Abstract
The promoter of p53 induced gene 3 (PIG3) contains a variable number of tandem repeats (VNTRs) of pentanucleotides (TGYCC)n that is known as a p53 binding site. In this study, we investigated whether other potential molecules could bind to this PIG3 promoter (TGYCC)n motif. Ligand-chromatography combined with liquid chromatography–tandem mass spectrometry analyses indicated direct interactions of prohibitin and/or prohibiton with the (TGYCC)15 motif, which was confirmed by electrophoretic mobility shift assay and super-gel shift analysis with anti-prohibitin and anti-prohibiton antibodies. Using the chromatin immunopercipipation assay, we further demonstrated that prohibitin and prohibiton associated with the (TGYCC)15 motif in vivo regardless of the p53 status and apoptotic stress. We also found that prohibitin and prohibiton up-regulated PIG3 transcription independent of p53, although p53 obviously enhanced this process, and that the knock-down of prohibitin and prohibiton inhibited camptothecin-induced apoptosis. Taken together, our findings suggest that prohibitin and prohibiton contribute to PIG3-mediated apoptosis by binding to the PIG3 promoter (TGYCC)15 motif.
Keywords: PIG3, Prohibitin, Prohibiton, (TGYCC)n motif
1. Introduction
The p53-inducible gene 3 (PIG3), is a down-stream target of p53 involved in p53-initiated apoptosis [1]. The PIG3 promoter contains the sequence (TGYCC)n, a variable number of tandem repeats (VNTR) that contains a biding site for p53; and up to now, p53 is the only known molecule that binds to the PIG3 promoter (TGYCC)n [2]. Our recent study indicated that (TGYCC)15, the most common wild-type allele, led to the most effective transcriptional activity of the promoter, compared to the other three variant (TGYCC)n motifs [3], but a previous study observed a direct linear correlation between the expression levels and the number of the (TGYCC)n motifs [2]. Furthermore, we has demonstrated that the (TGYCC)15 within the PIG3 promoter is associated with a decreased risk of squamous cell carcinoma of the head and neck [3]. However, this finding does not agree with the previous findings from smaller association studies of breast cancer and lung cancer [4] as well as bladder cancer [5].
Inspired by the inconsistent findings for the role of the PIG3 promoter (TGYCC)n motif in modifying transcriptional activity and cancer susceptibility [2–5], we initiated the present study to screen for other potential molecules that may bind to the PIG3 promoter (TGYCC)15 motif and to assess their role in the transcriptional regulation of PIG3. The identification of novel molecules interacting with this PIG3 promoter (TGYCC)15 motif may provide the underlying molecular mechanisms to help explain the reported inconsistent findings and add to our knowledge of mechanisms regulating PIG3 and cancer risk associated with the PIG3 promoter (TGYCC)15 [2–5].
2. Materials and methods
2.1. Cell lines, vectors, and transfection
The UM-SCC-17B, UM-SCC-22B, and MDA886 cell lines were from the collection in the Department of Head and Neck Surgery, The University of Texas M. D. Anderson Cancer Center, Houston, TX, USA [6]. These cell lines were grown in DMEM, medium supplemented with 10% fetal bovine serum and antibiotics. The HCT116 human colon cancer cell lines (p53+/+ and p53−/−) were generously provided by Dr. Bert Vogelstein (Johns Hopkins University). The cells were grown in McCoy’s 5A medium supplemented with 10% fetal bovine serum and antibiotics at 37 °C in a humidified incubator containing 5% CO2. For transfection, the cell lines were seeded into 24-well plates at 0.5 × 105 cells per well (BD Biosciences, Bedford, MA), and 24 h after plating, the cells were co-transfected with the FuGENE HD reagent (Roche Applied Science, Indianapolis, IN).
