Abstract
Little is known about the genetic factors that contribute to familial colorectal cancer type X (FCCX), characterized by hereditary nonpolyposis colorectal carcinoma with no mismatch repair defects. Genetic linkage analysis, exome sequencing, tumor studies, and functional investigations of 4 generations of a FCCX family led to the identification of a truncating germline mutation in RPS20, which encodes a component (S20) of the small ribosomal subunit and is a new colon cancer predisposition gene. The mutation was associated with a defect in pre–ribosomal RNA maturation. Our findings show that mutations in a gene encoding a ribosomal protein can predispose individuals to microsatellite-stable colon cancer. Evaluation of additional FCCX families for mutations in RPS20 and other ribosome-associated genes is warranted.
Keywords: Colon Cancer, Hereditary Nonpolyposis Colorectal Cancer, Ribosome, Exome Sequencing
Abbreviations used in this paper: FCCX, hereditary nonpolyposis colorectal cancer type X; rRNA, ribosomal RNA
See Covering the Cover synopsis on page 547; see editorial on page 554.
Hereditary nonpolyposis colorectal cancer as defined by the Amsterdam criteria1, 2 includes 2 distinct entities with roughly comparable shares. Families with germline mutations in DNA mismatch repair genes represent Lynch syndrome (MIM 120435-6), with some 3000 unique predisposing mutations known.3 Familial colorectal cancer type X (FCCX) is a collective designation for families with no evidence of DNA mismatch repair deficiency, wherein type X refers to the as yet unknown nature of predisposition.4
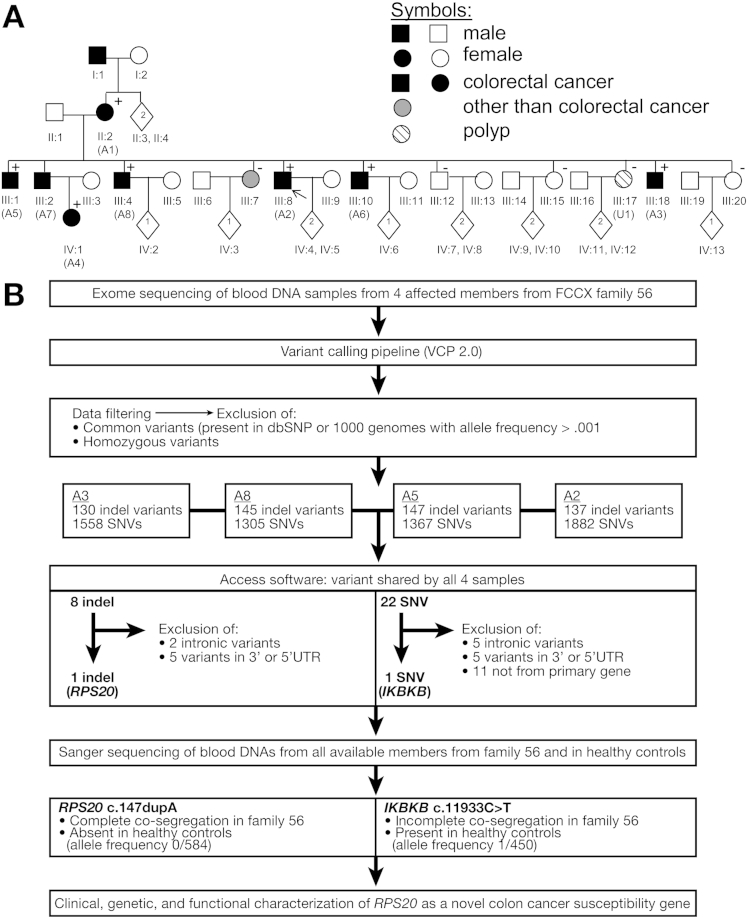
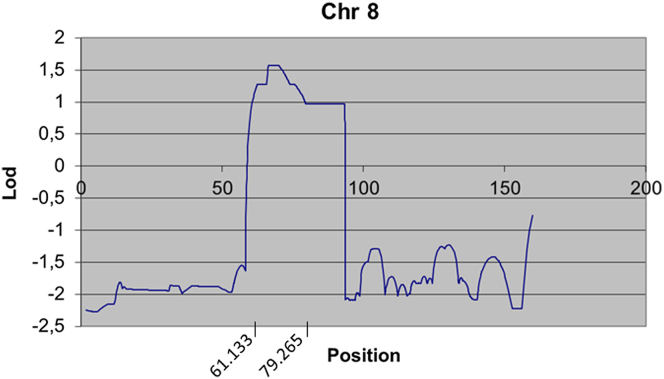
We recently discovered germline mutations in the gene for bone morphogenetic protein receptor type IA in 2 Amsterdam-positive families of 18 FCCX families investigated (11%).5 Among families with no bone morphogenetic protein receptor type IA mutations, family F56 fulfilling the Amsterdam criteria (Figure 1A) was chosen for closer scrutiny by genetic linkage analysis, exome sequencing, and tumor investigations. The mean age at colorectal cancer diagnosis was 52.3 years, with a 6–8 ratio of proximal to distal cancers. Genome-wide linkage analysis of the core pedigree resulted in the highest multipoint lod score (1.6) for D8S507 (Genethon) and D8S1115 (Marshfield), both of which reside in the area of linkage between D8S255 and D8S1718 on chromosome 8p11-8q12 (Supplementary Materials and Methods and Supplementary Figure 1).
