Abstract
Background
LIN28 is an RNA-binding protein that not only plays key roles in multiple cellular developmental processes and tumorigenesis, but also is involved in tissue inflammatory response. However, no published study has investigated associations between genetic variants in LIN28 and radiation-induced pneumonitis (RP) in patients with non-small cell lung cancer (NSCLC) treated with definitive radiation therapy.
Methods
We genotyped eight potentially functional single nucleotide polymorphisms (SNPs) of LIN28A (rs11247946 T>C, rs3811464 C>T, rs11581746 T>C, and rs12728900 G>A), and LIN28B (rs314280 A>G, rs12194974 G>A, rs17065417 A>C and rs314276 C>A) in 362 patients with NSCLC, who received definitive radio (chemo) therapy. The associations between RP risk and genotypes were assessed by hazards ratio (HR) in Cox proportional hazards regression analysis with time to event considered with and without adjustment for potential confounders.
Results
Multivariate analyses found that patients carrying LIN28B rs314280 AG and AA/AG or rs314276 AC and AA/AC genotypes had a higher risk of grade ≥3 RP (for rs314280 AG and AA/AG versus GG, adjusted HR= 2.97 and 2.23, 95% CI, 1.32–6.72 and 1.01–4.94, P = 0.009 and 0.048, respectively; for rs314276 AC and AA/AC versus CC, adjusted HR = 2.30 and 2.00, 95% CI, 1.24–4.28 and 1.11–3.62, and P = 0.008 and 0.022, respectively). Further stratified analyses showed a more profound risk in the subgroups of the age <65 years subjects, males, stage III/IV, ever smokers and MLD ≥19.0 Gy.
Conclusion
Genetic variants of LIN28B, but not LIN28A, may be biomarkers for susceptibility to severe RP in NSCLC patients. Large, prospective studies are needed to confirm our findings.
Keywords: LIN28, polymorphisms, inflammation, Hazards ratio, Non–small cell lung cancer, radiation pneumonitis
1. Introduction
Lung cancer is the leading cause of cancer death in both men and women in the United States, with an estimated 228,190 new cancer cases and 159,480 deaths in 2013 [1], Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer cases [2]. Radiotherapy, alone or in combination with concurrent or sequential chemotherapy, is the standard treatment approach for patients with inoperable or unresectable NSCLC, and such a definitive therapy improves local control and overall survival. However, radiation pneumonitis (RP), which can occur after radiation therapy as a result of inflammation of normal lung tissues injured by radiation, has been identified as one of the most common dose-limiting complications of thoracic radiation; in these NSCLC patients, approximately 10–20% have experienced severe RP (grade ≥3), and almost half of the patients who developed severe RP died of this radiation complication [3]. Previous studies have demonstrated an association between RP risk and multiple therapeutic and patient-related factors, such as Karnofsky performance status (KPS), dosimetric parameters, smoking status and plasma inflammatory cytokine levels [4]. Recently, some genetic variants, such as single nucleotide polymorphisms (SNPs) of several genes, are also shown to be associated with an increased risk of severe RP in patients with NSCLC [5, 6], suggesting that genetic factors may play an important role in RP development.
Studies have indicated that microRNAs (miRNAs), a class of approximately 19–23 nucleotide long RNA molecules in the genome, play critical roles in posttranscriptional gene regulation [7, 8]. miRNAs can negatively regulate target gene transcription through hybridization to incomplete, complementary sequences in the 3’ untranslated region (UTR) of their target messenger RNAs (mRNAs). This process results in either the degradation of target mRNAs or repression of their translation. Studies have found that LIN28 (including LIN28A and LIN28B) is an important regulator of miRNAs and mRNAs [9, 10]. For example, LIN28 not only binds to the non-coding miRNAs but also binds directly to thousands of mRNAs. Therefore, LIN28 functions to regulate not only the translation of mRNAs important for cell growth and metabolism, but also the biogenesis of miRNAs, particularly the let-7 family miRNAs [11, 12]. More recently, some studies have reported that let-7 overexpression significantly reduced IL-6 expression, which is a major mediator of the tissue inflammatory response [13, 14]. Although the inflammation mechanism of Lin28 is still largely unknown, it is believed that LIN28B plays a key role in regulation of inflammation by regulating IL-6 expression. Indeed, it has been identified that the nuclear factor-kB (NF-kB) regulates the expression of a wide range of genes involved in inflammation and that NF-kB activates LIN28B but inhibits let-7 miRNA expression, resulting in higher levels of IL-6 by NF-kB activation [13–15].
Functional experiments have demonstrated that LIN28 regulates radiosensitivity of human lung cancer cells [16]. Several studies, including genome-wide association studies (GWAS), also have reported that genetic variants of the LIN28 gene were associated with type 2 diabetes mellitus and some cancer susceptibility [17–21]. But no published study has evaluated the associations between genetic variants of the LIN28 gene and risk of RP in cancer patients who had received radiotherapy. Considering the LIN28/let7 might play important roles in the mechanism of inflammation induced by irradiation, we hypothesized that SNPs of the LIN28 gene are associated with risk of RP in patents with NSCLC and thus are biomarkers for predicting RP risk of NSCLC patients undergoing definitive radiation therapy. To test this hypothesis, we evaluated associations between LIN28 genotypes and severe RP in a well-defined cohort of NSCLC patients with detailed information about the definitive radiotherapy they have received.
2. Materials and Methods
2.1. Study populations
The characteristic details of subjects used in the present study have been previously reported [5, 6]. Briefly, the study population included 393 patients who had DNA samples available and follow-up data from a dataset of NSCLC patients treated with definitive radiation at a single institution between March 1998 and June 2009. Among these 393 patients, except for 31 who developed recurrent diseases or underwent surgical resection before radiotherapy, the remaining 362 patients had both radiation dosimetric data and documented information on RP with the complete follow-up data. We interviewed each of the 362 eligible patients to obtain data on tobacco smoking. Those who had smoked <100 cigarettes in their lifetime were considered “never smokers”, and all others were considered “ever smokers”. Techniques for treatment planning and delivery changed considerably during the study period. For example, the 3-dimensional (3D) CT-based simulation and 3D conformal radiation therapy (3D-CRT) was used before July 2004, and the 4-dimensional CT-based simulation with respiratory motion management and intensity modulation radiation therapy (IMRT) or proton beam radiation (PBT) was used thereafter [22]. The characteristics of patients, tumors, and treatment are described in Table 1, which are consistent with our previous reports [6]. Severe RP (grade ≥3) was defined according to the National Cancer Institute’s Common Terminology Criteria for Adverse Events, version 3.0. Symptomatic RP that interfered with daily activities or required the administration of oxygen was scored as grade 3. Briefly, RP had been diagnosed by reviewing all available radiographic images, including thoracic x-ray, CT, or positron emission tomography (PET)/CT scans; severity was established and confirmed by reviewing dictated clinical notes. During radiation therapy, patients were seen at least weekly; thereafter, patients were followed up at one to three months after therapy and then every three months. Follow-up evaluations included an interval history and physical examination and chest CT or PET/CT, pulmonary functional tests, and routine blood tests. The time to RP development was calculated from the start of radiation therapy; patients not experiencing either endpoint were censored at the date of the last follow-up or death [5]. The present study was approved by The University of Texas M.D. Anderson Cancer Center institutional review board, and the Health Insurance Portability and Accountability Act (HIPAA) regulations were strictly followed.
