Abstract
Aim:
The non-neuronal acetylcholine system (NNAS) in endothelial cells participates in modulating endothelial function, vascular tone, angiogenesis and inflammation, thus plays a critical role in cardiovascular diseases. In this study, we used a proteomic approach to study potential downstream receptor-effectors of NNAS that were involved in regulating cellular function in endothelial cells.
Methods:
Human umbilical vein endothelial cells were incubated in the presence of acetylcholine, oxotremorine, pilocarpine or nicotine at the concentration of 10 μmol/L for 12 h, and the expressed proteins in the cells were separated and identified with two-dimensional electrophoresis (2-DE) and LC-MS. The protein spots with the largest changes were identified by LC-MS. Biowork software was used for database search of the peptide mass fingerprints.
Results:
Over 1200 polypeptides were reproducibly detected in 2-DE with a pH range of 3–10. Acetylcholine, oxotremorine, pilocarpine and nicotine treatment caused 16, 9, 8 and 9 protein spots, respectively, expressed differentially. Four protein spots were identified as destrin, FK506 binding protein 1A (FKBP1A), macrophage migration inhibitory factor (MIF) and profilin-1. Western blotting analyses showed that treatment of the cells with cholinergic agonists significantly decreased the expression of destrin, FKBP1A and MIF, and increased the expression of profilin-1.
Conclusion:
A set of proteins differentially expressed in endothelial cells in response to cholinergic agonists may have important implications for the downstream biological effects of NNAS.
Keywords: non-neuronal acetylcholine system, endothelial cell, cholinergic agonist, destrin, FK506 binding protein 1A, macrophage migration inhibitory factor, profilin-1, proteomics, cardiovascular disease
Introduction
Endothelial cells play a critical role in regulating vascular tone, angiogenesis, and vascular intimal inflammation1. Endothelial dysfunction is commonly observed in patients with atherosclerosis, hypertension, chronic heart failure, and other conditions, and has been recognized as a driving force in the development of cardiovascular diseases2,3. Targeting the endothelium to maintain endothelial function is considered to be a new strategy for prevention and therapy for several cardiovascular diseases4,5,6. Recently, evidence has indicated that the non-neuronal acetylcholine system (NNAS) described in endothelial cells participates in modulating the production and release of endothelial autacoids, eg, nitric oxide (NO) and prostaglandins, subsequently affecting cell function7,8. It has been shown that there are also two major types of cholinergic receptors in endothelial NNAs: the non-neuronal muscarinic receptors (NNMR) and non-neuronal nicotinic receptors (NNNR)9. Acetylcholine, an endogenous agonist of both muscarinic receptors and nicotinic receptors, can induce endothelium-dependent vascular relaxation by enhancing the bioavailability of NO10. Despite that the so-called “endothelium-dependent vascular relaxation by acetylcholine” is used as a golden standard to measure endothelial function, the underlying physiological and/or pathophysiological role of NNAs in endothelial cells is still not fully understood. For example, it was well documented that stimulation of endothelial NNNRs with nicotine can induce endothelial dysfunction, vascular intimal inflammation, and accelerate atherosclerosis11. In contrast, treatment of the cholinergic system with acetylcholinesterase inhibitors, where the increased availability of acetylcholine stimulates two types of cholinergic receptors, significantly attenuates atherogenesis12. Therefore, it is unclear whether stimulation of endothelial NNAs is atherogenic or atheroprotective. Our previous studies showed that pharmacological characteristics of NNMRs were different from those of neuronal muscarinic receptors13. Pilocarpine, an agonist of nonselective neuronal muscarinic receptors, could not induce endothelium-dependent vascular relaxation14. However, another commonly studied muscarinic receptor agonist, arecoline, was able to stimulate NNMRs to produce endothelium-dependent vascular relaxation and to inhibit the development of atherosclerosis. Moreover, arecoline afforded potent protection against endothelial dysfunction induced by multiple atherogenic risk factors that contain high D-glucose and homocysteine15,16,17. The intracellular mechanism after activation of NNMRs that regulates signaling events to oppose risk factors that induce endothelial dysfunction is largely unknown. Therefore, it is essential to uncover the biological effectors after stimulation of NNAs in endothelial cells.
Proteomics is a discovery tool that allows previously unrecognized associations to be identified. It is a useful tool for studying the signal transduction mechanisms of cells and the effects of new drugs on cell function. In the present study, using two-dimensional gel electrophoresis (2-DE) and liquid chromatography/mass spectrometry (LC-MS)18,19, we analyzed and identified the protein changes before and after stimulation with four different cholinergic agonists (acetylcholine bromide, oxotremorine, pilocarpine and nicotine) in cultured human umbilical vein endothelial cells (HUVECs). The four drugs used to treat the cells represent different agonists corresponding to different types of acetylcholine receptors (Table 1). Acetylcholine is able to activate the non-neuronal nicotinic and muscarinic receptors; oxotremorine can activate the non-neuronal muscarinic receptors but not non-neuronal nicotinic receptors; pilocarpine does not have the ability to activate non-neuronal nicotinic or muscarinic receptors; nicotine is another acetylcholine receptor agonist that activates only non-neuronal nicotinic receptors. We found sixteen, eight, eight and nine proteins changed after the stimulation of endothelial cells by acetylcholine, oxotremorine, pilocarpine and nicotine. In all of these proteins, Destrin, FKBP1A, MIF and Profilin-1 may have important roles in mediating downstream biological effects after stimulation of NNAs in endothelial cells. These data will contribute to a better understanding of the signaling events that are relevant to NNAs activation.
Table 1. Summary of the different types of acetylcholine receptors of the four different agonists.
| Drug | NNMR | NNNR |
|---|---|---|
| ACh | + | + |
| Oxo | + | − |
| Pilo | − | − |
| Nico | − | + |
ACh, Acetylcholine; Oxo, Oxotremorine; Pilo, Pilocarpine; Nico, Nicotine; NNMR, non-neuronal muscarinic receptor; NNNR, non-neuronal nicotinic receptor. +, activation; −, no activation.
Materials and methods
Culture of human umbilical vein cells
HUVECs were obtained from Cascade Biologics (Portland, OR, USA). Cells from the third to fourth passage were used for experiments. HUVECs were cultured in PMRI-1640 supplemented with 10% FBS and in a water-saturated atmosphere with 5% CO2 at 37 °C in a 75-cm2 polystyrene culture flask. Full confluent HUVECs were digested with 0.25% trypsin for passage.
Cell treatment and protein sample preparation
Four cholinergic agonists, acetylcholine bromide (ACh), oxotremorine methiodide (Oxo), pilocarpine nitrate salt (Pilo) and (−)-nicotine hydrogen tartrate salt (Nico) were all purchased from Sigma-Aldrich (St Louis, MO, USA). The cells were incubated with ACh, Oxo, Pilo, and Nico at the concentration of 10 μmol/L for 12 h. After treatment, the cells were digested with trypsin and washed three times with buffer solution (20 mmol/L Tris-HCl, pH 7.5, 250 mmol/L mannitol). After centrifugation at 1000×g at 4 °C for 2 min, the cells were resuspended in sample lysis buffer for 1 h with shaking. Then, the supernatant was obtained after centrifugation at 10 000×g at 4 °C for 1 h and directly applied to a 180-mm immobilized pH gradient (IPG) strip for isoelectric focusing (IEF). Protein concentration was measured with a BCA protein assay kit (Pierce, Rockford, IL, USA), and the samples were stored at -70 °C until further use.