2.2. Preparation of PIG3 promoter (TGYCC)15 binding protein by DNA-ligand chromatography
A 150 bp-DNA fragment corresponding to the 15 repeats-allele of PIG3 was prepared by PCR amplification with the forward primer 5′-TGCTCCGCGAGGATACAGCG-3′ and the biotin-labeled reverse primer 5′-CCCTGCAGTGCACGGCTAACATATTG-3′ in the UM-SCC-17B cell line. This DNA fragment was used as the binding ligand and a chromatography column was prepared by coupling it with TetraLink? tetrameric Avidin Resin (Promega Co., Madison, WI). Binding reaction of nuclear extracts was conducted in 1x binding buffer [1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris–HCl (pH 7.5)]. A series of buffers were made by mixing 1x binding buffer [1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris–HCl (pH 7.5)] with different final concentrations of NaCl ranging from 0.25 to 3.25 M. These NaCl/binding buffers were used as the washing buffer or the eluting buffer. The eluted solutions were precipitated with 3 vol of cold-acetone, and the generated protein pellet was desalted with 75% ethanol two times. After electrophoresis on 12% (v/v) SDS–polyacrylamide gel, the protein was stained with Coomassie Brilliant Blue R250 and the corresponding protein band was cut out and digested with trypsin. The digested peptides were analyzed on a MALDI mass spectrometer and identified with ProFound software (http://prowl.rockefel-ler.edu/cgi-bin/ProFound).
2.3. Extraction of nuclear protein and electrophoretic mobility shift assay
Nuclear protein extracts were prepared from cell lines UM-SCC-17B (p53 proficient) and MDA886 (p53 deficient) cultured with a modified Dignam’s method [19]. Briefly, the cells were grown in monolayer culture to 80–90% confluence and were directly scraped off into a tube after washing with PBS. The cell pellet was re-suspended with 500 μl buffer-A (10 mM KCl, 1.5 mM MgCl2, 10mM DTT, 0.1% Nonidet-P40, and 10 mM Hepes, pH 7.9 at 4 °C) containing 1 mM PMSF and 1x Complete™ protease inhibitor cocktail (Roche Diagnostics GmbH/Roche Molecular Biochemicals, Mannheim, Germany). After incubated for 30 min on ice with frequent agitation, the crude nuclear pellet was recovered by centrifugation, and this procedure was repeated three times. The crude nuclear pellet was then resuspended in 100 μl buffer-C (0.42 M NaCl, 0.2 mM EDTA, 25% glycerol, and 20 mM Hepes, pH 7.9 at 4 °C) containing 5 mM DTT and 10 mM PMSF and centrifuged to collect nuclear extracts. The nuclear extracts were dialyzed overnight against 0.1x PBS containing 1 mM DTT and 1 mM PMSF.
Four single-strand oligo-probes corresponding to the pentanucleotide repeat numbers of 15 were prepared by PCR amplification with the forward primer 5′-TGCTCCGCG AGGATACAGCG-3′ and the reverse primer 5′-CCCTGCAGTGCACGGCTAACATATTG-3′. The size of the probe corresponding to the pentanucleotide repeat number of 15 was 150 bp. To label the probes, a phosphorylation reaction mixture was assembled with 0.5 μg of the probe, 1x T4 kinase buffer, 1.5 μg of Ci γ-32P-ATP and 20 U of T4 polynucleotide kinase (New England Biolabs Inc., Beverly, MA). After incubation for 10 min at 37 °C, water was added to 100 μl total volume and the unlabelled γ-32P-ATP was removed using a G-25 spin column. The components of the electrophoretic mobility shift assay (EMSA) reaction included 1x binding buffer (20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl and 50 mM Tris–HCl, pH 7.5), the dialyzed nucleic extracts, 5 μg of salmon sperm DNA and ~20 ng of 32P-lablled probes. After incubation at room temperature for 20 min, the reaction mixture was electrophoresed on a 4% non-denaturing polyacrylamide gel at 1000 V for 2.5 h in 0.5x TBE and the dried gel was exposed to an X-ray film. Signals were quantified by densitometry with the AlphaEaseFC™ software (VERSION 3.2.1; Alpha Innotech Corporation). Super-gel Shift Analysis was conducted with anti-PHB1 and anti-PHB2 antibodies, respectively. Other antibodies used included: Anti-BAP37 (6118), a polyclonal antibody against recombinant C-terminal of human BAP37 (PHB2) (Biolegend) and Anti-prohibitin (CP34), a monoclonal antibody against purified recombinant prohibitin (PHB1) (Calbiochem).