Figure 1.
(A) Pedigree of FCCX family 56. Numbers below the symbols are patient identifiers; key members also are marked with a letter code A1–A8 for affected and U1 for unaffected. Carrier status for the c.147dupA in RPS20 is shown (+, mutation carrier, -, noncarrier). Arrow denotes the index person. Clinical diagnoses are specified in Supplementary Table 1. Nonessential pedigree features were omitted or modified to protect confidentiality. (B) Exomic sequencing of blood DNAs from individuals A2, A3, A5, and A8 (see Supplementary Materials and Methods for details). The stepwise reduction in the number of insertions or deletions and single-nucleotide variants (SNV) remaining for consideration is shown, ultimately resulting in 2 exonic alterations shared by the 4 affected members. The RPS20 insertion or deletion (indel) alteration fulfilled the prerequisites of a predisposing mutation and was characterized fully in this investigation whereas the available evidence (incomplete co-segregation, occurrence in healthy controls, equivocal pathogenicity by predictions, as well as other data detailed in the Supplementary Materials and Methods) suggested that the inhibitor of κ light polypeptide gene enhancer in B cells, kinase β (IKBKB) SNV alteration was unlikely to explain the colorectal cancer susceptibility of F56 and was excluded from further consideration. U, noncarrier.
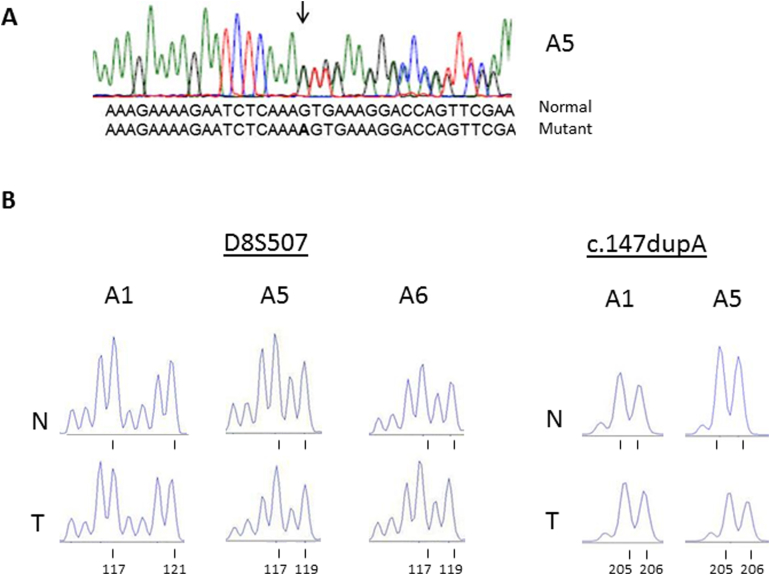
However, because a few other chromosomal regions also showed lod scores greater than 1, we opted for exome sequencing and chose 4 siblings with colorectal cancer from F56 to be included in the analysis (Figure 1B). A single truncating alteration of RPS20 (c.147dupA, RefSeq NM_001023.3) (Supplementary Figure 2A), a ribosomal protein gene, turned out to be shared by all 4 affected members investigated. It leads to frameshift and premature truncation (p.Val50SerfsX23). RPS20 is located on 8q12.1 in the region identified by genetic linkage analysis. The alteration showed a full co-segregation with microsatellite-stable colorectal cancer in F56 (Figure 1A), yielding a lod score of 3.0 for segregation. The sequence change was absent in healthy controls (allele count 0 of 584); moreover, it has not been reported in 4300 European Americans and 2203 African Americans (Exome Variant Server; available: http://evs.gs.washington.edu/EVS/; date