Two-dimensional gel electrophoresis
For IEF, 300 μg of protein was mixed with a rehydration buffer containing 7 mol/L urea, 2 mol/L thiourea, 2% (w/v) CHAPS, 0.3% (w/v) DTT, and 0.5% (v/v) IPG buffer up to 450 μL total volume. This mixture was used to rehydrate the immobilized 24 cm, linear Ready-StripTM IPG Strips (pH 3–10, GE Healthcare, Piscataway, NJ, USA) for 12 h with a constant voltage (30 V) applied across the gel strips, which were placed in the IPGphor focusing tray. The rehydrated IPG gel strips were electrophoresed at 200 V for 1 h, 500 V for 1 h, subjected to a linear voltage ramp from 1000 to 10 000 V for 1 h, and finally rapidly ramped up from 10 000 V to a total of 60 000 kVh. The maximum current was 50 μA per gel strip.
After IEF, the IPG strips were placed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) equilibration buffer containing 6 mol/L Urea, 75 mmol/L Tris-HCl (pH 8.8), 30% glycerol, 2% SDS, 1% DTT for 15 min. This procedure was repeated with SDS-PAGE equilibration buffer with 2.5% (w/v) iodoacetamide for an additional 15 min. After equilibration, the IPG gel was transferred onto a 10% polyacrylamide gel and the second-dimension of electrophoresis (SDS-PAGE) was performed in the Ettan DALT six (GE Healthcare, USA), and performed at 1 W per gel for 1 h, followed by 15 W per gel for 5 h. Finally, silver staining of gels was performed as described previously20. Briefly, the gels were fixed for 30 min with 50% methanol containing 5% acetic acid. After several alternating washes of 50% methanol and water, the gels were sensitized by incubation in 0.2% sodium thiosulfate followed by several washes with water. The gels were incubated in 0.2% silver nitrate for 20 min followed by thorough rinsing with water. The gels were developed in 0.04% formalin in 2% sodium carbonate and the reaction was terminated with 5% acetic acid. For mass spectrometry-based identification, 1.5 mg of protein was resolved by following the protocol used for large analytical gels, and the gels were then fixed with 50% ethanol containing 3% phosphoric acid for 30 min and stained with 0.02% Coomassie Brilliant Blue (CBB)-G250, 3% phosphoric acid, 17% ammonium sulfate, and 34% ethanol. Relative molecular weights were determined by simultaneously running standard protein markers (PageRuler™ Prestained Protein Ladder Plus, #SM0671, Fermentas). The isoelectric point (PI) values used were those given by the supplier of the immobilized pH gradient strips.
Gel imaging and analysis
Gels were digitally scanned using a LabScan scanner. Mell files of the gel images were transferred to a Sun Workstation for analysis with ImageMaster™ 2D Platinum software. Polypeptide spots in each gel image were delineated and the total intensity of pixels within each spot (the integrated intensity) was determined by the software. The integrated intensity of each spot was then expressed as a percent of the total integrated intensity over all spots within the region of analysis of the gel. This normalizes the amount of any given polypeptide spot to the largest possible number of polypeptides, and reports abundance as a relative value within each sample. The 2-DE analysis was performed for four replicates.
Liquid chromatography-mass spectrometry (LC-MS) and database search
Spots from the 2-DE gel were excised into approximately 1-mm3 cubes, transferred into a microcentrifuge tube (Axygen, USA) and spun down on a bench-top microcentrifuge. The gel plugs were destained with 100 μL of 100 mmol/L ammonium bicarbonate/acetonitrile [1:1 (v/v)] and incubated with occasional vortexing for 30 min. After destaining, 100 μL of neat acetonitrile was added and incubated at room temperature with occasional vortexing until the gel pieces became white and shrunken. Then, gel pieces were reduced with 20 mmol/L DTT at 56 °C for 30 min and dehydrated with neat acetonitrile once before treatment with 55 mmol/L iodoacetamide in the dark for 20 min at room temperature. The gel pieces were again dehydrated with neat acetonitrile and enough trypsin buffer [approx 50 μL, 13 ng/μL trypsin in 25 mmol/L ammonium bicarbonate containing 10% (v/v) acetonitrile] was added to cover the dry pieces, allowing the buffer to be absorbed into the gel pieces at 4 °C. The trypsin buffer was replenished if the buffer was completely absorbed into the gel to keep the gel pieces from drying. The tubes containing trypsin buffer were then placed into an air circulation thermostat and incubated overnight at 37 °C. When the digestion was completed, the gel pieces were extracted with 100 μL of extraction buffer [1:2 (v/v) of 5% formic acid/acetonitrile] for 15 min at 37 °C in a shaker. The volume of extracted solution was reduced in a SpeedVac (Thermo Fisher, USA) to approximately 30 μL. The extracted solution of tryptic peptides from each of the gel pieces was analyzed by a LCQ Deca XP mass spectrometer (Thermo Electron Corp, CA, USA). Spectra of a survey scan followed by 5 CID events were recorded by mass spectrometry. The “peak list” was created automatically by the EXTRACTMS program provided in the Xcalibur software package (Bioworks v3.3). The MS2 spectra with a total ion current (TIC) higher than 104 were used to search for matches against a maize protein sequence database ipi.HUMAN.v3.66.fasta (86867 entries) using the TurboSEQUEST algorithm. The search parameters were based on the TurboSEQUEST algorithm. The degree of completeness of the b- and y-ion series for each TurboSEQUEST result was manually checked for every protein identified.
Western blotting analysis
HUVECs cells treated with the four cholinergic agonists as described above were collected, and the proteins were extracted with a sample lysis buffer that includes a protease inhibitor cocktail (Roche, Germany). Protein concentration was determined with a BCA protein assay kit (Pierce, USA). Individual samples containing 20 μg of protein from the control and each agonist-treated HUVECs cells were loaded onto a 12% SDS-PAGE gel and separated by electrophoresis, and then electrophoretically transferred onto pieces of PVDF membranes for Western blotting. The membranes were blocked with 3% non-fat milk powder in TBST buffer (pH 7.6) composed of 150 mmol/L NaCl; 10 mmol/L Tris-HCl and 0.1% Tween-20 for 2 h at room temperature. Then, the membranes were incubated with specific primary antibodies against Destrin (Abcam, UK; ab95298, 1:500 dilution), FKBP1A (Thermo Scientific, USA; 1:200 dilution), Profilin-1 (Cell Signaling Technology, USA; 1:1000 dilution), or MIF (Abcam, UK; abab7207, 1:2000 dilution), in TBST (0.1% Tween-20) containing 5% nonfat milk powder overnight at 4 °C, or 1 h at room temperature. Then, membranes were washed three times with TBST buffer (0.1% Tween-20) for 5 min each and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at a dilution of 1:10 000 for 1 h at room temperature. After washing with TBST buffer, the chemiluminescence reaction in which HRP reacts with SuperSignal® West Pico Chemiluminescent substrate (Pierce, USA) was performed to detect signals. At the same time, GAPDH was used as a loading control to verify the amount and integrity of the whole proteins.