2.4. Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assay was performed using the ChIP Assay kit according to manufacturer instructions (Cat# 17-295, Upstate Cell Signaling Solutions, Lake placid, NY 12946). One confluent plate of HCT 116 p53+/+ and p53−/− cells (about 1 × 106 cells plate) was used for each ChIP reaction. Anti-PHB (Catalog No. LS-C146959) and Anti-PHB2 (Catalog No. LS-C90864-100) were purchased from LifeSpan BioSciences. The sequences of the PCR primers used in the PCR reactions were as follows: forward: GGGCGCTGCGGTGCCAGCCTGAG, reverse: ACCTT CAGGAGGACTTCACC for PIG3 (Genbank accession No. AF010309); forward: TCACCCACACTGTGCCCATCTACGA, reverse: CAGCGGAACC GCTCATTGCCAATGG for beta-actin gene (Genbank accession No. M10277). PCR products were electrophoresed in 2% agarose gel and visualized by ethidium bromide (AB information).
2.5. Western blot analysis
Total protein was extracted from 2 × 107 cells and used for Western blot analysis of PHB, PHB2, p53 and p21 protein expression. Briefly, cells were washed in PBS and lysed with lysis buffer containing 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulphate (SDS), 1% Nonidet-P40, 10 mM Dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF). After incubation on ice for 15 min and centrifugation at 12,000 rpm for 10 min, the supernatants were collected and the protein concentration was determined using a Protein Assay Kit (Bio-Rad, Hercules, CA). Proteins (30 μg) were separated on a 10% SDS–polyacrylamide gel and transferred to a nitrocellulose membrane by electroblotting. Membranes were probed with antibodies, and antibody binding was detected using an enhanced chemiluminescence (ECL) kit (Amersham Life Science, Arlington Heights, IL) according to the manufacturer’s instructions. The primary monoclonal antibody, anti-p53 (sc-263, Santa Cruz Biotechnology, Inc., Santa Cruz, CA 95060), anti-PHB (ab-1, Cat# CP34, Calbiochem, San Diego, CA92121), anti-PHB2 (Cat# 61180, Biolegend, San Diego, CA), and anti-p21 (Cat# OP64, Oncogene Research Products, Cambridge, MA) were diluted 1:1000, 1:500, 1:500, and 1:500, respectively, for western blotting. After probing with an HRP-conjugated anti-mouse or rabbit secondary antibody (diluted 1:1000), the signals were visualized with the enhanced chemiluminescence (ECL) system (Amersham Life Sciences). Membrane was stripped and re-probed for β-actin with a goat polyclonal antibody (diluted 1:500, sc-1616, Santa Cruz Biotechnology, Inc., Santa Cruz, CA 95060). The signals were visualized with an HRP-conjugated anti-goat secondary antibody (diluted 1:1000, sc-2350, Santa Cruz Biotechnology Inc., Santa Cruz, CA) and ECL system.
2.6. Real-time reverse transcription PCR
For RT-PCRs, 50 ng of total RNA from each sample was used. The primers and probes for detecting PIG3 and GAPDH cDNA sequences were designed by using Primers Express software (Perkin-Elmer Applied Biosystems, Foster City, CA). Forward: CATGGAGGCCAAC AAGAACA, reverse: CTGCCCCCATCCTCCTTC, and probe FAM-CAA GATCGTCCTGGAACT-TAMRA for PIG3; and forward: GAAGGTGA AGGTCGGAGTC, reverse: GAAGATGGTGATGGGATTTC, and probe FAM-CAAGCTTCCCGTTCTCAGCC-TAMRA for GAPDH (used as an internal control for relative quantification). Reverse transcription-PCR was done using the TaqMan One-Step Reverse Transcriptase-PCR Master Mix Reagents with an ABI PRISM 7700 Sequence Detection System according to the protocol of the manufacturer (Perkin-Elmer Applied Biosystems, Foster City, CA) as previously prescribed [7]. Each sample was measured in triplicate and the means of the three values were calculated for statistical analysis. About 10% of the samples were retested using the same RNA samples, and the measurements were found to be consistent between repeats.
2.7. siRNA transfection
PHB siRNA (M-010530-00), PHB2 siRNA (M-018703-00) and control siRNA (D-001050-01-05) were purchased from Dharmacon, Denver, Co, USA. This siRNA was transfected into HCT 116 p53+/+ and HCT 116 p53−/− cell lines with SMART pool reagents (Dharmacon, Denver, CO, USA) following the manufacturer’s instructions. Briefly, a total of 5 ×105 cells were plated in 6-well plates and transfected using 200 pmol siRNA per well. After 24 h of incubation, 250 nM of camptothecin was added to the cells. Following 24 h of incubation, the cells were harvested for analysis.