Table 1.
Characteristics(n = 362) of NSCLC patients received definitive radiation therapy
| Parameter | No.(%) | HR | 95%CI | P* |
|---|---|---|---|---|
| Sex | ||||
| Male | 200(55.0) | 1.00 | ||
| Female | 162(45.0) | 0.85 | 0.50–1.44 | 0.536 |
| Age(years) | ||||
| <65 | 181(50.0) | 1.00 | ||
| ≥65 | 181 950.0) | 0.96 | 0.57–1.63 | 0.886 |
| Race | ||||
| White | 297(82.0) | 1.00 | ||
| Black | 65(18.0) | 0.99 | 0.50–1.97 | 0.984 |
| KPS | ||||
| <80 | 74(20.4) | 1.00 | ||
| ≥80 | 288(79.6) | 0.62 | 0.34–1.12 | 0.111 |
| Stage groups | ||||
| I, II | 54(14.9) | 1.00 | ||
| III, IV | 307(84.8) | 0.82 | 0.42–1.63 | 0.580 |
| Missing | 1(0.3) | |||
| Histology | ||||
| Adenocarcinoma | 137(37.8) | 1.00 | ||
| Squamous cell | 127(35.1) | 0.99 | 0.53–1.85 | 0.981 |
| Other | 98(27.1) | 1.08 | 0.56–2.06 | 0.825 |
| Smoking status | ||||
| Never | 32(9.1) | 1.00 | ||
| Ever | 330(90.9) | 1.18 | 0.43–3.27 | 0.746 |
| Chemotherapy | ||||
| No | 33(8.8) | 1.00 | ||
| Yes | 329(90.9) | 1.30 | 0.47–3.59 | 0.616 |
| Mean lung dose(Gy) | ||||
| <19.0 | 181(50.0) | 1.00 | ||
| ≥19.0 | 181(50.0) | 3.12 | 1.73–5.64 | <0.001 |
| Radiation dose(Gy) | ||||
| <66 | 173(47.8) | 1.00 | ||
| ≥66 | 189(52.2) | 1.00 | 0.59–1.69 | 0.999 |
P values were calculated by Cox proportional model using univariate analysis.
NSCLC: non-small cell lung cancer.
2.2. Selection of SNP and Genotyping
The LIN28A and LIN28B genes have been mapped to chromosomes, 1p36.11 and 6q21, respectively. LIN28A encodes an approximately 25-kDa protein, whereas LIN28B has a similar molecular structure and biologic functions as LIN28A [18, 21, 23]. We searched the National Institute of Environmental Health Sciences Genome Program’s SNPs database (http://www.ncbi.nlm.nih.gov/projects/SNP) and related literature to identify all potentially functional SNPs of LIN28 that met at least two of the following four criteria: (1) a minor allele frequency ≥0.05 in Caucasians, (2) affecting transcription factor binding site (TFBS) activity in the putative promoter or intron regions (FuncPred, http://snpinfo.niehs.nih.gov/snpfunc.htm), (3) previously reported associations with diseases [18–21], and (4) not in high linkage disequilibrium (LD), i.e., LD ≥ 0.8. With assistance of the online searching tools of HapMap database (http://hapmap.ncbi.nlm.nih.gov), we selected and genotyped eight potentially functional SNPs of the LIN28A (rs11247946 T>C, rs3811464 C>T, rs11581746 T>C, and rs12728900 G>A), and LIN28B (rs314280 A>G, rs12194974 G>A, rs17065417 A>C and rs314276 C>A) genes using the TaqMan assay with the Sequence Detection Software on an ABI-Prism 7900 instrument according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA).
We extracted genomic DNA from the buffy coat fraction of the blood samples by using a blood DNA mini kit (Qiagen, Inc.) according to the manufacturer’s instructions. The DNA purity and concentrations were determined by spectrophotometer measurement of absorbance at 260 and 280 nm. Primers and probes were supplied by Applied Biosystems. Each plate included four negative controls (no DNA), duplicated positive controls, and eight repeat samples. Amplification was done under the following conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95 °C for 15 s and 60°C for 1 min. For all genotypes, the assay success rate was >99%, and the repeated sample’s results were 100% concordant.
2.3. Real-time RT-PCR for expression levels of LIN28B mRNA
The expression levels of LIN28B mRNA were examined by quantitative RT-PCR using the total RNA that was isolated from peripheral blood mononuclear cells (PBMCs) of 105 cancer-free controls with the TRIzol reagent (Invitrogen™, Carlsbad, CA). LIN28B mRNA expression levels were detected by using TaqMan gene expression assays with the master mix reagent (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Real-time RT-PCR was performed using the ABI-Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Each sample was analyzed in duplicate and the expression level of LIN28 mRNA was calculated relative to that of 18S.
2.4. Statistical Analysis
Patients were grouped by their genotypes. All statistical analyses were performed with SAS software (version 9.1.3; SAS Institute, Inc., Cary, NC), unless stated otherwise. Cox proportional hazards regression analysis was performed to evaluate effect of genotypes on RP risk by calculating the hazard ratios (HRs) and 95% confidence intervals (CIs). Kaplan-Meier analysis was performed to assess the effect of different genotypes on cumulative probability of RP. In addition, a multivariate Cox regression analysis was performed to adjust for other covariates. Likelihood ratio tests were also performed for each multivariate Cox regression model to assess the goodness of fit. A 2-sided test with a P value ≤ 0.05 was considered statistically significant. We also used the false-positive report probability (FPRP) to test for false positive associations [24]. For all significant genetic effects observed in our study, we calculated FPRP with prior probabilities of 0.0001, 0.001, 0.01, 0.1, and 0.25. The HR was set close to the observed value in our study, and a probability <0.2 was considered a noteworthy. All tests were two sided and P < 0.05 was considered significant.