Results
Two-dimensional protein maps of endothelial cells stimulated by cholinergic agonists
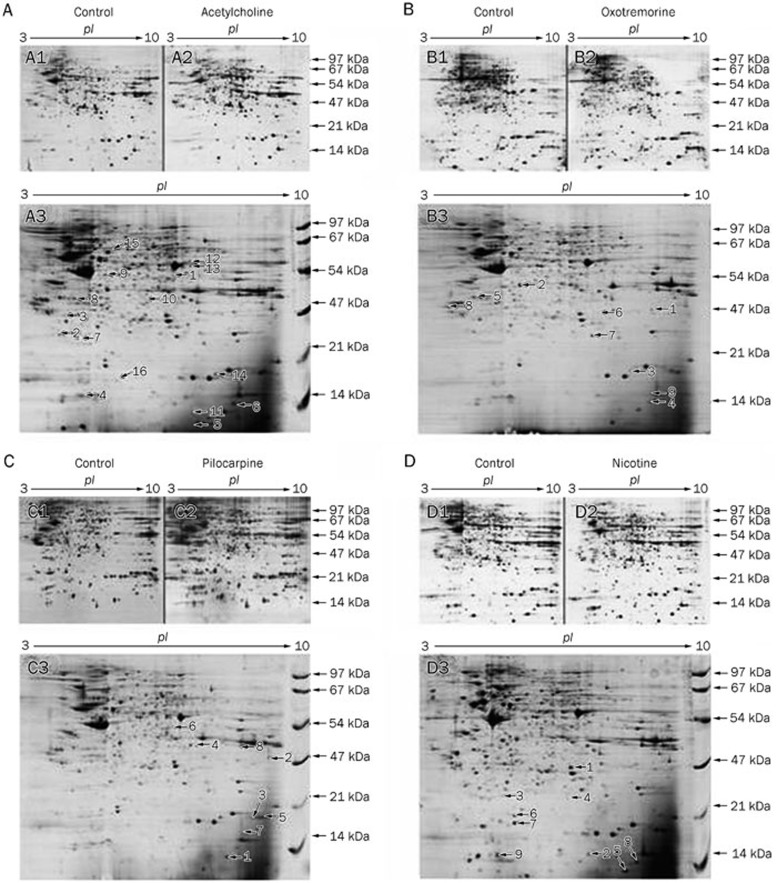
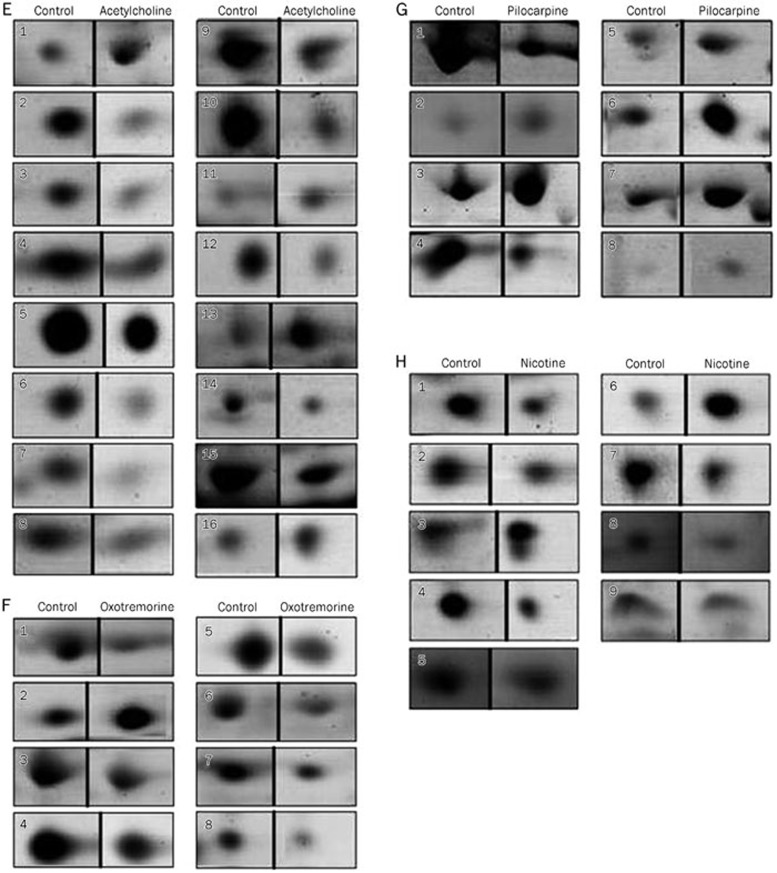
In this work, we established the 2-DE protein patterns of the control group and the endothelial cells activated by ACh, Oxo, Pilo, and Nico. More than 1200 protein spots were detected on the 2-DE gels that showed similar protein patterns with silver staining of 300 μg of total proteins. Figure 1 displays the overall 2-DE patterns of protein extracts of control/ACh (Figure 1A), control/Oxo (Figure 1B), control/Pilo (Figure 1C), and control/Nico (Figure 1D). Based on analysis with Image Master 2D Platinum software, among all the detected spots, 16, 8, 8 and 9 spots were separately changed in cells treated with ACh, Oxo, Pilo and Nico, showing detectable changes above 2-fold and the match rate was above 60% (Figure 1E–1H).
Figure 1A–1D.
Two-dimensional gel showing the proteome of cultured HUVECs at a pH range of 3–10 after stimulation with cholinergic agonists. The cells were incubated with different agonists at pharmacological concentration of 10 μmol/L for 12 h. Positions of the differentially expressed protein spots with corresponding spot numbers are indicated with arrows. These results are representative of at least four independent experiments. A1/A2: before/after acetylcholine treatment, protein maps were stained with silver; A3: differentially expressed spots between the control and acetylcholine group, spots in A3 were visualized with Coomassie Brilliant Blue staining. The expression-changed proteins with spot numbers were identified and they are listed in Table 2. B1/B2: before/after oxotremorine treatment, silver staining; B3: differentially expressed protein spots between control and oxotremorine, Coomassie Brilliant Blue staining. The expression-changed proteins with the spot numbers are listed in Table 3. C1/C2: before/after pilocarpine treatment, silver staining; C3: differentially expressed protein spots between control and pilocarpine, Coomassie Brilliant Blue staining. The protein expression changes with the spot numbers were listed in Table 4. D1/D2: before/after nicotine treatment, silver staining; D3: differentially expressed protein spots between control and nicotine, Coomassie Brilliant Blue staining. The protein expression changes with the spot numbers are listed in Table 5. (E–H) show magnified views of some differentially expressed proteins between the control and each acetylcholine receptor agonist treatment group. (E) differentially expressed proteins between the control and acetylcholine treatment group. (F) between the control and oxotremorine treatment group. (G) between the control and pilocarpine treatment group. (H) between the control and nicotine treatment group.
Figure 1E–1H.
Two-dimensional gel showing the proteome of cultured HUVECs at a pH range of 3–10 after stimulation with cholinergic agonists. The cells were incubated with different agonists at pharmacological concentration of 10 μmol/L for 12 h. Positions of the differentially expressed protein spots with corresponding spot numbers are indicated with arrows. These results are representative of at least four independent experiments. A1/A2: before/after acetylcholine treatment, protein maps were stained with silver; A3: differentially expressed spots between the control and acetylcholine group, spots in A3 were visualized with Coomassie Brilliant Blue staining. The expression-changed proteins with spot numbers were identified and they are listed in Table 2. B1/B2: before/after oxotremorine treatment, silver staining; B3: differentially expressed protein spots between control and oxotremorine, Coomassie Brilliant Blue staining. The expression-changed proteins with the spot numbers are listed in Table 3. C1/C2: before/after pilocarpine treatment, silver staining; C3: differentially expressed protein spots between control and pilocarpine, Coomassie Brilliant Blue staining. The protein expression changes with the spot numbers were listed in Table 4. D1/D2: before/after nicotine treatment, silver staining; D3: differentially expressed protein spots between control and nicotine, Coomassie Brilliant Blue staining. The protein expression changes with the spot numbers are listed in Table 5. (E–H) show magnified views of some differentially expressed proteins between the control and each acetylcholine receptor agonist treatment group. (E) differentially expressed proteins between the control and acetylcholine treatment group. (F) between the control and oxotremorine treatment group. (G) between the control and pilocarpine treatment group. (H) between the control and nicotine treatment group.