2.8. Statistical analysis
All data are presented as the mean ± SD. Experiments were conducted in triplicate. Significance of differences was determined using two-tailed unpaired Student’s t test. A value P < 0.05 was considered statistically significant.
3. Results
3.1. Identification of nuclear proteins interacting with the PIG3 (TGYCC)15 motif
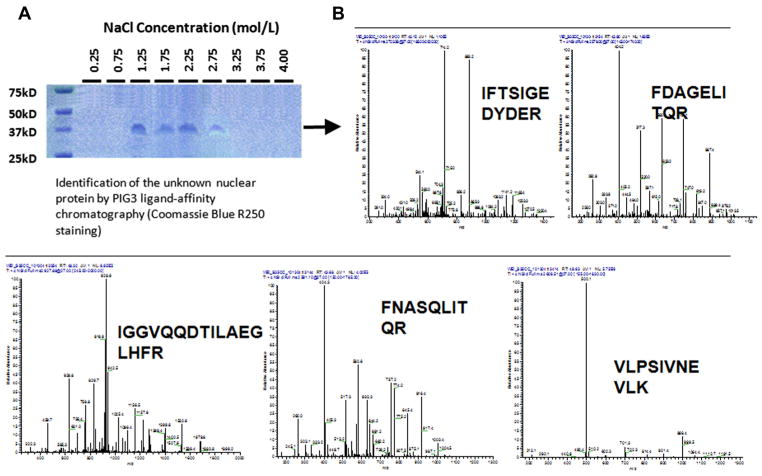
Nuclear protein eluted from the PIG3 (TGYCC)15 ligand chromatography column was stained with Coomassie Brilliant Blue R250 after electrophoresis on SDS–polyacrylamide gel. As shown in Fig. 1a, we detected a corresponding protein band with the molecular size of about 37 kD. Protein specifically pull-downed by the PIG3 (TGYCC)15 ligand was then analyzed on a MALDI mass spectrometer and identified with ProFound software (Fig. 1b). The 12 identified proteins are listed in Table 1, which include five histone subunits (i.e., H1C, H2AD, H2BA, H4D, and H3H), Claudin-2 (an integral membrane proteins of tight junctions), voltage-dependent anion channel 1 (VDAC1, interacts with Bcl-2 family proteins in modulating the mitochondrial permeability during apoptosis), ADP/ATP translocase (T2), ATP synthase subunit d (ATP5D), calmodulin 2 (CALM2, assigned to chromosomes 2), prohibitin (PHB), and prohibiton (PHB2). Of these 12 identified proteins, the most relevant protein that may interact with the PIG3 (TGYCC)15 motif appeared to be PHB and PHB2, because their molecular sizes are about 37 kD [8,9]. In addition, PHB is also closely associated with PHB2 in the form of a heterodimer [10] that was also identified in the pull-down complex as well.
Fig. 1.
Identification of nuclear proteins interacting with the PIG3 (TGYCC)15 motif. Nuclear protein eluted from the PIG3 (TGYCC)15 ligand chromatography column was stained with Coomassie Brilliant Blue R250 after electrophoresis on SDS–polyacrylamide gel. A corresponding protein band with the molecular size of about 37 kD was detected (A). The band was then analyzed on a MALDI mass spectrometer and identified with ProFound software, of the 12 identified proteins, the most relevant protein that may interact with the PIG3 (TGYCC)15 motif appeared to be PHB and PHB2 (B).
Table 1.
List of the proteins ID identified by MS/MS analysis.