3. Results
3.1. Patient Characteristics and Risk of severe RP (grade ≥3)
As shown in Table 1, there were 200 men and 162 women, with a median age of 65 years (range, 35 to 88 years) in the pooled group. Of these patients, 297 (82.0%) were Caucasians, 85% had stage III/IV diseases according to the 6th edition of the AJCC stage grouping criteria, and 90.9% were treated with a combination of chemotherapy and radiotherapy. The median radiation dose received by patients was 66 Gy (range, 50.0–87.5 Gy) with a median mean lung dose (MLD) of 19.0 Gy (range, 2.7–30.6 Gy). The median follow-up time for assessment of RP after radiation therapy was 20.6 months (range, 1.0 to 157.6 months). The overall incidence of severe RP was 15.5%, and the median occurrence time for severe RP (grade ≥3) was 3.6 months (95% CI, 2.1–10.1 months). We evaluated the association between RP and clinic-pathologic characteristics to identify any confounding factors, including age, sex, ethnicity, KPS, disease stage, tumor histology, smoking status, use of chemotherapy, radiation dose and MLD. In the univariate analysis, we found that only MLD was significantly associated with severe RP (grade ≥3) (MLD ≥19.0 Gy vs. MLD <19.0 Gy, crude HR = 3.12, 95% CI 1.73–5.64, P < 0.001) (Table 1 and Fig. 1a). No other clinic-pathologic characteristics were found to be associated with severe RP risk in this study population.
Figure 1.
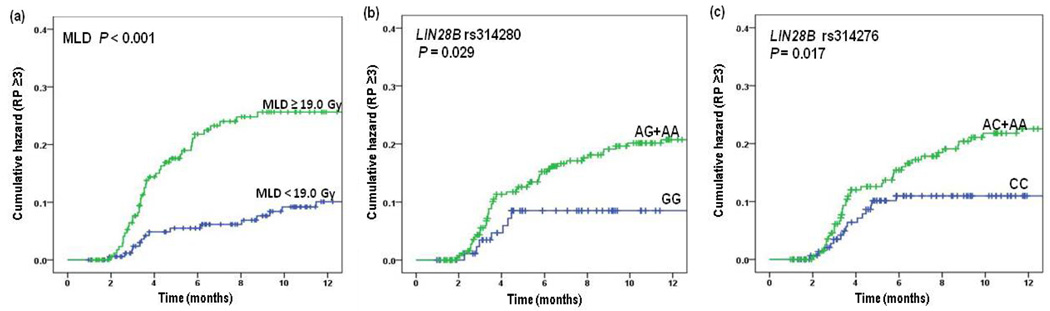
The risk of radiation pneumonitis by selected clinical factors and polymorphisms of the LIN28B gene is illustrated according to (a) mean lung dose (MLD). Gy indicates grays; (b) LIN28B rs314280 AG+AA versus GG; and (c) LIN28B rs314276 AC+AA versus CC genotypes.
3.2. Association between severe RP (grade ≥3) and LIN28 SNPs
Table 2 shows the results of univariate and multivariate analyses of the associations between LIN28 genotypes and severe RP (grade ≥3) using the Cox proportional hazards regression analysis. We found that the LIN28B rs314280 A>G and rs314276 C>A SNPs were significantly associated with hazards of severe RP (grade ≥3). Compared with the rs314280 GG genotype, the variant AG and AA/AG genotypes were associated with more than 2-fold increased hazards of severe RP (AG and AA/AG versus GG; crude HR = 2.87 and 2.42; 95% CI, 1.27–6.45 and 1.10–5.34; P = 0.011 and 0.029, respectively) (Fig. 1b). Similar results were observed in multivariate analyses with adjustment for disease stage, chemotherapy history, smoke status and MLD (Table 2) that were considered potential confounding factors of RP [4, 5]. In addition, The LIN28B rs314276A variant allele (i.e., AC and AA/AC) was also associated with significantly increased hazards of grade ≥3 RP in both univariate and multivariate analyses. Specifically, compared with the rs314276 CC genotype, the variant AC and AA/AC genotypes were associated with a statistically significantly increased hazards of severe RP (AC and AA/AC versus CC; crude HR = 2.14 and 2.05; 95% CI, 1.15–3.96 and 1.14–3.71; P = 0.016 and 0.017, respectively; adjusted HR = 2.30 and 2.00; 95% CI, 1.24–4.28 and 1.11–3.62; P = 0.008 and 0.022, respectively) (Table 2 and Fig. 1c). In contrast, no associations were observed between the genotypes of other SNPs of LIN28B (rs12194974 G>A and rs17065417 A>C) and LIN28A (rs11247946 T>C, rs3811464 C>T, rs11581746 T>C, and rs12728900 G>A) and hazards of severe RP (Table 2).
Table 2.