Identification of differentially expressed proteins by LC-MS and database search
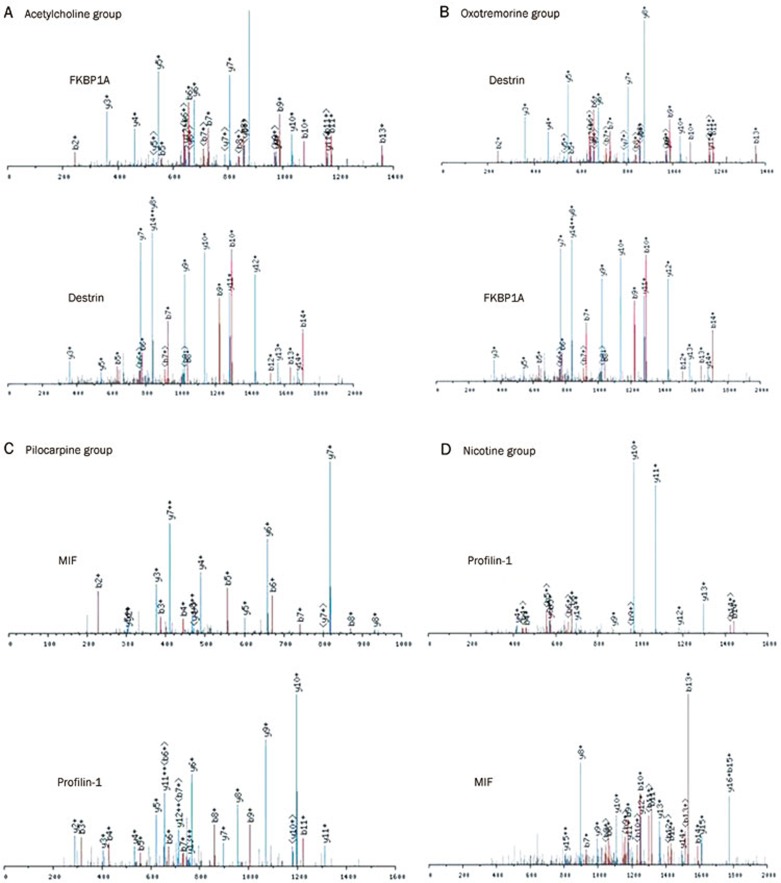
To further identify these differentially expressed proteins, while keeping the silver-staining gels as a reference, we excised the differentially expressed proteins from hot Coomassie blue stained 2-DE gels and digested them with trypsin. These markedly changed proteins were successfully identified by LC-MS. Several peptide mass fingerprints (PMF) were successfully obtained. A few of the selected PMFs are displayed in Figure 2. Database searches of all the PMFs were performed with Bioworks to identify the protein spots. The identified proteins in the ACh, Oxo, Pilo and Nico group are listed separately in Table 2,3,4,5.
Figure 2.
Peptide mass fingerprints of a few of selected protein spots obtained from MALDI-TOF-MS. (A) protein spot 6 (FKBP1A) and 14 (Destrin) from Figure 1A3. (B) protein spot 3 (Destrin) and 4 (FKBP1A) from Figure 1B3. (C) protein spot 1 (MIF) and 7 (Profilin-1) from Figure 1C3. (D) protein spot 1 (Profilin-1) and 4 (MIF) from Figure 1D3.
Table 2. List of differentially expressed proteins found in the most significant network 12 h after acetylcholine treatment using Ingenuity Pathway Analysis (Figure 1A1–1A3). The proteins were identified by 2-DE and LC-MS. The spots identified by 2-DE are indicated with a corresponding number on the gel (↑, the tprotein up-regulated; ↓, the protein down-regulated).
| Spot No | Protein name | Gene names | Accession | Mw (kDa) | pI | Change | Protein function |
|---|---|---|---|---|---|---|---|
| 1 | Aspartate aminotransferase, cytoplasmic | GOT1 | P17174 | 46116.31 | 6.57 | ↑ | Plays a role in amino acid metabolism and the urea and tricarboxylic acid cycles. |
| 2 | Tumor protein, translationally-controlled 1 | TPT1 | P13693 | 21525.66 | 5.34 | ↓ | Involved in calcium binding and microtubule stabilization. |
| 3 | Calpain small subunit 1 | CAPNS1 | P04632 | 28315.71 | 5.05 | ↓ | Regulatory subunit of the calcium-regulated non-lysosomal thiol-protease which catalyzes limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction. |
| 4 | Galectin-1 | LGALS1 | P09382 | 14584.51 | 5.34 | ↓ | Regulate apoptosis, cell proliferation and cell differentiation. Binds beta-galactoside and a wide array of complex carbohydrates. Inhibits CD45 protein phosphatase activity and therefore the dephosphorylation of Lyn kinase. |
| 5 | Protein S100-A10 | S100A10 | P60903 | 11071.91 | 7.3 | ↓ | A regulator of protein phosphorylation in that the ANXA2 monomer is the preferred target (in vitro) of tyrosine-specific kinase. |
| 6 | FKBP1A protein | FKBP1A | P62942 | 15688.97 | 9.26 | ↓ | Plays a role in modulation of ryanodine receptor isoform-1 (RYR-1); catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. |
| 7 | Lactoylglutathione lyase | GLO1 | Q04760 | 20646.52 | 5.12 | ↓ | Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. |
| 8 | Microtubule-associated protein | MAPRE1 | Q15691 | 29867.89 | 5.02 | ↓ | Involved in microtubule polymerization, and spindle function by stabilizing microtubules and anchoring them at centrosomes. May play a role in cell migration. RP/EB family member 1 |
| 9 | Selenide, water dikinase 1 | SEPHS1 | P49903 | 42910.58 | 5.65 | ↓ | Synthesizes selenophosphate from selenide and ATP. |
| 10 | Phosphatidylinositol transfer protein (PITP) alpha isoform | PITPNA | Q00169 | 31675.13 | 6.13 | ↓ | Catalyzes the transfer of PtdIns and phosphatidylcholine between membranes. |
| 11 | D-dopachrome decarboxylase | DDT | P30046 | 12580.57 | 7.25 | ↑ | Tautomerization of D-dopachrome with decarboxylation to give 5,6-dihydroxyindole (DHI). |
| 12 | Isoform 1 of platelet-activating factor acetylhydrolase IB subunit alpha | PAFAH1B1 | P43034 | 46506.78 | 7.03 | ↓ | Required for proper activation of Rho GTPases and actin polymerization at the leading edge of locomoting cerebellar neurons and postmigratory hippocampal neurons in response to calcium influx triggered via NMDA receptors. |
| 13 | Isoform mitochondrial of fumarate hydratase, mitochondrial | FH | B1ANK7 | 45494.82 | 7.27 | ↑ | Fumarate hydratase activity; cell junction. |
| 14 | Destrin | DSTN | P60981 | 18374.53 | 8.12 | ↓ | Actin-depolymerizing protein. Severs actin filaments (F-actin) and binds to actin monomers (G-actin). Acts in a pH-independent manner. |
| 15 | Copine-1 | CPNE1 | Q99829 | 59058.79 | 5.52 | ↓ | May function in membrane trafficking. Exhibits calcium-dependent phospholipid binding properties. |
| 16 | Ubiquitin-conjugating enzyme E2N | UBE2N | P61088 | 17006.63 | 6.18 | ↑ | Plays a role in the control of progress through the cell cycle and differentiation. Plays a role in the error-free DNA repair pathway and contributes to the survival of cells after DNA damage. |
Table 3. List of differentially expressed proteins found in the most significant network 12 h after oxotremorine treatment using Ingenuity Pathway Analysis (Figure 1B1–1B3). The proteins were identified by 2-DE and LC-MS. The spots identified by 2-DE are indicated with a corresponding number on the gel (↑, the protein up-regulated; ↓, the protein down-regulated).