accessed: April 1, 2014). We subsequently screened RPS20 for mutations in blood DNA from 25 other FCCX families from Finland and in tumor DNA from 61 primary colorectal cancers and cancer cell lines (Supplementary Materials and Methods); no RPS20 mutations were detected. Based on COSMIC (http://cancer.sanger.ac.uk) and TCGA (http://cancergenome.nih.gov/) databases (date accessed: May 28, 2014), at least 11 unique missense variants of RPS20 in cancer are known (mutation frequency of up to 2.6% depending on tumor type), with pathogenicity varying from benign to deleterious by in silico predictions. At least one colon cancer case with a somatic missense change (R79C) is included.6
Tumors from mutation carriers showed no loss of the wild-type allele (Supplementary Figure 2B), arguing against Knudson’s 2-hit mechanism for tumor-suppressor genes.7 The absence of loss of heterozygosity complies with observations from zebrafish showing that ribosomal protein genes act as haploinsufficient suppressors of tumorigenesis.8
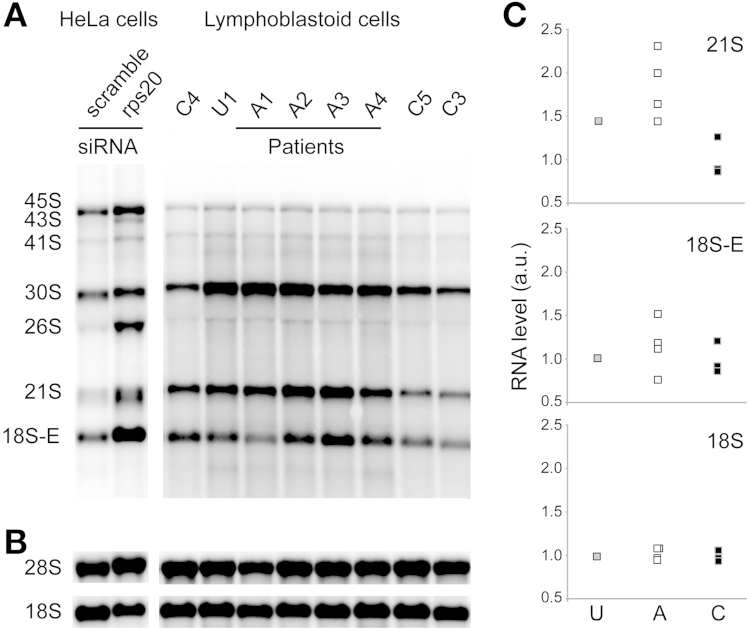
RPS20 is required during the late steps of 18S ribosomal RNA (rRNA) formation.9 Indeed, Northern blot analysis showed that small interfering RNA depletion of RPS20 in HeLa cells led to a significant increase of 21S pre-rRNAs (which are distributed in 2 close bands in this cell type), as well as an accumulation of 18S-E pre-rRNAs (Figure 2A). This was accompanied by a strong decrease of the 18S/28S ratio (Figure 2B). Patients carrying the RPS20 c.147dupA mutation (A1–A4) showed a marked increase of 21S pre-rRNAs compared with healthy unrelated controls (C1–C3), while the 18S-E pre-rRNA level was in the same range in control, noncarrier, and patient samples (Figure 2C). The 18S/28S ratios were unchanged in patient cells compared with controls and a noncarrier. Altogether, these results show a late pre-rRNA processing defect in mutation carrier cells consistent with RPS20 haploinsufficiency. Polysome analysis showed a slight increase in the 60S peak relative to the 40S peak in mutation carriers compared with a noncarrier and a healthy unrelated control (Supplementary Figure 3). Collectively, RNA results suggest that the RPS20 mutation disturbs ribosome biogenesis by affecting the equilibrium between the different pre-rRNA species and the formation of mature 18S rRNA.
Figure 2.