Univariate and multivariate analyses of associations between LIN28 genotypes and severe RP(grade ≥3) in NSCLC patients received definitive radiation therapy
| Genotypes | Patient No. |
Event No. |
Crude |
Pa | Adjusted |
Pb | ||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |||||
| LIN28A(rs11247946) | ||||||||
| TT | 166 | 24 | 1.00 | 1.00 | ||||
| CT | 147 | 23 | 1.10 | 0.62–1.94 | 0.751 | 1.08 | 0.61–1.92 | 0.784 |
| CC | 49 | 9 | 1.37 | 0.64–2.94 | 0.424 | 1.28 | 0.59–2.77 | 0.526 |
| CC/CT | 196 | 32 | 1.16 | 0.68–1.97 | 0.580 | 1.33 | 0.67–1.93 | 0.644 |
| LIN28A(rs3811464) | ||||||||
| CC | 92 | 17 | 1.00 | 1.00 | ||||
| CT | 177 | 24 | 0.66 | 0.36–1.23 | 0.194 | 0.66 | 0.36–1.24 | 0.199 |
| TT | 93 | 15 | 0.84 | 0.42–1.68 | 0.618 | 0.92 | 0.45–1.85 | 0.805 |
| TT/CT | 270 | 39 | 0.72 | 0.41–1.27 | 0.259 | 0.74 | 0.42–1.32 | 0.305 |
| LIN28A(rs11581746) | ||||||||
| TT | 255 | 41 | 1.00 | 1.00 | ||||
| CT | 97 | 14 | 0.89 | 0.48–1.63 | 0.703 | 0.86 | 0.47–1.58 | 0.625 |
| CC | 10 | 1 | 0.61 | 0.08–4.45 | 0.627 | 0.62 | 0.08–4.49 | 0.632 |
| CC/CT | 107 | 15 | 0.86 | 0.48–1.56 | 0.624 | 0.84 | 0.46–1.52 | 0.558 |
| LIN28A(rs12728900) | ||||||||
| GG | 195 | 31 | 1.00 | 1.00 | ||||
| AG | 133 | 22 | 1.05 | 0.61–1.81 | 0.861 | 1.04 | 0.60–1.80 | 0.896 |
| AA | 34 | 3 | 0.59 | 0.18–1.92 | 0.378 | 0.60 | 0.18–1.98 | 0.406 |
| AA/AG | 167 | 25 | 0.96 | 0.57–1.63 | 0.877 | 0.96 | 0.56–1.62 | 0.865 |
| LIN28B(rs314280) | ||||||||
| GG | 95 | 7 | 1.00 | 1.00 | ||||
| AG | 168 | 35 | 2.87 | 1.27–6.45 | 0.011 | 2.97 | 1.32–6.72 | 0.009 |
| AA | 99 | 14 | 1.74 | 0.70–4.32 | 0.231 | 1.36 | 0.54–3.38 | 0.512 |
| AA/AG | 267 | 49 | 2.42 | 1.10–5.34 | 0.029 | 2.23 | 1.01–4.94 | 0.048 |
| LIN28B(rs12194974) | ||||||||
| GG | 280 | 42 | 1.00 | 1.00 | ||||
| AG | 76 | 13 | 1.20 | 0.64–2.23 | 0.567 | 1.37 | 0.73–2.57 | 0.329 |
| AA | 6 | 1 | 1.10 | 0.15–7.96 | 0.929 | 2.16 | 0.29–16.26 | 0.455 |
| AA/AG | 82 | 14 | 1.19 | 0.65–2.18 | 0.571 | 1.40 | 0.76–2.59 | 0.281 |
| LIN28B(rs17065417) | ||||||||
| AA | 290 | 47 | 1.00 | 1.00 | ||||
| AC | 68 | 9 | 0.74 | 0.36–1.51 | 0.410 | 0.77 | 0.38–1.57 | 0.470 |
| CC | 4 | 0 | N/A | |||||
| CC/AC | 72 | 9 | 0.73 | 0.36–1.48 | 0.377 | 0.75 | 0.37–1.54 | 0.435 |
| LIN28B(rs314276) | ||||||||
| CC | 154 | 15 | 1.00 | 1.00 | ||||
| AC | 155 | 31 | 2.14 | 1.15–3.96 | 0.016 | 2.30 | 1.24-4.28 | 0.008 |
| AA | 53 | 10 | 1.83 | 0.82–4.07 | 0.139 | 1.42 | 0.63–3.19 | 0.395 |
| AA/AC | 208 | 41 | 2.05 | 1.14–3.71 | 0.017 | 2.00 | 1.11–3.62 | 0.022 |
| Combined effect of risk genotypes in rs314280 and rs314276 | ||||||||
| 0–1c | 211 | 29 | ||||||
| 2c | 95 | 27 | 2.07 | 1.22–3.49 | 0.007 | 2.55 | 1.50–4.35 | 0.0006 |
RP = radiation pneumonitis; NSCLC = non-small cell lung cancer; CI = confidence interval; HR = hazard ratio; N/A, not applicable.
P values were calculated by Cox proportional model using univariate analysis.
P values were calculated with adjustment for smoking status, disease stage, chemotherapy history, and mean lung dose.
Total number of unfavorable genotypes
The LD analysis showed that all SNPs were in low LD using our genotyping data (i.e. LIN28B polymorphisms, r2 = 0.14 for rs314280 G>A and rs12194974 G>A; r2 = 0.10 for rs314280 G>A and rs17065417 A>C; r2 = 0.35 for rs314280 G>A and rs314276 C>A; r2 = 0.01 for rs12194974 G>A and rs17065417 A>C; r2 = 0.05 for rs12194974 G>A and rs314276 C>A; r2 = 0.04 for rs17065417 A>C and rs314276 C>A; Fig. 2a), the LD results were consistent with these calculated by using the CEPH (Utah residents with ancestry from northern and western Europe) population of HapMap data (Rel 28 Phase II+ III) (Fig. 2b). These suggest that each SNP may have an independent effect. Furthermore, we examined the combined effects of rs314280 and rs314276 genotypes. As shown in Table 2, when we used “0–1” risk genotypes as the reference, we found that carriers with risk genotypes on both SNPs (AA-AA, AA-AC, AG-AA or AG-AC) had significantly increased hazards of RP (adjusted HR=2.55, 95% CI=1.50–4.35; P = 0.0006).
Figure 2.
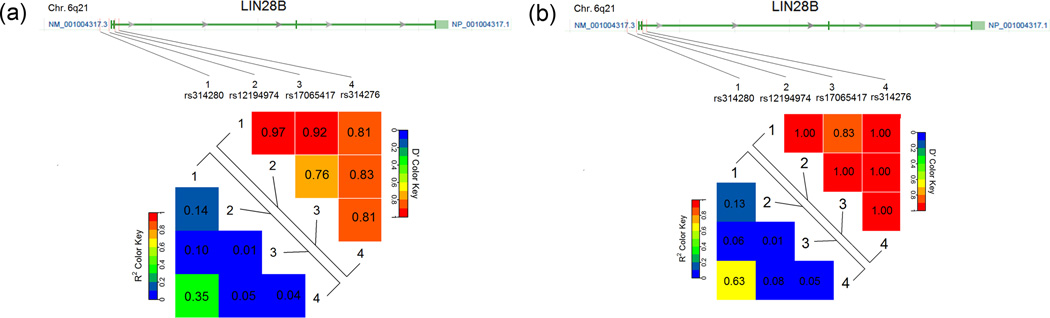
Positions of the four potentially functional single-nucleotide polymorphisms (SNPs) in LIN28B and pairwise linkage disequilibrium (LD) between them. The lower panel shows the LD plot of the LIN28B region, using the genotyping data from the present study (a) and CEPH (Utah residents with ancestry from northern and western Europe) population from the international HapMap project (Rel 28 Phase II + III) (b). Each number in grid represents the pairwise D’ and r2 between SNPs. All polymorphisms were in low LD with each other in both populations.
To further characterize biological significance of the LIN28B rs314280 and rs314276, we conducted correlation analysis between genotypes of rs314280 and rs314276 and mRNA expression levels of LIN28B in PBMCs from 105 cancer-free controls. As we expected from the literature, we found that there were no detectable expressions of LIN28B in these PBMCs (data not shown).