| Spot No | Protein name | Gene names | Accession | Mw (kDa) | pI | Change | Protein function |
|---|---|---|---|---|---|---|---|
| 1 | Isoform 1 of pyridoxal kinase | PDXK | O00764 | 35102.3 | 5.75 | ↓ | Required for synthesis of pyridoxal-5-phosphate from vitamin B6. |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | GNB1 | P62873 | 7245.77 | 5.6 | ↓ | Guanine nucleotide-binding proteins (G proteins) are involved as a modulator or transducer in various transmembrane signaling systems. | |
| 2 | Isoform 1 of calcyclin-binding protein | CACYBP | Q9HB71 | 26078.75 | 8.32 | ↑ | Involved in calcium-dependent ubiquitination and subsequent proteosomal degradation of target proteins; serves as a molecular bridge in ubiquitin E3 complexes; participates in the ubiquitin-mediated degradation of beta-catenin (CTNNB1). |
| 3 | Destrin | DSTN | P60981 | 18374.53 | 8.12 | ↓ | Actin-depolymerizing protein. Severs actin filaments (F-actin) and binds to actin monomers (G-actin). Acts in a pH-independent manner. |
| 4 | FKBP1A protein | FKBP1A | P62942 | 15688.97 | 9.26 | ↓ | Plays a role in modulation of ryanodine receptor isoform-1 (RYR-1); catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. |
| 5 | Annexin A5 | ANXA5 | P08758 | 35805.58 | 4.94 | ↓ | This protein is an anticoagulant protein that acts as an indirect inhibitor of the thromboplastin-specific complex, which is involved in the blood coagulation cascade. |
| 6 | Serine/threonine-protein kinase 38-like | STK38L | Q9Y2H1 | 53871.65 | 6.38 | ↓ | Involved in the regulation of structural processes in differentiating and mature neuronal cells. |
| 7 | ADP-ribosylation factor-like protein 3 | ARL3 | P36405 | 20324.33 | 6.94 | ↓ | Small GTP-binding protein which cycles between an inactive GDP-bound and an active GTP-bound form. |
| 8 | 14-3-3 Protein epsilon | YWHAE | P62258 | 29173.9 | 4.63 | ↓ | Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathway. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner. |
| 9 | Isoform 4 of abhydrolase domain-containing protein 11 | ABHD11 | Q8NFV4 | 34689.95 | 9.5 | ↓ | Located in the Williams-Beuren syndrome (WBS) critical region; arise as a consequence of unequal crossing over between highly homologous low-copy repeat sequences flanking the deleted region. |
Table 4. List of differentially expressed proteins found in the most significant network 12 h after pilocarpine treatment using Ingenuity Pathway Analysis (Figure 1C1–1C3). The proteins were identified by 2-DE and LC-MS. The spots identified by 2-DE are indicated with a corresponding number on the gel (↑, the protein up-regulated; ↓, the protein down-regulated).
| Spot No | Protein name | Gene names | Accession | Mw (kDa) | pI | Change | Protein function |
|---|---|---|---|---|---|---|---|
| 1 | Macrophage migration inhibitory factor | MIF | P14174 | 12345.11 | 8.24 | ↓ | Pro-inflammatory cytokine. Involved in the innate immune response to bacterial pathogens; as mediator in regulating the function of macrophages in host defense. Counteracts the anti-inflammatory activity of glucocorticoids. |
| 2 | NAD(P)H dehydrogenase [quinone] 1 | NQO1 | P15559 | 30867.65 | 8.91 | ↑ | Serves as a quinone reductase in connection with conjugation reactions of hydroquinons involved in detoxification pathways as well as in biosynthetic processes. |
| 3 | Isoform 1 of nucleoside diphosphate kinase B | NME2 | P22392 | 17166.84 | 8.55 | ↑ | Major role in the synthesis of nucleoside triphosphates other than ATP; negatively regulates Rho activity by interacting with AKAP13/LBC; acts as a transcriptional activator of the MYC gene; binds DNA non-specifically. Exhibits histidine protein kinase activity. |
| Mesencephalic astrocyte-derived neurotrophic factor | MANF | P55145 | 21143.7 | 9.13 | ↑ | Selectively promotes the survival of dopaminergic neurons of the ventral mid-brain; Inhibits cell proliferation and endoplasmic reticulum (ER) stress-induced cell death. | |
| 4 | Pyrroline-5-carboxylate reductase 1 isoform 2 | PYCR2 | B3KMB7 | 33340.6 | 6.3 | ↓ | Oxidation-reduction process; pyrroline-5-carboxylate reductase activity. |
| 5 | Putative RNA-binding protein 3 | RBM3 | P98179 | 17170.37 | 8.86 | ↓ | Enhances global protein synthesis at both physiological and mild hypothermic temperatures; reduces the relative abundance of microRNAs, when overexpressed. Enhances phosphoryaltion of translation initiation factors and active polysome formation. |
| Isoform 1 of nucleoside diphosphate kinase B | NME2 | P22392 | 17166.84 | 8.55 | ↓ | Major role in the synthesis of nucleoside triphosphates other than ATP; negatively regulates Rho activity by interacting with AKAP13/LBC; acts as a transcriptional activator of the MYC gene; exhibits histidine protein kinase activity. | |
| 6 | 26S proteasome non-ATPase regulatory subunit 7 | PSMD7 | P51665 | 65907.54 | 8.15 | ↑ | Acts as a regulatory subunit of the 26S proteasome which is involved in the ATP-dependent degradation of ubiquitinated proteins. |
| 7 | Profilin-1 | PFN1 | P07737 | 14923.04 | 8.47 | ↑ | Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG. |
| 8 | L-lactate dehydrogenase A-like 6B | LDHAL6B | Q9BYZ2 | 41942.92 | 8.88 | ↓ | Higher expression level in adult testis as compared to 6-week-old fetal testis. |
Table 5. List of differentially expressed proteins found in the most significant network 12 h after nicotine treatment using Ingenuity Pathway Analysis (Figure 1D1–1D3). The proteins were identified by 2-DE and LC-MS. The spots identified by 2-DE are indicated with a corresponding number on the gel (↑, the protein up-regulated; ↓, the protein down-regulated).
| Spot No | Protein name | Gene names | Accession | Mw (kDa) | pI | Change | Protein function |
|---|---|---|---|---|---|---|---|
| 1 | Profilin-1 | PFN1 | P07737 | 14923.04 | 8.47 | ↓ | Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG. |
| 2 | Adenine phosphoribosyl-transferase | APRT | P07741 | 19476.58 | 5.79 | ↓ | Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis. |
| 3 | Profilin-1 | PFN1 | P07737 | 14923.04 | 8.47 | ↑ | Same to spot1. |
| 4 | Macrophage migration inhibitory factor | MIF | P14174 | 12345.11 | 8.24 | ↓ | Pro-inflammatory cytokine. Involved in the innate immune response to bacterial pathogens; as mediator in regulating the function of macrophages in host defense. Counteracts the anti-inflammatory activity of glucocorticoids. |
| 5 | Superoxide dismutase [Cu-Zn] | SOD1 | P00441 | 15804.55 | 5.7 | ↓ | Destroys radicals which are normally produced within the cells and which are toxic to biological systems |
| 6 | Stathmin | STMN1 | P16949 | 17171.32 | 5.77 | ↑ | Involved in the regulation of the microtubule (MT) filament system by destabilizing microtubules. Prevents assembly and promotes disassembly of microtubules. |
| 7 | FKBP1A protein | FKBP1A | P62942 | 15688.97 | 9.26 | ↓ | Plays a role in modulation of ryanodine receptor isoform-1 (RYR-1); catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. |
| 8 | Adenine phosphoribosyl-transferase | APRT | P07741 | 19476.58 | 5.79 | ↓ | Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis |
| 9 | Putative RNA-binding protein 3 | RBM3 | P98179 | 17170.37 | 8.86 | ↓ | Enhances global protein synthesis at both physiological and mild hypothermic temperatures; reduces the relative abundance of microRNAs, when overexpressed. Enhances phosphoryaltion of translation initiation factors and active polysome formation. |
Bioinformatics analysis of effectors after endothelial NNAs activation
After comprehensive analysis of the effects of acetylcholine receptor agonists on endothelial cells (Table 6), we found that acetylcholine, which can activate non-neuronal nicotinic and muscarinic receptors, down-regulated Destrin expression in endothelial cells, and oxotremorine, which can activate the non-neuronal muscarinic receptors but not the non-neuronal nicotinic receptors, had a similar effect. However, neither pilocarpine, which cannot activate non-neuronal nicotinic or muscarinic receptors, nor nicotine, another acetylcholine receptor agonist that only activates non-neuronal nicotinic receptors, could induce changes to Destrin expression. These results suggested that Destrin might have a relationship to non-neuronal muscarinic receptor effects. Acetylcholine, oxotremorine, and nicotine were able to induce changes in the expression of FKBP1A, but pilocarpine could not affect it. These results suggested that FKBP1A might be related to the downstream effects of non-neuronal acetylcholine receptors. Nicotine, an agonist of non-neuronal nicotinic receptors, induced expression changes of MIF and Profilin-1 in endothelial cells. This result suggested that MIF and Profilin-1 might be the effectors of non-neuronal nicotinic receptors. However we found two spots (spot 1 and spot 3 in Table 5) that were both identified as Profilin-1, but the expression changes were in different directions. This result was unexpected and needs further validation. Pilocarpine affected the expression of MIF and Profilin-1, which was also affected by nicotine. This result suggested that there might be a new target of pilocarpine on endothelial cells. This target might be similar to the non-neuronal nicotinic receptors (Table 6).
