NMR structure for the Antennapedia homeodomain-DNA complex. Nat. Struct. Biol. 1998;5:692–697. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- 42.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]