3.3. Stratified analyses
We then performed stratified analyses to evaluate the effects of variant genotypes on the hazards of RP by age, sex, race, KPS, stage, tumor histology, smoking status, and chemotherapy status, and MLD and radiation dose (Table 3). The results showed that the increased hazards associated with the variant A allele of rs314276 and risk genotypes (AA/AG and AA/AC) of the combined rs314280 and rs314276 were more evident in subjects who were <65 years old (adjusted HR = 2.74 and 3.79, 95% CI = 1.11–6.73 and 1.76–8.13, P = 0.029 and 0.0006, respectively), males (adjusted HR = 2.41and 3.65, 95% CI = 1.12–5.22 and 1.79–7.44, P = 0.025 and 0.0004, respectively), or having stage III and IV (adjusted HR = 3.11, 95% CI = 1.74–5.59, P = 0.0001 for combined) or squamous cell carcinomas (adjusted HR = 3.24 and 3.54, 95% CI = 1.05–10.05 and 1.39–9.01, P = 0.041 and 0.008, respectively), ever smokers (adjusted HR = 1.99 and 2.68, 95% CI = 1.08–3.68 and 1.54–4.65, P = 0.028 and 0.0005, respectively), and having radio-chemotherapy (adjusted HR = 1.92 and 2.61, 95% CI = 1.04–3.54 and 1.50–4.52, P = 0.037 and 0.0006, respectively), MLD ≥19.0Gy (adjusted HR = 3.76, 95% CI = 2.00–7.07, P < 0.0001 for combined), and radiation dose ≥66 (adjusted HR = 2.89 and 5.20, 95% CI = 1.08–7.70 and 2.13–12.67, P = 0.034 and 0.0003, respectively). For rs314280, the increased hazards associated with the variant A allele of rs314280 was more evident only for stage III and IV (adjusted HR = 3.11, 95% CI = 1.74–5.59, P = 0.017), MLD ≥19.0Gy (adjusted HR = 3.76, 95% CI = 2.00–7.07, P = 0.009), and radiation dose ≥66 (adjusted HR = 3.19, 95% CI = 1.01–10.03, P = 0.047) (Table 3).
Table 3.
Stratification analysis for associations between LIN28B variant genotypes and severe RP(grade ≥3) in NSCLC patients received definitive radiation therapy
| Variables | rs314280 (patient/event) |
Adjusted HRa(95%CI) | rs314276 (patient/event) |
Adjusted HRa(95%CI) | Combined effect of risk genotypes b |
Adjusted HRa(95%CI) | |||
|---|---|---|---|---|---|---|---|---|---|
| GG | AA/AG | CC | AA/AC | 0–1 | 2 | ||||
| Age, yr | |||||||||
| <65(median) | 43/5 | 109/24 | 2.20(0.83–5.82) | 73/6 | 79/23 | 2.74(1.11–6.73) | 109/11 | 43/18 | 3.79(1.76–8.13) |
| ≥65(median) | 42/9 | 112/18 | 1.16(0.51–2.62) | 66/9 | 88/18 | 1.49(0.67–3.34) | 102/18 | 52/9 | 1.46(0.64–3.33) |
| Gender | |||||||||
| Males | 45/6 | 120/27 | 2.31(0.94–5.71) | 83/9 | 82/24 | 2.41(1.12–5.22) | 119/15 | 46/18 | 3.65(1.79–7.44) |
| Females | 40/8 | 101/15 | 1.11(0.47–2.66) | 56/6 | 85/17 | 1.65(0.65–4.20) | 92/14 | 49/9 | 1.68(0.70–3.99) |
| Race | |||||||||
| White | 51/10 | 220/36 | 1.35(0.66–2.76) | 111/13 | 140/33 | 1.79(0.94–3.41) | 160/23 | 91/23 | 2.21(1.23–3.98) |
| Black | 34/4 | 21/6 | 3.68(0.93–14.61) | 28/2 | 27/8 | 3.13(0.63–15.69) | 51/6 | 4/4 | 6.41(1.76–23.34) |
| KPS | |||||||||
| <80 | 72/11 | 175/30 | 1.68(0.83–3.39) | 108/11 | 139/30 | 1.78(0.89–3.56) | 173/22 | 74/19 | 2.48(1.33–4.63) |
| ≥80 | 13/3 | 46/12 | 1.37(0.37–5.17) | 31/4 | 28/11 | 2.90(0.91–9.28) | 38/7 | 21/8 | 2.42(0.84–6.95) |
| Stage group | |||||||||
| I, II | 10/6 | 34/4 | 0.37(0.10–1.36) | 20/2 | 24/8 | 3.64(0.74–17.89) | 28/8 | 16/2 | 0.84(0.16–4.45) |
| III, IV | 75/8 | 186/38 | 2.54(1.18–5.49) | 119/13 | 142/33 | 1.80(0.94–3.42) | 183/21 | 78/25 | 3.11(1.74–5.59) |
| Histology | |||||||||
| Adenocarcinoma | 32/5 | 84/16 | 1.34(0.49–3.70) | 52/5 | 64/16 | 2.32(0.84–6.42) | 76/10 | 40/11 | 2.11(0.89–5.01) |
| Squamous cell | 30/6 | 78/13 | 1.57(0.57–4.34) | 52/4 | 56/15 | 3.24(1.05–10.05) | 78/10 | 30/9 | 3.54(1.39–9.01) |
| Other | 23/3 | 59/13 | 4.28(1.11–16.57) | 35/6 | 47/10 | 0.70(0.24–2.03) | 57/9 | 25/7 | 2.30(0.83–6.38) |
| Smoking status | |||||||||
| Never | 7/2 | 21/2 | 0.55(0.07–4.69) | 10/1 | 18/3 | 3.45(0.21–57.42) | 17/3 | 11/1 | 1.62(0.14–19.30) |
| Ever | 78/12 | 200/40 | 1.82(0.94–3.50) | 129/14 | 149/38 | 1.99(1.08–3.68) | 194/26 | 84/26 | 2.68(1.54–4.65) |
| Chemotherapy | |||||||||
| No | 6/2 | 22/2 | 3.00(0.20–44.40) | 15/1 | 13/3 | 1.79(0.14–22.47) | 19/3 | 9/1 | 7.40(0.27–204.10) |
| Yes | 79/12 | 198/40 | 1.78(0.93–3.42) | 123/14 | 154/38 | 1.92(1.04–3.54) | 191/26 | 86/26 | 2.61(1.50–4.52) |
| Mean lung dose | |||||||||
| <19.0 Gy | 31/6 | 135/9 | 0.43(0.15–1.23) | 75/4 | 91/11 | 2.15(0.68–6.82) | 102/10 | 64/5 | 0.87(0.30–2.57) |
| ≥19.0 Gy | 54/8 | 86/33 | 2.82(1.29–6.19) | 64/11 | 76/30 | 1.91(0.96–3.82) | 109/19 | 31/22 | 3.76(2.00–7.07) |
| Radiation dose | |||||||||
| <66 | 42/4 | 105/22 | 3.19(1.01–10.03) | 67/5 | 80/21 | 2.89(1.08–7.70) | 102/9 | 45/17 | 5.20(2.13–12.67) |
| ≥66 | 43/10 | 116/20 | 1.19(0.54–2.62) | 72/10 | 87/20 | 1.47(0.69–3.15) | 109/20 | 50/10 | 1.49(0.68–3.26) |
Adjusted for smoking status, disease stage, chemotherapy history, and mean lung dose.