Table 6. Analysis of endothelial non-neuronal acetycholine receptors effectors.
| Drug | Destrin | FKBP1A | MIF | Profilin-1 |
|---|---|---|---|---|
| ACh | + | + | − | − |
| Oxo | + | + | − | − |
| Pilo | − | − | + | + |
| Nico | − | + | + | + |
ACh, Acetylcholine; Oxo, Oxotremorine; Pilo, Pilocarpine; Nico, Nicotine; NNMR, non-neuronal muscarinic receptor; NNNR, non-neuronal nicotinic receptor. +, activation; −, no activation.
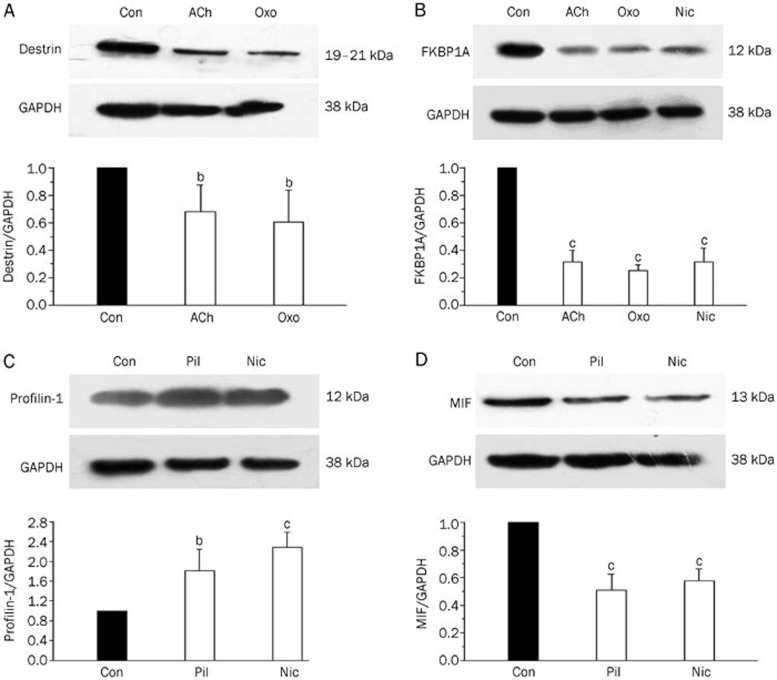
Confirmation of protein expression differences using Western blotting analysis
To corroborate the expression data derived from proteomic analysis between the control group and the acetylcholine receptor agonist treatment group, Western blotting was conducted against four proteins: Destrin, FKBP1A, MIF, and Profilin-1 (Figure 3). The Western blots showed the same trend as the proteomic analyses did. Regarding the confusing spots, the results showed that nicotine up-regulated Profilin-1 expression in endothelial cells, validating the result of spot 3 (Table 5).
Figure 3.
Western blotting was performed to corroborate the results derived from proteomic analysis. The Western blots show that Destrin decreased when the cells were treated with acetylcholine or oxotremorine (A); FKBP1A decreased when the cells were treated with acetylcholine, oxotremorine or nicotine (B); Profilin-1 increased when the cells were treated with pilocarpine or nicotine (C); MIF decreased when the cells were treated with pilocarpine or nicotine (D). GAPDH was used to normalize protein loading. Values are expressed as the mean±SD. n=3. bP<0.05, cP<0.01 vs control.
Discussion
Cardiovascular disease remains the most common cause of death in the developed world and is predicted to kill ∼20 million people worldwide each year until at least 2015 according to the World Health Organization21. Endothelial dysfunction is responsible for many cardiovascular diseases, such as atherosclerosis, hypertension, and hyperlipidemia2,3. Proteomics technologies are rapidly evolving and provide the opportunity to identify and characterize protein profiles of samples at different time and conditions. Many studies have applied proteomics technologies to endothelial cells to investigate cardiovascular disease. In this study, we investigated the effects of four different types of acetylcholine receptor agonists in endothelial cells by comparing the altered protein profiles between the control group and the treatment group that was treated with acetylcholine, oxotremorine, pilocarpine or nicotine at the pharmacological concentration of 10 μmol/L for 12 h. We identified 16, 9, 8 and 9 protein spots in the groups that were separately treated with acetylcholine, oxotremorine, pilocarpine or nicotine. According to the characteristics of the four types of agonist, we focused our attention on Destrin, FKBP1A, MIF and Profilin-1. Destrin might have a relationship with non-neuronal muscarinic receptor effects; FKBP1A might be related to non-neuronal acetylcholine receptor effects; and MIF and Profilin-1 might be the effectors of non-neuronal nicotinic receptors. Pilocarpine affected the expression of MIF, and Profilin-1 expression patterns suggested that there might be a new target of pilocarpine on endothelial cells and that it might behave similarly to non-neuronal nicotinic receptors.
Destrin is known to regulate actin dynamics by enhancing the depolymerization of F-actin into G-actin and promoting filament severing22,23. Remodeling of the actin cytoskeleton through actin dynamics is essential for numerous basic biological processes, including cell polarization, contractile force generation, cell migration, cell division, endocytosis and exocytosis24,25,26,27. In addition to its structural role, the status of the actin cytoskeleton has been shown to affect a diverse array of intracellular signaling processes. Destrin belongs to the actin-binding proteins ADF (actin-depolymerizing factor)/cofilin family, which participate in the actin-dependent vesicular trafficking of postsynaptic receptors28. Postsynaptic regulation of receptors by vesicular trafficking is an important basic mechanism of synaptic plasticity, and actin plays a key role in this process. ADF/cofilin-mediated actin dynamics regulate the vesicular trafficking of acetylcholine receptors in space and time and the targeting of acetylcholine receptors to cell surfaces of neonatal postsynaptic sites29,30. Compared with the passive diffusion mechanism, active transport mechanisms play a more important role in the vesicular trafficking of acetylcholine receptors from cluster generation to redistribution to the postsynaptic sites. In this process, ADF/cofilin may be the first to localize in generating sites to regulate the actin cytoskeleton local dynamic positioning and to assist in maintaining vesicle fusion and acetylcholine receptors in recycling. ADF/cofilin can thus more quickly transport cell signals, such as acetylcholine, into cells and play the messenger function, thereby further triggering cell polarization and migration. In this study, we found that acetylcholine and oxotremorine, agonists of non-neuronal muscarinic receptors, affected Destrin expression in endothelial cells. This is the first report that Destrin may have a relationship with non-neuronal muscarinic receptor effectors, which may be helpful in explaining the role of non-neuronal muscarinic receptors in slowing the atherosclerosis progress.