(A) Northern blot analysis of total RNAs from HeLa cells treated for 48 hours with a scramble small interfering RNA (siRNA) or a small interfering RNA targeting RPS20 messenger RNA, and lymphoblastoid RNAs from controls (C1–C3), a noncarrier (U), and affected mutation carriers (A1–A4). Precursor rRNAs were detected with a 5’ internal-transcribed spacer 1 probe.9 (B) Mature rRNAs detected with 18S and 28S probes. (C) Quantification of pre-rRNA species by phosphorimaging after normalization to 28S rRNA. For each species, the value of the mean of the 3 control samples arbitrarily was set to 1.
All RPSs are essential in human cells, except RPS25.9 The ribosomal protein gene family comprises 80 genes,8 at least 11 of which are known to be mutated in Diamond–Blackfan anemia, a dominantly inherited form of pure red cell aplasia, growth retardation, and congenital anomalies.10, 11 No such features were present in colon cancer patients from F56. Why is the RPS20 mutation associated with colorectal cancer susceptibility, while mutations in 11 other ribosomal protein genes cause predisposition to Diamond–Blackfan anemia? Haploinsufficiency for RPS19 or RPS20 in mice was shown to stabilize p53, which in turn had different effects in different cell types.12 Mouse findings make it tempting to speculate that cell type–specific effects of RPS20 haploinsufficiency might play a role in RPS20-associated colon tumorigenesis in human beings, with disturbed ribosome biogenesis, altered p53 dosage, or various downstream events as possible mediators. Among ribosomal proteins, “detector” and “effector” types have been distinguished based on contribution to p53 stress response.13 RPS20 was proposed to be primarily of the detector type, with reduction perturbing ribosomal biogenesis9 (Figure 2 and Supplementary Figure 3), leading to stabilization of p5313 (Supplementary Figure 4). Conversely, the constant activation of p53 consecutive to ribosomal stress induced by RPS20 mutation could favor, in the long run, the selection of cells that escape regulation by p53.
In summary, we show that inactivating germline mutation of RPS20 is associated with a dominant predisposition to colorectal cancer. This report links germline mutation of RPS20 to human disease. Future investigations are necessary to establish the prevalence of RPS20 mutations in FCCX families worldwide as well as the exact tumorigenic mechanisms and the basis of apparent tumor-type specificity. Finally, our study encourages investigations into the possible involvement of other ribosomal protein genes in colon cancer susceptibility.
Acknowledgments
The authors thank Saila Saarinen for expert technical assistance and Tuula Lehtinen and Kirsi Pylvänäinen for help in collecting clinical data. The authors also thank Dr Hanna Gazda for helpful discussions.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the Paulo Foundation (T.T.N.), Suomen Akatemia (257795 to P.P.), The Finnish Cancer Organizations (P.P., J.-P.M.), the Sigrid Juselius Foundation (P.P., H.L., J.-P.M.), the Päivikki and Sakari Sohlberg Foundation (P.P.), Nordic Cancer Union (P.P.), Biocentrum Helsinki (P.P.), European Research Council (FP7-ERC-232635 to P.P.), Kuopio University Hospital (B13-104 and B14-01 to J.-P.M.), Agence Nationale pour la Recherche (ANR-10-BLAN-1115-1 to P.-E.G.), Association pour la Recherche sur le Cancer (PJA20131200432 to M.-F.O.’D.), and Genome Canada through the Ontario Genomics Institute (S.W.S. and A.D.P.). The Centre for Applied Genomics provided infrastructure support.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2014.06.009.
Supplementary Materials and Methods
Patients and Samples
This study was based on 26 FCCX families from Finland, 15 of which fulfilled the Amsterdam criteria1, 2 and 11 fulfilled the Bethesda criteria.3 The families had no DNA mismatch repair defects in tumor tissue or in the germline4; furthermore, germline mutations in bone morphogenetic protein receptor type IA were excluded.5 In addition, 11 colorectal cancer cell lines including 7 with microsatellite instability (HCA7, HCT115, HCT116, KM12, LIM1215, LoVo, and RKO) and 4 microsatellite-stable (T84, SW480, SW837, and CACO2), 50 primary colorectal cancers6 (18 with microsatellite instability and 32 microsatellite stable), as well as 197 healthy blood donors from Finland and 95 from the United Kingdom were screened for RPS20 mutations. The Institutional Review Board of the Helsinki University Central Hospital (Helsinki, Finland) approved this study.