The number represents the numbers of unfavorable genotypes for each SNP.
NSCLC: non-small cell lung cancer.
Because most of the significant findings were in the subgroup analyses, we calculated the FPRP values for all the observed significant associations. As shown in (Table 4), when the assumption of a prior probability was 0.25, the association with the combined LIN28B (AA/AG and AA/AC) genotypes was still noteworthy for all subjects (FPRP = 0.009), age <65 years (FPRP = 0.036), males (FPRP = 0.022), stage III and IV (FPRP = 0.006), ever smokers (FPRP = 0.009), radio-chemotherapy (FPRP = 0.011) as well as for MLD >19.0Gy (FPRP = 0.005); the association with LIN28B (rs314276 AA/AC) genotypes was only noteworthy for all subjects (FPRP = 0.007). When the assumption of prior probability was 0.1, the association with the combined LIN28B (AA/AG and AA/AC) genotypes was still noteworthy for all subjects (FPRP = 0.028), stage III and IV (FPRP = 0.019), ever smokers (FPRP = 0.027), radio-chemotherapy (FPRP = 0.031) as well as for MLD >19.0 Gy (FPRP = 0.014).
Table 4.
False-positive report probability (FPRP) values for associations between severe RP (grade >3) and frequencies of rs314280, rs314276 and combined genotypes in NSCLC patients received definitive radiation therapy
| Genotype | Positive HR 95%CIa |
Pb | Statistical powerc |
Prior probability |
||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs314280: G>A | GG vs. AA/AG | |||||||
| All subjects | 2.23 (1.01–4.94) | 0.048 | 0.394 | 0.268 | 0.523 | 0.924 | 0.992 | 0.999 |
| Stage III,IV | 2.54 (1.18–5.49) | 0.018 | 0.272 | 0.164 | 0.371 | 0.866 | 0.985 | 0.998 |
| Histology Other | 4.28 (1.11–16.57) | 0.035 | 0.135 | 0.439 | 0.701 | 0.963 | 0.996 | 1.000 |
| MLD ≥19.0 | 2.82 (1.29–6.19) | 0.009 | 0.196 | 0.130 | 0.309 | 0.831 | 0.980 | 0.998 |
| Radiation dose <66 | 3.19 (1.01–10.03) | 0.047 | 0.212 | 0.400 | 0.667 | 0.957 | 0.996 | 1.000 |
| rs314276: C>A | CC vs. AA/AC | |||||||
| All subjects | 2.00 (1.11–3.62) | 0.022 | 0.500 | 0.007 | 0.284 | 0.814 | 0.978 | 0.998 |
| Age < 65 | 2.74 (1.11–6.73) | 0.028 | 0.246 | 0.254 | 0.505 | 0.918 | 0.991 | 0.999 |
| Males | 2.41 (1.12–5.22) | 0.026 | 0.318 | 0.195 | 0.421 | 0.889 | 0.988 | 0.999 |
| Histology Squamous cell | 3.24 (1.05–10.05) | 0.042 | 0.202 | 0.383 | 0.651 | 0.954 | 0.995 | 1.000 |
| Ever Smokers | 1.99 (1.08–3.68) | 0.028 | 0.506 | 0.143 | 0.334 | 0.847 | 0.982 | 0.998 |
| Chemotherapy Yes | 1.92 (1.04–3.54) | 0.037 | 0.552 | 0.166 | 0.374 | 0.868 | 0.985 | 0.998 |
| Radiation dose <66 | 2.89 (1.08–7.70) | 0.034 | 0.231 | 0.305 | 0.569 | 0.935 | 0.993 | 0.999 |
| Combined | 0–1 vs. 2 | |||||||
| All subjects | 2.55 (1.50–4.35) | <0.001 | 0.186 | 0.009 | 0.028 | 0.239 | 0.760 | 0.969 |
| Age < 65 | 3.79 (1.76–8.13) | <0.001 | 0.050 | 0.036 | 0.100 | 0.550 | 0.925 | 0.992 |
| Males | 3.65 (1.79–7.44) | <0.001 | 0.049 | 0.022 | 0.063 | 0.426 | 0.882 | 0.987 |
| White | 2.21 (1.23–3.98) | 0.008 | 0.370 | 0.063 | 0.167 | 0.668 | 0.957 | 0.996 |
| Black | 6.41 (1.76–23.34) | 0.005 | 0.039 | 0.273 | 0.530 | 0.925 | 0.992 | 0.999 |
| KPS <80 | 2.48 (1.33–4.63) | 0.004 | 0.250 | 0.050 | 0.136 | 0.633 | 0.946 | 0.994 |
| Stage III,IV | 3.11 (1.74–5.59) | <0.001 | 0.070 | 0.006 | 0.019 | 0.174 | 0.680 | 0.955 |
| Histology Squamous cell | 3.54 (1.39–9.01) | 0.008 | 0.115 | 0.172 | 0.384 | 0.873 | 0.986 | 0.999 |
| Ever Smokers | 2.68 (1.54–4.65) | <0.001 | 0.149 | 0.009 | 0.027 | 0.232 | 0.753 | 0.968 |
| Chemotherapy Yes | 2.61 (1.50–4.52) | <0.001 | 0.171 | 0.011 | 0.031 | 0.263 | 0.783 | 0.973 |
| MLD ≥19.0 | 3.76 (2.00–7.07) | <0.001 | 0.025 | 0.005 | 0.014 | 0.135 | 0.611 | 0.940 |
| Radiation dose <66 | 2.89 (1.08–7.70) | 0.034 | 0.231 | 0.305 | 0.569 | 0.935 | 0.993 | 0.999 |
The adjusted HR.
The omnibus chi-square test of the genotype frequency distributions.