FKBP1A, also known as FKBP12, belongs to the immunophilin protein family, which plays a role in immunoregulation and basic cellular processes involving protein folding and trafficking. FKBP1A interacts with several intracellular signal transduction proteins including the type I TGF-beta receptor and also plays a role in intracellular calcium regulation by associating with three types of calcium release channel complexes, cardiac and skeletal ryanodine receptors and the inositol 1,4,5-trisphosphate receptor31,32,33. In endothelial cells, non-neuronal muscarinic receptors are G protein-coupled receptors, which bind extracellular signal molecules of this signaling pathway, activate phospholipase C (PLC-β) on the plasma membrane, cause hydrolysis from 4,5-phosphatidylinositol bisphosphate (PIP2) into 1,4,5-trisphosphate (IP3) and diacylglycerol (DG), and the conversion of extracellular signals into intracellular signals. IP3 binds to the endoplasmic reticulum IP3 ligand-gated calcium channels, which open calcium channels, and causes the increase of intracellular the Ca2+ level, and regulates the secretion of NO and vascular contraction and relaxation. In the present study, we found that acetylcholine, oxotremorine and nicotine induced expression changes of FKBP1A but pilocarpine did not affect FKBP1A expression. This result suggested that FKBP1A might be related to non-neuronal acetylcholine receptor effectors. Therefore, the non-neuronal acetylcholine system, which can induce calcium release through the IP3 pathway, and further influence the secretion of NO in endothelial cells, might be regulated by FKBP1A.
MIF (Macrophage migration inhibitory factor), encoded by the MIF gene, is a structurally unique cytokine, which is a critical mediator of acute and chronic inflammatory diseases such as atherosclerosis, septic shock, Crohn's disease and rheumatoid arthritis34,35,36,37,38. MIF plays its role in influencing the regulation of macrophages in the host defense function by suppressing the anti-inflammatory effects of glucocorticoids39. Contrary to its historically based name, MIF has been recognized to exhibit chemokine-like properties40,41 and is corroborated as a non-cognate ligand of the chemokine receptors CXCR2 and CXCR4. MIF is highly expressed in different types of macrophages and endothelial cells in atherosclerotic plaques, which suggests that it is closely associated with atherosclerosis, and functional studies established the contribution of MIF to lesion progression and plaque inflammation42. Studies have shown that non-neuronal nicotinic receptors are involved in angiogenesis during many pathological processes such as inflammation, ischemia, cancer and atherosclerosis, so that the protein is very important for the study of endothelial cells non-neuronal nicotinic receptors in inflammation and atherosclerosis. Profilin-1, which belongs to the fiber protein family, is an actin monomer binding protein which is encoded by PFN1 genes. A role for Profilin-1 in promoting actin filament formation has been suggested by several studies in lots of organisms43. Overexpression of Profilin-1 in a human endothelial cell line indicate potential roles for Profilin-1 in stabilizing dynamic actin filaments or directing filament assembly44, and its silence can inhibit endothelial cell proliferation and migration. Profilin-1 serves as an essential regulator of hypertension and atherosclerosis45,46. In the current study, we found that nicotine, an agonist of non-neuronal nicotinic receptors, induced expression changes of MIF and Profilin-1 in endothelial cells. This suggested that MIF and Profilin-1 might be the effectors of non-neuronal nicotinic receptors. Pilocarpine affected the expression of MIF and Profilin-1, which was also affected by nicotine, suggesting that there might be a new target of pilocarpine on endothelial cells. This target might be similar to non-neuronal nicotinic receptors. Therefore, the MIF and Profilin-1 signaling pathway has a very significant role in the study of the non-neuronal acetylcholine system and the discovery of new drug targets.
In conclusion, we used the 2-DE proteomic approach with highly sensitive mass spectrometry to analyze changes in the protein expression of HUVECs induced by cholinergic agonists. Although it would not be scrupulous to declare that we had characterized all the protein changes after stimulation of NNAs, it was not possible to comment here on all the detected proteins that had remarkable changes. The above examples were sufficient enough to manifest that our experiments revealed some proteins that were directly associated with cell function and the subsequent cellular signal transduction pathway of non-neuronal acetylcholine receptors. It appears that Destrin might be a non-neuronal muscarinic receptors effector; FKBP1A might be an effector of non-neuronal acetylcholine receptors; and MIF and Profilin-1 might be associated with the activation of non-neuronal nicotinic receptors. Pilocarpine regulation of the expression of MIF and Profilin-1 suggested that there might be a new target of pilocarpine on endothelial cells that is similar to non-neuronal nicotinic receptors. Further study should be performed to facilitate understanding of non-neuronal acetylcholine receptors.
Perspectives
The non-neuronal acetylcholine system in endothelial cells plays a physiological/pathophysiological role in regulating endothelial function, vascular tone, angiogenesis, and inflammation. However, the identity of the mediators of intracellular signal transduction in endothelial cells after stimulation of the non-neuronal acetylcholine system remains unclear. In the current study, we analyzed the underlying effectors after cholinergic agonist activation of the non-neuronal acetylcholine system in cultured human umbilical vein endothelial cells. The identification of the differentially expressed proteins Destrin, FKBP1A, MIF, and Profilin-1 provided new clues toward further study of the influences of non-neuronal acetylcholine system activation on endothelial function in cardiovascular diseases.
Author contribution
Hai WANG designed the research; Yuan-yuan ZHANG, Wei SHEN, Wen-yu CUI, Yan-fang ZHANG, and Chao-liang LONG performed the research; Yuan-yuan ZHANG, Lian-cheng ZHANG, and Zhi-yuan PAN analyzed the data and wrote the paper.
Abbreviations
2-DE, two-dimensional gel electrophoresis; ACh, acetylcholine; DTT, Dithiothreitol; FKBP1A, FK506 binding protein 1a; HUVEC, human umbilical vein endothelial cells; LC-MS, liquid chromatography–mass spectrometry; MIF, macrophage migration inhibitory factor; Nico, nicotine; NNAs, non-neuronal acetylcholine system; NNMR, non-neuronal muscarinic receptors; Oxo, oxotremorine; Pilo, pilocarpine; SDS-PAGE, sodium dodecyl sulfate (SDS)-Polyacrylamide gel electrophoresis.
Acknowledgments
The present study was supported by grants from the State Key Research Project of China (AWS11J003) and Tianjing Key Technologies Research and Development Program, China (05ZHGCGX01300).