| No. | Genbank No. | Protein name | Symbol | Location | Amino acids | MW*(kD) | PI |
|---|---|---|---|---|---|---|---|
| 1 | NP_005311 | H1 histone family, member 3; histone H1c | HIST1H1D | 6p21.3 | 220 | 22.2 | 11.0 |
| 2 | NP_009204 | Repressor of estrogen receptor activity; B-cell associated protein | PHB2 | 12p13 | 299 | 33.3 | 9.8 |
| 3 | AAA35579 | ADP/ATP carrier protein T2 | SLC25A5 | Xq24–26 | 297 | 32.8 | 9.8 |
| 4 | NP_003365 | Voltage-dependent anion channel 1 | VDAC1 | 5q31 | 282 | 30.6 | 8.6 |
| 5 | NP_002625 | Prohibitin | PHB | 17q21 | 272 | 29.8 | 5.6 |
| 6 | NP_066409 | Histone 1, H2ad; histone H2AD; H2A histone H2AD;H2A histone family, member G | HIST1H2AD | 6p21.3 | 129 | 14.0 | 10.9 |
| 7 | AAH66241 | HIST1 H2BA protein | HIST1H2BA | 6p22.2 | 126 | 14.0 | 10.3 |
| 8 | AAH67496 | HIST1 H4D protein | HIST1H4D | 6p21.3 | 102 | 11.2 | 11.4 |
| 9 | AAP97236 | Claudin-2 | CLDN1 | 3q28-q29 | 211 | 22.7 | 8.4 |
| 10 | NP_001678 | ATP synthase, H + transporting, mitochondrial F1 complex, delta subunit | ATP5D | 19q13.3 | 168 | 17.5 | 5.4 |
| 11 | NP_001734 | Calmodulin 2; phosphorylase kinase delta | CALM2 | 2p21 | 148 | 16.7 | 4.1 |
| 12 | AAH67493 | HIST1 H3H protein | HIST1H3I | 6p22-p21.3 | 138 | 15.6 | 11.1 |
3.2. Validation of PHB and PHB2 as the novel nuclear proteins binding to the PIG3 (TGYCC)15 motif
To validate the interaction of PHB and PHB2 with PIG3 (TGYCC)15, we performed EMSA with native sequences of PIG3 (TGYCC)15 as the probe to identify nuclear proteins that may bind to the PIG3 (TGYCC)15 motif in two HNSCC cell lines without environmentally induced stress; one cell line with detectable p53 expression (UMSCC-17B) and the other one without p53 (MDA-886). Fig. 2a shows consistent nuclear protein banding regardless of the presence of p53 expression in both UMSCC-17B and MDA-886 cell lines. Super-shift analysis with anti-PHB and anti-PHB2 antibodies further confirmed that both PHB and PHB2 interact with the PIG3 (TGYCC)15 motif (Fig. 2b). Furthermore, we treated HCT116 p53+/+ and HCT116 p53−/− with camptothecin, a topoisomerase 1 inhibitor and an effective apoptotic stressor [9], and then we conducted a ChIP assay to determine the in vivo status of the interaction between PHB/PHB2 and PIG3 (TGYCC)15. As shown in Fig. 3, both PHB and PHB2 were specifically associated with the PIG3 (TGYCC)15 motif regardless of camptothecin stimulation, with the same findings observed in both p53+/+ and p53−/− HCT116 cell lines. However, it appeared that p53 only bound to the PIG3 (TGYCC)15 motif in the presence of apoptotic stimulation (Fig. 3). These findings suggested that PHB and PHB2 could bind to the (TGYCC)15 motif in vivo regardless of p53 status and apoptotic stress.
Fig. 2.
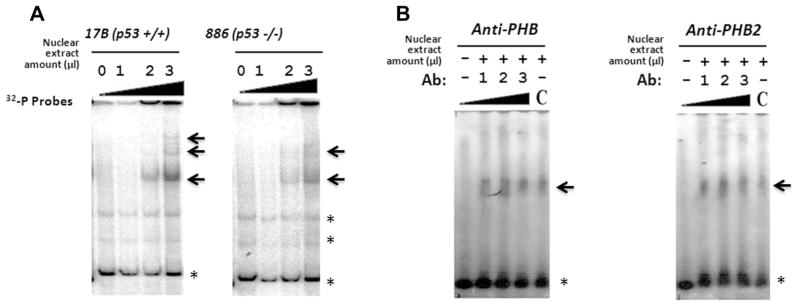
Validation of the interaction of PHB and PHB2 with the PIG3 (TGYCC)15 motif. Electrophoretic mobility shift assay (EMSA) showed consistent nuclear protein bands regardless of p53 expression in both UMSCC-17B and MDA-886 cell lines (The arrows indicate the specific bands; asterisks indicate non-specific band or free probes) (A). Super-shift analysis with anti-PHB and anti-PHB2 antibodies confirmed that both proteins interact with the PIG3 (TGYCC)15 motif (B).
Fig. 3.