Calculated using study subjects to detect a HR of 2.00 with the common genotype used as the reference.
NSCLC: non-small cell lung cancer.
4. Discussion
In the present study, we evaluated the associations between genetic variants of LIN28A and LIN28B genes and risk of severe RP in patients with NSCLC treated with definitive radiotherapy. We found that LIN28B rs314280 variant A genotypes were associated with a significantly increased risk of grade ≥3 RP, compared with the GG genotype. Furthermore, we found that the AA/AC genotypes of LIN28B rs314276 were also associated with a significantly increased risk of grade ≥3 RP, compared with its common CC genotype. Stratified analyses showed that the increased risk was more evident in subgroups of subjects of <65 years, males, having squamous cell carcinoma, ever smokers, and having radio-chemotherapy. Although multiple testing had been performed in the present study, the results of FPRP indicated that the combined effect of risk genotypes was less likely to be false positive. Therefore, our data indicated that LIN28B (rs314280 and rs314276) genetic variants may independently and jointly have an effect on the risk of severe RP in NSCLC patients treated with radiotherapy, suggesting that these variants may be useful biomarkers for RP susceptibility in lung cancer patients. Our data also indicated that patients at highest risk of severe RP were those who were carrying risk genotypes on both SNPs (AA/AG and AA/AC) and received MLD >19.0 Gy. These findings suggest that the MLD should be controlled as low as possible for patients with the rs314280 AA/AG or rs314280 AA/AG genotypes to avoid high risk of severe RP. Because MLD is known before the radiation has begun, genotyping LIN28B at rs314280 and rs314276 could be used as a predictive biomarker for prescribing highly personalized radiation (chemo) therapy once our results are validated in clinical trials.
Experimental and clinical as well as epidemiological studies have suggested a strong association between inflammation and radiation pneumonitis [25–27]. Inflammation is a physiological response to cellular and tissue damage, including radiation-induced damage. Appropriate response to this damage is tightly regulated through a balance between pro-inflammatory and anti-inflammatory cytokines and signaling molecules [28, 29]. Inflammatory cytokines are the key to modulating and ameliorating the inflammation-related effects following lung irradiation as the observed RP. Indeed, lung irradiation induces the expression of pro-inflammatory cytokines; for example, interleukin (IL-6) levels were significantly higher during and after thoracic irradiation therapy in lung cancer patients who developed RP [30, 31], whereas let-7 miRNA overexpression significantly reduced IL-6 expression. Recently, some published studies have indicated that LIN28 links inflammation to cell transformation [13, 14], LIN28B functions as a post-transcriptional regulator of let-7 miRNA biogenesis by inhibiting the microprocessor-mediated cleavage of pri-let-7 miRNAs [12], and NF-kB activates LIN28B and inhibits let-7 miRNA expression, leading to overexpression of IL-6 by NF-kB activation [13–15]. It is well known that LIN28B is a homologue of LIN28A [20], and both LIN28A and LIN28B also pay an important role during multiple cellular development processes and tumorigenesis in humans. For example, overexpression of LIN28B has been shown to be more frequent in human cancers, such as colon cancer, hepatocellular carcinomas, and NSCLC [10, 23] as well as to be able to stimulate tumor growth [32]. More recently, one study has found that LIN28 regulates radiosensitivity of human lung cancer cells [16], in which, by using LIN28-siRNA, LIN28 was found to be specifically inhibited, leading to upregulation of the let-7a expression but downregulation of the K-Ras protein expression and resulting in the radio-sensitization of lung cancer cells. Taken together, it is confirmed that LIN28 functions to regulate not only the biogenesis of miRNAs, particularly the let-7 family miRNAs, but also the translation of mRNAs important for cell growth and metabolism [9]. Our data further implied that LIN28B may be involved in inflammatory response. However, the exact inflammation mechanism of LIN28, especially in the RP development, is still unclear.
Several GWAS studies have identified some genetic variants of LIN28B (i.e., re314276, rs314277 and rs314280) that were associated with age at menarche and age at natural menopause as well as the timing of puberty [18, 19, 33]. In a GWAS of 15,297 Icelandic women subjects, Sulem et al. reported that the LIN28 rs314280 T allele was associated with earlier age at menarche [18]. In another GWAS of 21,087 European women population, it was shown that LIN28B rs314276 C allele was associated with the timing of puberty [19]. To date, however, no published study has investigated the association between genetic variants of LIN28 and radiation treatment-related toxicities. More recently, only one study investigated the associations between genetic variants of the LIN28 gene and risk of type 2 diabetes mellitus (T2DM) [17]. In a hospital-based association study of 588 T2DM patients and 588 age and sex matched controls, Zhang et al. demonstrated that rs3811463 polymorphism was associated with increased risk of T2DM in a Chinese population [17].
In the present study, we found that among NSCLC patients, severe RP was more likely to occur in those carrying LIN28B rs314276 AA/AC and rs314280 AA/AG variant genotypes than in those carrying the LIN28B rs314276 CC and rs314280 GG genotypes, respectively. These two SNPs are potentially functional, because they are located either in intron 2 (rs314276) or at the 5’ end (rs314280) of the LIN28B gene [18] that are predicted to affect the binding activity at the transcription factor binding sites (TFBS); thereby, we speculated that these LIN28B variants may result in altered gene expression or regulation of signaling function, which may be the mechanisms underlying the increased radiosensitivity of the irradiated tissues or RP susceptibility, as we observed in the current study. Therefore, we further tested for correlation between these two SNPs and expression levels of LIN28B mRNA in peripheral blood mononuclear cells (PBMCs) of 105 cancer-free controls. However, by using RT-PCR, as reported in a previous study [18], we did not detect expression of LIN28B in these cells, which is consistent with the reports that LIN28B expression was undetectable or detected at very low levels in most normal adult tissue [9, 11, 34]; furthermore, we did not have lung cancer tumor tissues for the detection of LIN28B expression. Therefore, we were not able to correlate LIN28B expression with the RP-associated genetic variants in the target tissues in the present study. However, our genotype results are consistent with the function of LIN28 that is involved in the inflammation mechanism.
To the best of our knowledge, the present study provided for the first time the evidence that potentially functional SNPs of the LIN28B gene may predict risk of severe RP in lung cancer patients who have received definitive radiotherapy. However, the present study has several limitations. Firstly, the exact molecular mechanisms by which the LIN28B SNPs influence the risk of RP were not fully understood, as they were beyond the scope of the current study. Secondly, we used the candidate SNP/gene approach that allowed us to focus on potentially common, functional SNPs in the LIN28 gene, but this approach did not include all representative or tagging SNPs in the entire gene. Some important SNPs but with a low frequency might have been missed or the observed associations may have been due to genetic linkages with other untyped SNPs. Therefore, additional investigations of the tagged or functional SNPs are warranted and will be the subject of future studies.