References
- Triggle CR, Samuel SM, Ravishankar S, Marei I, Arunachalam G, Ding H. The endothelium: influencing vascular smooth muscle in many ways. Can J Physiol Pharmacol. 2012;90:713–38. doi: 10.1139/y2012-073. [DOI] [PubMed] [Google Scholar]
- Butt M, Dwivedi G, Blann A, Khair O, Lip GY. Endothelial dysfunction: methods of assessment & implications for cardiovascular diseases. Curr Pharm Des. 2010;16:3442–54. doi: 10.2174/138161210793563383. [DOI] [PubMed] [Google Scholar]
- Grassi D, Desideri G, Ferri C. Cardiovascular risk and endothelial dysfunction: the preferential route for atherosclerosis. Curr Pharm Biotechnol. 2011;12:1343–53. doi: 10.2174/138920111798281018. [DOI] [PubMed] [Google Scholar]
- Evora PR, Evora PM, Celotto AC, Rodrigues AJ, Joviliano EE. Cardiovascular therapeutics targets on the NO-sGC-cGMP signaling pathway: a critical overview. Curr Drug Targets. 2012;13:1207–14. doi: 10.2174/138945012802002348. [DOI] [PubMed] [Google Scholar]
- Lee R, Channon KM, Antoniades C. Therapeutic strategies targeting endothelial function in humans: clinical implications. Curr Vasc Pharmacol. 2012;10:77–93. doi: 10.2174/157016112798829751. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zeng FH, Long CL, Pan ZY, Cui WY, Wang RH, et al. The novel ATP-sensitive potassium channel opener iptakalim prevents insulin resistance associated with hypertension via restoring endothelial function. Acta Pharmacol Sin. 2011;32:1466–74. doi: 10.1038/aps.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154:1558–71. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K, Fujii T. Basic and clinical aspects of non-neuronal acetylcholine: overview of non-neuronal cholinergic systems and their biological significance. J Pharmacol Sci. 2008;106:167–73. doi: 10.1254/jphs.fm0070073. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CJ, Bittinger F, Nozadze K, Wessler I. Expression and function of the non-neuronal cholinergic system in endothelial cells. Life Sci. 2003;72:2111–6. doi: 10.1016/s0024-3205(03)00069-9. [DOI] [PubMed] [Google Scholar]
- Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–6. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- Lee J, Cooke JP. The role of nicotine in the pathogenesis of atherosclerosis. Atherosclerosis. 2011;215:281–3. doi: 10.1016/j.atherosclerosis.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanaga K, Ichiki T, Miyazaki R, Takeda K, Hashimoto T, Matsuura H, et al. Acetylcholinesterase inhibitors attenuate atherogenesis in apolipoprotein E-knockout mice. Atherosclerosis. 2010;213:52–8. doi: 10.1016/j.atherosclerosis.2010.07.027. [DOI] [PubMed] [Google Scholar]
- Yu M, Chen DM, Hu G, Wang H. Proteomic response analysis of endothelial cells of human coronary artery to stimulation with carbachol. Acta Pharmacol Sin. 2004;25:1124–30. [PubMed] [Google Scholar]
- Shan LM, Wang H. Pharmacological characteristics of the endothelial target for acetylcholine induced vascular relaxation. Life Sci. 2002;70:1285–98. doi: 10.1016/s0024-3205(01)01506-5. [DOI] [PubMed] [Google Scholar]
- Shi CG, Hu G, Wang H. Protective effect of arecoline on expression of inflammatory molecules in endotheial cells injuried by oxLDL. Chin J Cardiol. 2004;32:650. [Google Scholar]
- Duan ZB, Wang H. Regulation effect of arecoline on excess expression of adhesive molecules in endothelial cells injuried with high concentration of D-glucose. Chin J Clin Pharmacol Ther. 2006;11:27–32. [Google Scholar]
- Duan ZB, Wang H. Protective effect of compounds on endothelial cells injuried with homocysteine. Chin Pharm Bull. 2006;22:537–42. [Google Scholar]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–46. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Panpac DI, Shahrokh Z. Mass spectrometry innovations in drug discovery and development. Pharm Res. 2001;18:131–45. doi: 10.1023/a:1011049231231. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–5. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Edwards AVG, White MY, Cordwell SJ. The role of proteomics in clinical cardiovascular biomarker discovery. Mol Cell Proteomics. 2008;7:1824–37. doi: 10.1074/mcp.R800007-MCP200. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, et al. Actin depolymerizing factor (ADF/cofilin)enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–81. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–11. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Abe H, Nagaoka R, Obinata T. Cytoplasmic localization and nuclear transport of cofilin in cultured myotubes. Experimental Cell Res. 1993;206:1–10. doi: 10.1006/excr.1993.1113. [DOI] [PubMed] [Google Scholar]
- Lee CW, Han J, Bamburg JR, Han L, Lynn R, Zheng JQ. Regulation of acetylcholine receptor clustering by ADF/cofilin-directed vesicular trafficking. Nat Neurosci. 2009;12:848–56. doi: 10.1038/nn.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Gohla A, Bokoch GM. 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol. 2002;12:1704–10. doi: 10.1016/s0960-9822(02)01184-3. [DOI] [PubMed] [Google Scholar]
- Bultynck G, De Smet P, Rossi D, Callewaert G, Missiaen L, Sorrentino V, et al. Characterization and mapping of the 12 kDa FK506-binding protein (FKBP12)-binding site on different isoforms of the ryanodine receptor and of the inositol 1,4,5-trisphosphate receptor. Biochem J. 2001;354:413–22. doi: 10.1042/0264-6021:3540413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultynck G, Rossi D, Callewaert G, Missiaen L, Sorrentino V, Parys JB, et al. The conserved sites for the FK506-binding proteins in ryanodine receptors and inositol 1,4,5-trisphosphate receptors are structurally and functionally different. J Biol Chem. 2001;276:47715–24. doi: 10.1074/jbc.M106573200. [DOI] [PubMed] [Google Scholar]
- MacMillan D, Currie S, Bradley KN, Muir TC, McCarron JG. In smooth muscle, FK506-binding protein modulates IP3 receptor-evoked Ca2+release by mTOR and calcineurin. J Cell Sci. 2005;118:5443–51. doi: 10.1242/jcs.02657. [DOI] [PubMed] [Google Scholar]
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug target network. Nat Biotechnol. 2007;25:1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RA, Bucala R. Tumor growth-promoting properties of macrophage migration inhibitory factor (MIF) Semin Cancer Biol. 2000;10:359–366. doi: 10.1006/scbi.2000.0328. [DOI] [PubMed] [Google Scholar]
- Morand EF, Leech M, Bernhagen J. MIF: a new cytokine link between rheumatoid arthritis and atherosclerosis. Nat Rev Drug Discov. 2006;5:399–410. doi: 10.1038/nrd2029. [DOI] [PubMed] [Google Scholar]
- Zernecke A, Bernhagen J, Weber C. Macrophage migration inhibitory factor in cardiovascular disease. Circulation. 2008;117:1594–602. doi: 10.1161/CIRCULATIONAHA.107.729125. [DOI] [PubMed] [Google Scholar]
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. New Engl J Med. 2005;353:1711–23. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–96. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- Kamir D, Zierow S, Leng L, Cho Y, Diaz Y, Griffith J, et al. A Leishmania ortholog of macrophage migration inhibitory factor modulates host macrophage responses. J Immunol. 2008;180:8250–61. doi: 10.4049/jimmunol.180.12.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober A, Bernhagen J, Weber C. Chemokine-like functions of MIF in atherosclerosis. J Mol Med (Berl) 2008;86:761–70. doi: 10.1007/s00109-008-0334-2. [DOI] [PubMed] [Google Scholar]
- Yah J, Hearer BK. Profilin-1 is required for the normal timing of actin polymerization in response to thermal-stress. FEBS Lett. 1996;398:303–7. doi: 10.1016/s0014-5793(96)01259-8. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. The faces of profilin. Cell. 1993;75:835–8. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- Moustafa-Bayoumi M, Alhaj MA, El-Sayed O, Wisel S, Chotani MA, Abouelnaga ZA, et al. Vascular hypertrophy and hypertension caused by transgenic overexpression of profilin 1. J Biol Chem. 2007;282:37632–9. doi: 10.1074/jbc.M703227200. [DOI] [PubMed] [Google Scholar]
- Romeo GR, Moulton KS, Kazlauskas A. Attenuated expression of profilin-1 confers protection from atherosclerosis in the LDL receptor null mouse. Circ Res. 2007;101:357–67. doi: 10.1161/CIRCRESAHA.107.151399. [DOI] [PubMed] [Google Scholar]