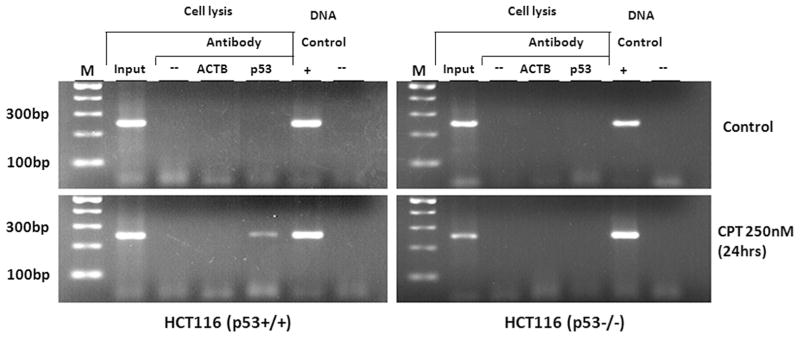
Chromatin immunoprecipitation (ChIP) assay. The results indicated that PHB and PHB2 can specifically bind to the PIG3 (TGYCC)15 motif in vivo regardless of p53 status and camptothecin treatment.
3.3. Role of PHB and PHB2 in PIG3-mediated apoptosis pathway
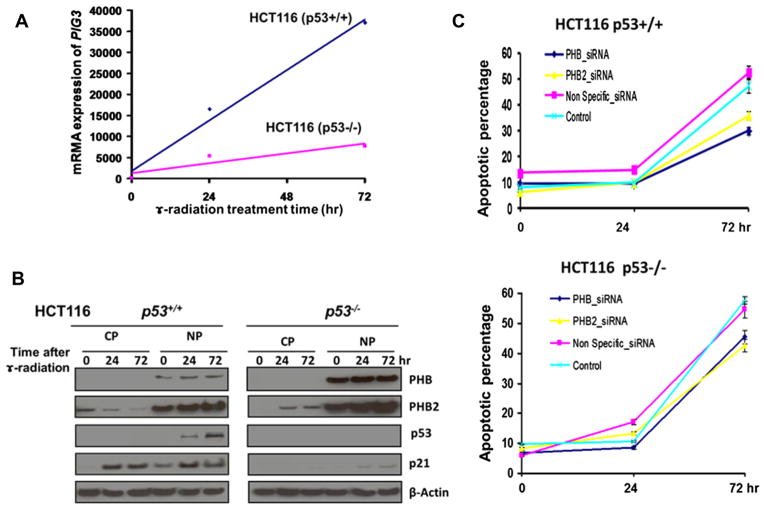
It is likely that PHB and PHB2 are involved in p53-PIG3-mediated apoptosis. To further substantiate our findings, we investigated the roles of PHB and PHB2 in apoptosis induced by camptothecin and γ-irradiation. As shown in Fig. 4a, there was a direct linear correlation between PIG3 mRNA expression levels and the exposure time of a 12 Gy γ-irradiation regardless of the p53 status, although mRNA expression was clearly higher in the presence of p53 (HCT116 p53+/+ cells) than that in the absence of p53 (HCT116 p53−/− cells). Consistent with this finding, protein expression levels of PHB2, but not PHB, appeared to be increased in the nucleus overtime after irradiation. Interestingly, the protein levels of both PHB and PHB2 were up-regulated in the absence of p53, but the p21 protein, a p53-dependent down-stream target, was detected only in the presence of p53, but not detectable in the absence of p53 (Fig. 4b). Furthermore, PHB was exclusively expressed in the nucleus, and the expression levels did not change noticeably over a period of 72 h in response to an incident exposure of a dose of 12 Gy γ-irradiation, whereas PHB2 appeared to be present in both the cytoplasm and the nucleus. To investigate the roles of PHB and PHB2 in the PIG3-mediate apoptosis, we examined the effects of knock-down of PHB and PHB2 by siRNA on the camptothecin-induced apoptosis. As expected, the percentage of apoptosis was significantly decreased in both p53+/+ and p53−/− HCT116 cell lines that were treated with PHB and PHB2 siRNA, compared with those cells treated with control siRNA (Fig. 4c). The results suggested that knock-down of PHB and PHB2 inhibited the camptothecin-induced apoptosis independent of p53, although p53 obviously enhanced the apoptotic process.
Fig. 4.