In summary, the present study demonstrated that the selected potentially functional SNPs of LIN28B (rs314280 and rs314276) may independently and jointly affect the development of severe RP in NSCLC patients treated with radiotherapy. Further large, prospective studies are essential to confirm our findings. Once validated, LIN28B SNPs may be useful biomarkers for susceptibility to RP.
Supplementary Material
Acknowledgements
We thank Margaret Lung and Jessica Fiske for their assistance in recruiting the subjects and gathering the questionnaire information and Min Zhao, Jianzhong He and Kejing Xu for their laboratory assistance.
Funding
This study was supported in part by National Institutes of Health Grants R01 ES 11740 and R01 CA 131274 (to Q. W.) and by Cancer Center Core Grant P30 CA016672 to The University of Texas MD Anderson Cancer Center.
Abbreviations
- RP
Radiation-induced pneumonitis
- NSCLC
Non-small cell lung cancer
- SNPs
Single nucleotide polymorphisms
- MLD
mean lung dose
- HR
Hazards ratio
- 95% CIs
95% Confidence intervals
Footnotes
Conflict of interest statement
None declared.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Gandara D, Narayan S, Lara PN, Jr, Goldberg Z, Davies A, Lau DH, et al. Integration of novel therapeutics into combined modality therapy of locally advanced non-small cell lung cancer. Clin Cancer Res. 2005;11(13 Pt 2):5057s–5062s. doi: 10.1158/1078-0432.CCR-05-9012. [DOI] [PubMed] [Google Scholar]
- 3.Roach M, 3rd, Gandara DR, Yuo HS, Swift PS, Kroll S, Shrieve DC, et al. Radiation pneumonitis following combined modality therapy for lung cancer: analysis of prognostic factors. J Clin Oncol. 1995;13(10):2606–2612. doi: 10.1200/JCO.1995.13.10.2606. [DOI] [PubMed] [Google Scholar]
- 4.Provatopoulou X, Athanasiou E, Gounaris A. Predictive markers of radiation pneumonitis. Anticancer Res. 2008;28(4C):2421–2432. [PubMed] [Google Scholar]
- 5.Yuan X, Liao Z, Liu Z, Wang LE, Tucker SL, Mao L, et al. Single nucleotide polymorphism at rs1982073:T869C of the TGFbeta 1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J Clin Oncol. 2009;27(20):3370–3378. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong H, Liao Z, Liu Z, Xu T, Wang Q, Liu H, et al. ATM polymorphisms predict severe radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys. 2013;85(4):1066–1073. doi: 10.1016/j.ijrobp.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip Rev RNA. 2012;3(4):483–494. doi: 10.1002/wrna.1112. [DOI] [PubMed] [Google Scholar]
- 10.King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71(12):4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3(6):719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 12.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan HW, Lappas M, Yee SW, Vaswani K, Mitchell MD, Rice GE. The expression of the let-7 miRNAs and Lin28 signalling pathway in human term gestational tissues. Placenta. 2013;34(5):443–448. doi: 10.1016/j.placenta.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76(1):5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Zhang L, Fan R, Guo N, Xiong C, Wang L, et al. The polymorphism in the let-7 targeted region of the Lin28 gene is associated with increased risk of type 2 diabetes mellitus. Mol Cell Endocrinol. 2013;375(1–2):53–57. doi: 10.1016/j.mce.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41(6):734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 19.Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41(6):729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen AX, Yu KD, Fan L, Li JY, Yang C, Huang AJ, et al. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet. 2011;7(9):e1002259. doi: 10.1371/journal.pgen.1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71(11):3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao ZX, Komaki RR, Thames HD, Jr, Liu HH, Tucker SL, Mohan R, et al. Influence of technologic advances on outcomes in patients with unresectable, locally advanced non-small-cell lung cancer receiving concomitant chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2010;76(3):775–781. doi: 10.1016/j.ijrobp.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rube CE, Wilfert F, Palm J, Konig J, Burdak-Rothkamm S, Liu L, et al. Irradiation induces a biphasic expression of pro-inflammatory cytokines in the lung. Strahlenther Onkol. 2004;180(7):442–448. doi: 10.1007/s00066-004-1265-7. [DOI] [PubMed] [Google Scholar]
- 26.Miyake K, Tani K, Kakiuchi S, Suzuka C, Toyoda Y, Kishi J, et al. Epidermal growth factor receptor-tyrosine kinase inhibitor (gefitinib) augments pneumonitis, but attenuates lung fibrosis in response to radiation injury in rats. J Med Invest. 2012;59(1–2):174–185. doi: 10.2152/jmi.59.174. [DOI] [PubMed] [Google Scholar]
- 27.Flechsig P, Dadrich M, Bickelhaupt S, Jenne J, Hauser K, Timke C, et al. LY2109761 attenuates radiation-induced pulmonary murine fibrosis via reversal of TGF-beta and BMP-associated proinflammatory and proangiogenic signals. Clin Cancer Res. 2012;18(13):3616–3627. doi: 10.1158/1078-0432.CCR-11-2855. [DOI] [PubMed] [Google Scholar]
- 28.Dinarello CA. Proinflammatory cytokines. Chest. 2000;118(2):503–508. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 29.Opal SM, DePalo VA. Anti-inflammatory cytokines. Chest. 2000;117(4):1162–1172. doi: 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Rubin P, Williams J, Hernady E, Smudzin T, Okunieff P. Circulating IL-6 as a predictor of radiation pneumonitis. Int J Radiat Oncol Biol Phys. 2001;49(3):641–648. doi: 10.1016/s0360-3016(00)01445-0. [DOI] [PubMed] [Google Scholar]
- 31.Rube CE, Uthe D, Wilfert F, Ludwig D, Yang K, Konig J, et al. The bronchiolar epithelium as a prominent source of pro-inflammatory cytokines after lung irradiation. Int J Radiat Oncol Biol Phys. 2005;61(5):1482–1492. doi: 10.1016/j.ijrobp.2004.12.072. [DOI] [PubMed] [Google Scholar]
- 32.Lu L, Katsaros D, Shaverdashvili K, Qian B, Wu Y, de la Longrais IA, et al. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur J Cancer. 2009;45(12):2212–2218. doi: 10.1016/j.ejca.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 33.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41(6):724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22(1):51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