Genome-Wide Linkage Analysis
Genetic linkage analysis of the core pedigree of F56 was conducted at Ohio State University, taking advantage of microsatellite markers from the Genethon collection (ABI PRISM Linkage Mapping Set Version 2, Grand Island, NY)7 and at the University of Toronto using single-nucleotide polymorphisms and microsatellite markers from the Marshfield collection (Human Mapping 10K array V2; Affymetrix, Santa Clara, CA) (Supplementary Figure 1). Parametric multipoint analysis was performed with MERLIN v1.0-alpha (Ann Arbor, MI),8 using the following parameters: autosomal-dominant mode of inheritance, gene frequency of 0.001, 4 age-dependent liability classes,9 and 4 age-dependent phenocopy rates (0.00, 0.01, 0.03, and 0.05 corresponding to liability classes 1–4, respectively). Family members with a colorectal polyp as the only tumor were treated in 2 alternative ways, considering them either to have an unknown status or to be affected.
Exome Sequencing and Data Analysis
Exome sequencing was conducted at the Institute for Molecular Medicine Finland (Helsinki, Finland). Exome sequencing was performed on an Illumina HiSeq 2000 platform (San Diego, CA) with Roche NimbleGen SeqCap EZ Exome Library 2.0 (Roche, Basel, Switzerland). The probes cover a total size of 44.1 Mb, comprising the whole human exome and more than 20,000 genes. A variant calling pipeline10 version 2.0 for quality control, short read alignment, variant identification, and annotation were used for the primary analysis of sequence data. Paired-end reads were aligned to the GRCh37/Hg19 human genome build. An algorithm10 developed at the Institute for Molecular Medicine Finland was used for single-nucleotide variant calling, and Pindel11 was used for insertion or deletion calling. The minimum acceptable read depth was 7, and the required quality score was 20 for single-nucleotide variants and 50 for insertions or deletions. The coverage depth was 20. After filtering out low-quality variations as well as common and homozygous variants, the individual Excel (Microsoft, Redmond, WA) files were analyzed by Microsoft Access software to detect variants shared by all 4 family members. As an alternative to the scheme shown in Figure 1B with all 4 colorectal cancer patients expected to be carriers of the same mutation, we also analyzed the data allowing for any one of the individuals to be a possible noncarrier (phenocopy). The latter method showed no additional noteworthy candidates to be considered as susceptibility genes.
Exclusion of Inhibitor of κ Light Polypeptide Gene Enhancer in B Cells Kinase β as a Likely Susceptibility Gene for F56
The c.1933C>T (RefSeq NM_001556.2) alteration in inhibitor of κ light polypeptide gene enhancer in B cells kinase β that was shared by all 4 affected family members besides the RPS20 mutation (Figure 1B) constitutes a missense change in a gene encoding an inhibitor of the κ light polypeptide gene enhancer in B cells. Inhibitor of κ light polypeptide gene enhancer in B cells kinase β is located on 8p11.2 in an interval flanking the linked region, close to marker D8S255. The alteration results in an arginine to tryptophan substitution (p.R645W) and is suggested to be damaging by SIFT (Rockville, MD) and a polymorphism by MutationTaster (Berlin, Germany). Segregation analysis in the entire family 56 showed that the variant was absent in 1 member affected with a colorectal tumor (Figure 1A, IV:1) and present in the remaining colorectal cancer patients; in addition, it was present in 1 clinically unaffected member (Figure 1A, III:12) and absent in the remaining individuals not affected with colorectal cancer. The variant occurred in healthy controls with an allele count of 0.2% (1 of 450). Finally, inhibitor of κ light polypeptide gene enhancer in B cells kinase β has an established disease connection, with homozygous germline inactivation underlying severe combined immunodeficiency.12
Sanger Sequencing of RPS20
RPS20 was screened for point mutations by exon-specific sequencing with intronic primers from Sjoblom et al13 and for large rearrangements by long-range genomic polymerase chain reaction with primers 5’-ATTTTTGGTCCGCACGCTCCT-3’ (from the 5’ untranslated region) and 5’-CACTCTAAGATACCCATATATTCCACC-3’ (from the 3’ untranslated region). The c.147dupA mutation in exon 3 (Supplementary Figure 2A) was detected with primers from flanking introns (forward primer: 5’-CTCGTTAATGTTAGTGTAGAAGGTG-3’, reverse primer: 5’-GAACCTGAATTTAGTCAACATC-3’).