Role of PHB and PHB2 in PIG3-mediated apoptosis pathway. Direct linear correlation between PIG3 mRNA expression levels and the exposure time of a 12 Gy γ-irradiation regardless of p53 status, although the mRNA expression was clearly higher in the presence of p53 (HCT116 p53+/+ cells) than in the absence of p53 (HCT116 p53−/− cells) (A). PHB2, but not PHB, protein expression increased in the nucleus overtime after 12 Gy γ-irradiation. PHB was exclusively expressed in the nucleus, whereas PHB2 appeared to be present in both the cytoplasm and the nucleus (B). Knock-down of PHB and PHB2 by siRNA inhibited camptothecin-induced apoptosis independent of p53 (C).
4. Discussion
In the present study, we demonstrate for the first time that the polymorphic pentanucleotide microsatellite sequence (TGYCC)15 within the PIG3 promoter, a known p53 binding site, is a novel binding site of PHB and PHB2. The unexpected finding of PHB and PHB2 binding activities at the PIG3 (TGYCC)15 motif and their roles in the PIG3 mediated-apoptotic stimulation reveals a pivotal role for this DNA–protein binding site that provides awaited molecular insight into previously reported unexpected findings on the roles of PHB and PHB2 in p53-mediated cellular processes [8,11], as well as inconsistent findings on cancer risk (4–7).
The prohibitin gene (PHB), located at chromosome 17q21, encodes a protein of about 32 kDa molecular mass originally identified as an intracellular, antiproliferative protein [12] but later recognized as a novel link between oxidative stress and DNA damage response in cancer [13]. Its cellular location was found to be in the inner mitochondrial membrane [14] and co-expressed in physical interaction with PHB2 [15], another nuclear-encoded protein also known as B-cell associated protein BAP37 that is a molecular chaperone of PHB [12]. PHB and PHB2 also appear to be dependent on each other for stability, in that PHB cannot be detected in a PHB2 null yeast strain and PHB2 is absent in a PHB null yeast strain [16]. Functional studies have suggested that endogenous PHB contributes to the control of the G1 to S transition [17], which was later found to be associated with p53 [8]. PHB also physically interacts with Rb family proteins in repressing E2F-mediated transcription to inhibit cell proliferation [8,18]. It has also been shown that PHB co-localizes with Rb, E2F1, and p53 in the nucleus and is transported outside of the nucleus and the mitochondria upon apoptotic stimulation, facilitating recruitment of p53 to its downstream promoters, thus functioning as a possible tumor suppressor [8].
It was postulated that PHB may enhance the binding of p53 to the target sites either directly or through the mediation of other proteins [8]. Our data clearly show that PHB is dominantly present in the nucleus, at least in HNSCC cancer cell lines and HCT116 cells, where PHB binds to the PIG3 promoter (TGYCC)n sequence as an anchor protein that may facilitate the direct binding of p53 at the same site upon cellular stress. Indeed, as previously suggested, it is likely that PHB enhances the binding of p53 to the target sites either directly or through the mediation of other proteins [8]. Therefore, it is conceivable that immediate p53 binding at the PIG3 promoter and subsequent interaction with PHB and/or PHB2 together may facilitate the p53-meidiated PIG3 transcriptional regulation. Because of the nature of PHB as an acidic protein, the binding of PHB at the site of the PIG3 (TGYCC)n motif surrounded by histone subunits (i.e., H1C, H2AD, H2BA, H4D, and H3H) and PHB2 (here as a chaperone of PHB) [12] makes it possible for PHB to be stabilized in the basic environment in the nucleus. We confirm that PHB is a nuclear protein with a demonstrated interaction with p53 at the site of the PIG3 promoter (TGYCC)15 sequence. The present study, together with previous findings in other laboratories, provides new evidence that helps explain the elusive functions of PHB, which have been considered both p53-dependent and independent based on the origin of PHB as a cellular inner membrane protein [10]. It is biologically plausible that binding of PHB at the PIG3 promoter provides a molecular mechanism that prevents unwanted binding by p53 that may cause a major mobilization of a constellation of proteins, leading to dramatic changes in cellular functions.