Analysis of Loss of Heterozygosity in Tumor Tissue
Fluorescent fragment analysis was performed as described,14 taking advantage of 7 microsatellite markers used for haplotype and linkage analysis (from pter to qter: D8S255, D8S1828, D8S507, D8S260, D8S543, D8S1805, and D8S270). Microsatellite marker D8S507 (Supplementary Figure 2B) was investigated with forward primer 5’- CCCCTATTCCTTCTGCCTTT-3’, and reverse primer 5’-CAGCATTTTTCCTCAGAGCAG-3’. Loss of heterozygosity analysis with the germline mutation (RPS20 c.147dupA) as an intragenic marker (Supplementary Figure 2B) used forward primer 5’-GCAGAAGTGAAGGCCTTAA-3’ and reverse primer 5’-GAACCTGAATTTATGCAACATC-3’.
Analysis of the Effects of RPS20 Mutation on Pre-rRNA Processing by Northern Blot
Total RNAs were extracted by the TRIzol (Grand Island, NY) method (modified from Chomczynski15). Northern blot analysis of pre-rRNA species with probes hybridizing to the internal-transcribed spacer 1 or 2 was performed as described.16 HeLa cells treated for 48 hours with a scramble small interfering RNA or a small interfering RNA targeting RPS20 messenger RNA (5’-GGUGGCAAUUCACCGAAUUdTdT-3’) (Eurogentec, Liege, Belgium) and lymphoblastoid cells of individuals with or without the RPS20 c.147dupA mutation were included in the analysis. Precursor rRNAs were detected with a 5’ internal-transcribed spacer 1 probe, which is complementary to the junction of the 18S and the internal-transcribed spacer 1. Mature rRNAs were shown with 18S and 28S probes. Each pre-rRNA species was measured by phosphorimaging, quantified using MultiGauge (Tokyo, Japan) software, and normalized to 28S rRNA. For each species, the value of the mean of the 3 control samples arbitrarily was set to 1. RNA analyses were performed 3 times and led to similar results.
Analysis of Ribosomes by Sucrose Density Gradient Centrifugation
Polysome analysis was conducted by sucrose density gradient centrifugation of cytoplasmic fractions of cycloheximide-treated cells as described in detail previously.16
Western Blot Analysis of p53 Protein
Total proteins were extracted from lymphoblastoid cells, separated by electrophoresis on Novex (Carlsbad, CA) NuPAGE 4%–12% Bis-Tris Gels (1.0 mm), blotted on nitrocellulose membranes, hybridized with the primary and secondary antibodies, visualized with the Amersham (Buckinghamshire, UK) ECL Prime Western Blotting Detection reagents, and scanned with the CCE camera. P53 rabbit polyclonal antibody (9282; Cell Signaling Technology, Danvers, MA) with 1:1000 dilution was used as the primary antibody to detect p53 protein. The housekeeping protein glyceraldehyde-3-phosphate dehydrogenase served as a loading control and was detected with rabbit polyclonal antibody (1:1000 dilution, FL-335; Santa Cruz Biotechnology, Dallas, TX).
URL Addresses for Web Resources Used
The following online resources were used: Online Mendelian Inheritance in Man (OMIM), available at: http://www.ncbi.nlm.nih.gov/Omim; Entrez Gene, available at: http://www.ncbi.nlm.nih.gov/gene; Single Nucleotide Polymorphism database, available at: http://www.ncbi.nlm.nih.gov/SNP; 1000 Genomes database, available at: http://www.1000genomes.org; Sorting Intolerant from Tolerant (SIFT), available at: http://sift.bii.a-star.edu.sg/; MutationTaster, available at: http://www.mutationtaster.org/; Catalogue of Somatic Mutations in Cancer (COSMIC), available at: http://cancer.sanger.ac.uk; The Cancer Genome Atlas (TCGA), available at: http://cancergenome.nih.gov/; GeneCards, available at: http://www.genecards.org; and Exome Variant Server (EVS), available at: http://evs.gs.washington.edu/EVS/.
Author names in bold designate shared co-first authorship.
Supplementary Figure 1.
Results from genetic linkage analysis for chromosome 8 in F56, based on multipoint analysis with the program MERLIN v1.0-alpha (Supplementary Materials and Methods). The area between positions 61.133 and 79.265 on the Haldane marker map (interpolated from DeCode genetic map provided by Affymetrix) assuming no interference yielded lod scores over 1 (note that linkage analysis was based on a more concise pedigree compared to that shown in Fig. 1A, reflecting sample availability of the time).
Supplementary Figure 2.
Analysis of blood and tumor DNAs for the first (A) and second “hits” (B). A. Sequencing of RPS20 exon 3 with primers from flanking introns shows a duplication of A (c.147dupA) in blood DNA from affected individuals from family 56 (A5 given as an example). The duplication is marked with an arrow and highlighted in bold in the mutant sequence. B. Fluorescent fragment analysis of normal (N) and tumor (T) DNA from three affected family members (A1, A5, and A6) shows retention of heterozygosity at the flanking microsatellite marker locus D8S507 (located 2 Mb downstream of RPS20) in tumor DNA (left panel). The 117-bp fragment corresponds to the linked allele shared by all three individuals, whereas the 119-bp and 121-bp fragments represent the wild type allele. A similar analysis with the germline mutation (RPS20 c.147dupA) as an intragenic marker confirms the absence of LOH (right panel). The 205-bp fragment is derived from the wild-type allele and the 206-bp fragment from the mutant allele with the 1-bp insertion.
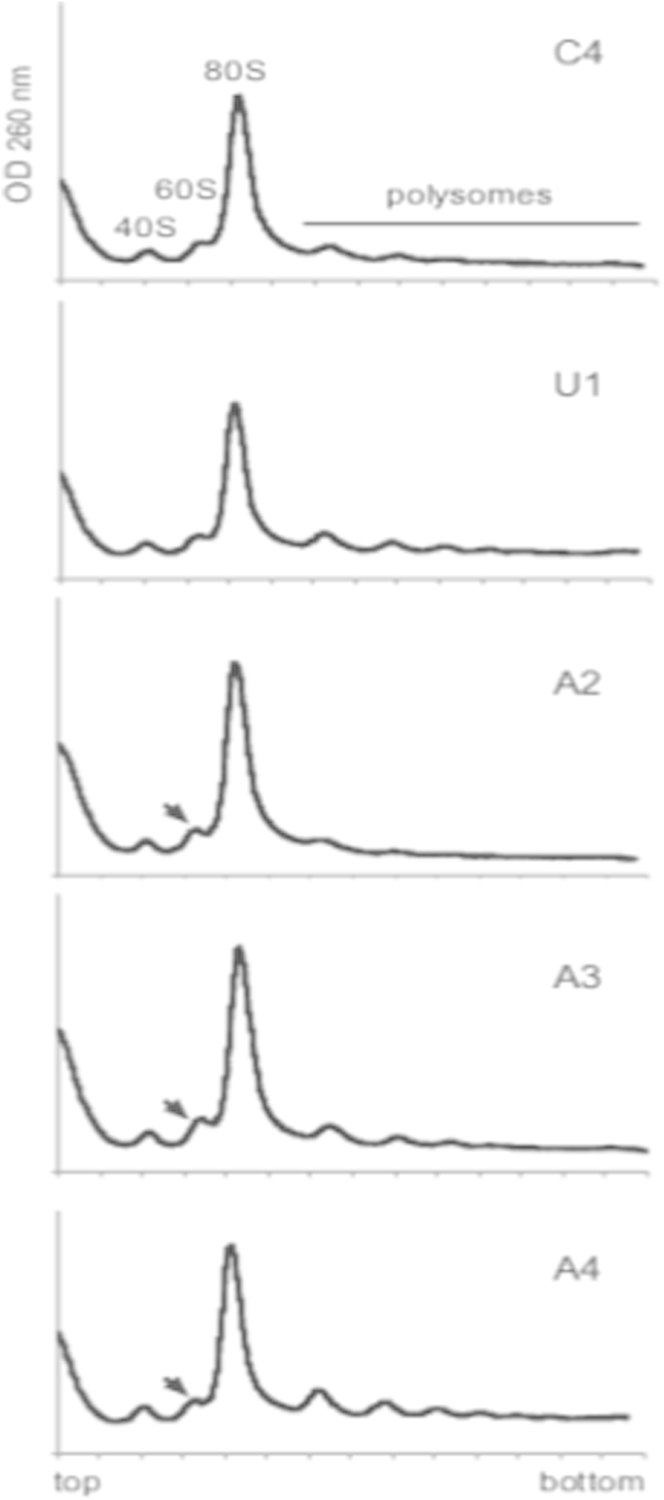
Supplementary Figure 3.