Based on our findings, we postulate that p53 is recruited to the PIG3 promoter (TGYCC)15 motif only upon apoptotic stimulation, whereas both PHB and PHB2 are most likely located at the PIG3 (TGYCC)15 motif both in the absence and presence of apoptotic stress. Although the exact molecular mechanisms of PHB and PHB2 binding to the PIG3 (TGYCC)15 are still unclear, our findings suggest that PHB and PHB2 might facilitate the recruitment of p53 proteins to the PIG3 promoter (TGYCC)15 motif, leading to the p53-induced PIG3 transcription and PIG3-mediated apoptosis. In addition, it is likely that PHB and PHB2 binding to the promoter (TGYCC)15 motif under apoptotic stress results in initiation of PIG3 transcription in the absence of p53. Other factors, such as p63 and p73 that have been confirmed to enhance the expression of PIG3 independent of p53 [19], may be another possible explanation for the PIG3-mediated, p53-independent apoptosis that we have observed in the HCT116 p53−/− cells, which needs additional mechanistic studies.
In summary, we characterized the unexpected findings of PHB and PHB2 binding at the p53 responsive element of the PIG3 promoter, the (TGYCC)n motif. This is the first time that PHB and PHB2 have been shown to mediate the induction of PIG3 expression through direct interaction with its promoter (TGYCC)n motif. Transcriptional activation of the PIG3 promoter mediated by these novel factors, PHB and PHB2, as well as p53, may affect the role of the PIG3 promoter (TGYCC)n motif in cancer susceptibility.
Acknowledgments
This work was funded by U.S. National Institutes of Health Grants R01 ES 11740 and R01 CA 131274 (to Q.W.), and by Grant from National Natural Science Foundation of China (No. 81272252, to X.G.).
References
- 1.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 2.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 3.Guan X, Liu Z, Wang L, Wang LE, Sturgis EM, Wei Q. Functional repeats (TGYCC)n in the p53-inducible gene 3 (PIG3) promoter and susceptibility to squamous cell carcinoma of the head and neck. Carcinogenesis. 2013;34:812– 817. doi: 10.1093/carcin/bgs388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Liloglou T, Sigala F, Korkolis D, Yannoukakos D, Papalambros E, Asimacopoulos PJ, Papavassiliou AG, Kotsinas A. Absence of association with cancer risk and low frequency of alterations at a p53 responsive PIG3 gene polymorphism in breast and lung carcinomas. Mutat Res. 2004;556:143–150. doi: 10.1016/j.mrfmmm.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Ito M, Nishiyama H, Watanabe J, Kawanishi H, Takahashi T, Kamoto T, Habuchi T, Ogawa O. Association of the PIG3 promoter polymorphism with invasive bladder cancer in a Japanese population. Jpn J Clin Oncol. 2006;36:116–120. doi: 10.1093/jjco/hyi225. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Wei S, Ma H, Zhao M, Myers JN, Weber RS, Sturgis EM, Wei Q. A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2011;32:1668–1674. doi: 10.1093/carcin/bgr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Wang LE, Wang L, Lu KH, Mills GB, Bondy ML, Wei Q. Methylation and messenger RNA expression of p15INK4b but not p16INK4a are independent risk factors for ovarian cancer. Clin Cancer Res. 2005;11:4968–4976. doi: 10.1158/1078-0432.CCR-04-2293. [DOI] [PubMed] [Google Scholar]
- 8.Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 9.Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem. 2006;281:2951–2959. doi: 10.1074/jbc.M508669200. [DOI] [PubMed] [Google Scholar]
- 10.Mishra S, Murphy LC, Nyomba BL, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol Med. 2005;11:192–197. doi: 10.1016/j.molmed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 12.Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Kohler G, Lamers MC. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994;13:3782–3792. doi: 10.1002/j.1460-2075.1994.tb06689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotsinas A, Aggarwal V, Tan EJ, Levy B, Gorgoulis VG. PIG3: a novel link between oxidative stress and DNA damage response in cancer. Cancer Lett. 2012;327:97–102. doi: 10.1016/j.canlet.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Ikonen E, Fiedler K, Parton RG, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 15.Coates PJ, Jamieson DJ, Smart K, Prescott AR, Hall PA. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol. 1997;7:607–610. doi: 10.1016/s0960-9822(06)00261-2. [DOI] [PubMed] [Google Scholar]
- 16.Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roskams AJ, Friedman V, Wood CM, Walker L, Owens GA, Stewart DA, Altus MS, Danner DB, Liu XT, McClung JK. Cell cycle activity and expression of prohibitin mRNA. J Cell Physiol. 1993;157:289–295. doi: 10.1002/jcp.1041570211. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 19.Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol. 2004;24:1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]