Analysis on sucrose gradient of cytoplasmic ribosomes isolated from control (C4), non-carrier (U1) or patient (A2-A4) lymphoblastoid cells. The profiles display well-separated 40S and 60S peaks, a high contribution of 80S peak, and polysomes. Arrowheads indicate a slight increase of the 60S peak in patient samples. The alteration is analogous to that seen in HeLa cells treated with RPS20 siRNA although less remarkable, in part reflecting a prominent 80S peak that may mask 60S changes in lymphoblastoid cells.
Supplementary Figure 4.

Western blot analysis of p53 protein expression in lymphoblastoid cells of two mutation carriers (A3 and A4), a proven non-carrier (U1) and a healthy unrelated control (C4). The housekeeping protein GAPDH was used as a loading control. The immunoblot shows an increased level of p53 in mutation carriers compared to individuals with no RPS20 mutation. The finding is analogous to the observation of elevated p53 dosage in RPL26 mutation carriers in DBA (ref. 10 in the main paper).
Supplementary Table 1.
Clinical Diagnoses of Members From F56 Tested for RPS20 c. 147dupA
| Individual ID | Mutation carrier status | Tumor diagnosis (age at diagnosis in years) |
|---|---|---|
| II:2 | Carrier | Carcinoma of sigmoid colon (75) |
| III:1 | Carrier | Carcinoma of ascending colon (24), carcinoma of transverse colon (60) |
| III:2 | Obligatory carrier | Carcinoma of transverse colon (52) |
| III:4 | Carrier | Carcinoma of ascending colon (64) |
| III:7 | Noncarrier | Carcinoma of breast (55) |
| III:8 | Carrier | Carcinoma of cecum (50), carcinoma of rectum (59) |
| III:10 | Carrier | Carcinoma of sigmoid colon (43), carcinoma of rectum (45) |
| III:12 | Noncarrier | – |
| III:15 | Noncarrier | – |
| III:17 | Noncarrier | Hyperplastic polyp of cecum (47), tubular adenoma of ascending colon (53) |
| III:18 | Carrier | Carcinoma of descending colon (54) |
| III:20 | Noncarrier | – |
| IV:1 | Carrier | Carcinoid tumor of rectum (33) |
References
- 1.Vasen H.F. Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 2.Vasen H.F. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 3.Thompson B.A. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindor N.M. Surg Oncol Clin N Am. 2009;18:637–645. doi: 10.1016/j.soc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieminen T.T. Gastroenterology. 2011;141:e23–e26. doi: 10.1053/j.gastro.2011.03.063. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knudson A.G., Jr. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amsterdam A. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donohue M.F., Choesmel V. J Cell Biol. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazda H.T. Hum Mutat. 2012;33:1037–1044. doi: 10.1002/humu.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landowski M. Hum Genet. 2013;132:1265–1274. doi: 10.1007/s00439-013-1326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGowan K.A. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daftuar L. PLoS One. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Vasen H.F. Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 2.Vasen H.F. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 3.Umar A. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renkonen E. J Clin Oncol. 2003;21:3629–3637. doi: 10.1200/JCO.2003.03.181. [DOI] [PubMed] [Google Scholar]
- 5.Nieminen T.T. Gastroenterology. 2011;141:e23–e26. doi: 10.1053/j.gastro.2011.03.063. [DOI] [PubMed] [Google Scholar]
- 6.Kuismanen S.A., Holmberg M.T. Am J Pathol. 2000;156:1773–1779. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J. Cancer Res. 2001;61:1619–1623. [PubMed] [Google Scholar]
- 8.Abecasis G.R. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 9.Peltomäki P., Aaltonen L.A. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 10.Sulonen A.M. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye K. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannicke U., Baumann B., Fuchs S., Henneke P. N Engl J Med. 2013;369:2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 13.Sjoblom T., Jones S., Wood L., Parsons D.W. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Rahman W.M. Oncogene. 2005;24:1542–1551. doi: 10.1038/sj.onc.1208387. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P. Biotechniques. 1993;15:532–534. 536–537. [PubMed] [Google Scholar]
- 16.O'Donohue M.F., Choesmel V. J Cell Biol. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]