Abstract
During 2010 and 2011, the Loreto region of Peru experienced a dengue outbreak of unprecedented magnitude and severity for the region. This outbreak coincided with the reappearance of dengue virus-2 (DENV-2) in Loreto after almost 8 years. Whole-genome sequence indicated that DENV-2 from the outbreak belonged to lineage II of the southeast Asian/American genotype and was most closely related to viruses circulating in Brazil during 2007 and 2008, whereas DENV-2 previously circulating in Loreto grouped with lineage I (DENV-2 strains circulating in South America since 1990). One amino acid substitution (NS5 A811V) in the 2010 and 2011 isolates resulted from positive selection. However, the 2010 and 2011 DENV-2 did not replicate to higher titers in monocyte-derived dendritic cells and did not infect or disseminate in a higher proportion of Aedes aegypti than DENV-2 isolates previously circulating in Loreto. These results suggest that factors other than enhanced viral replication played a role in the severity of this outbreak.
Introduction
Dengue fever (DF) is caused by four antigenically distinct but related dengue virus serotypes (DENV-1, -2, -3, and -4), each of which includes multiple genotypes that differ by at least 6% in nucleotide identity.1 DENV is transmitted by Aedes mosquitoes, primarily Ae. aegypti, in more than 100 countries and infects an estimated 390 million people annually.2 The clinical spectrum of dengue disease is broad. Most DENV infections are asymptomatic or present as a debilitating but self-limiting undifferentiated febrile illness; however, a small percentage of cases progress to the more severe forms of the disease: dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS).3 Worldwide, the burden of dengue disease is substantial, ranging from 150 to 1,300 disability-adjusted life years per million depending on the spatiotemporal attributes of epidemics.4–9 In many resource-poor countries, the costs associated with a DENV infection significantly exceed the average monthly income of the patient.5,9–18 Historically, southeast Asia has borne the brunt of severe dengue disease, but severe disease has also been on the rise in the Americas, where the number of DHF cases has increased more than eightfold over the last 30 years from 13,398 during the 1980s to 111,724 between 2000 and 2007.19
The increase in dengue disease severity in the Americas has been attributed to a shift from single-serotype endemism to hyperendemicity (cocirculation of multiple serotypes) of DENV coupled with the introduction of new genotypes with relatively high pathogenicity.20–22 After the eradication efforts of the Pan American Health Organization (PAHO) during the 1950s, DENV-2 was the only serotype detected in the Americas until the 1960s, and by the 1980s, all four serotypes of DENV were present.23,24 The circulation of multiple serotypes, which includes sequential novel serotype introduction into the same geographic area, results in the exposure of individuals to different serotypes over their lifetime, and secondary infection with a heterologous DENV serotype is a known risk factor for severe disease.25–27 Additionally, different genotypes within a given serotype and different lineages within a given genotype have been shown to vary in tendency to cause severe disease.28–30 For example, the DENV-2 viruses circulating in the Americas were of the American genotype until 1981, when DENV-2 viruses of the southeast Asian/American genotype were detected in the region.26 Infection with DENV-2 of the American genotype had limited association with severe disease in the Americas, although recent analyses indicate that outbreaks of this American genotype in the 1970s in Puerto Rico, Tahiti, New Caledonia, and Niue included severe dengue disease.30–34 Nonetheless, the invasion of the southeast Asian/American genotype DENV-2 resulted in the first major DHF epidemic in the Americas.35 Experimental studies revealed that the American genotype DENV-2 had lower replication capacity in cultured human cells and reduced infectivity for Ae. aegypti compared with DENV-2 of the southeast Asian/American genotype.1,22,36–38 Not surprisingly, given its higher fitness in both host and vector systems, southeast Asian/American DENV-2 has displaced American DENV-2 in many regions in the Americas.36,39
Since 2000, the US Naval Medical Research Unit No. 6 (NAMRU-6) in conjunction with the Ministry of Health of Peru has conducted surveillance for the etiologies of febrile illness in several regions of Peru.40 In October of 2010, this surveillance network detected the circulation of DENV-2 in Iquitos, Loreto, Peru. This finding was noteworthy, because DENV-2 had not been detected in this region since 2002. Furthermore, initial sequence analysis from the capsid/premembrane region indicated that the DENV-2 circulating was of a southeast Asian/American lineage that had never been identified in Loreto before. The months that followed the introduction of this DENV-2 were remarkable for both the absolute number and severity of dengue cases. In 2011, over 25,000 cases of dengue in the region of Loreto were reported to the Ministry of Health.41 This number was more than three times the highest number of cases that had been reported in any year over the last 8 years. Additionally, one of the largest hospitals in Iquitos experienced a vastly greater amount of dengue-related hospitalizations (654 in January of 2011) during this outbreak than for the outbreaks that occurred in 2008 (90 hospitalizations) and 2009 (40 hospitalizations) combined.42
This outbreak was associated with the reappearance of DENV-2 after an absence of almost 8 years. Before 2010, a few DENV-2 cases were identified in 2002 during the initial wave of a major DENV-3 epidemic, but the last period of significant DENV-2 transmission in Loreto ended in 1996.39,43 Analysis of the sequence of the envelope/non-structural region 1 region of eight DENV-2 isolates circulating during the 2010 and 2011 outbreak indicated that the DENV-2 strain responsible for the outbreak belonged to a lineage of DENV-2 that had not been detected in the region before.44 To better understand the viral factors that may have affected the severity of the 2010 and 2011 Loreto outbreak, we compared whole-genome sequences and phenotypic characteristics of DENV-2 isolates collected during the outbreak with isolates collected previously in Peru.
Materials and Methods
Ethics statement.
Study protocols NMRCD.2010.0010 and NMRCD.2007.0007 were approved by the NAMRU-6 Institutional Review Board (IRB) and study protocol NMRCD.2000.0006 was approved by the Naval Medical Research Center IRB in compliance with all US Federal Regulations governing the protection of human subjects. In addition, these study protocols were reviewed and approved by the Peruvian Ministry of Health. For study protocols NMRCD.2007.0007 and NMRCD.2000.0006, subjects 18 years or older provided written informed consent. For subjects under 18 years old, a parent or legal guardian provided written informed consent; children 8–17 years old also provided written assent. Study protocol NMRCD.2010.0010 involved no more than minimal risk of harm to subjects, and the following IRB-approved verbal consent process was used. On the identification of a subject that met the case definition, the contents of an information sheet were explained, and verbal consent was obtained from subjects 18 years or older; for subjects less than 18 years old, a parent or legal guardian provided verbal consent, and children from 8 to 17 years old also provided verbal assent. The date of verbal consent and/or assent with the subject's study identification number was maintained in a database.
Study protocol UTMB-IRB#13-149 was reviewed by the University of Texas Medical Branch IRB and used for blood collections from dengue-naive, healthy volunteers for the generation of the monocyte-derived dendritic cells (mo-DCs). Written consent to participate in the study was obtained from each subject enrolled in this study, and all data were handled anonymously and confidentially.
Study subjects.
Subjects for this study were identified by both passive and active surveillance. Passive surveillance was conducted as previously described.40 Briefly, patients 5 years of age or older who sought medical care at a participating outpatient clinic or hospital with an undifferentiated febrile illness (> 38°C for 5 days or less) were offered enrollment in the study. A standard questionnaire was used to obtain demographic and clinical information, and acute and convalescent blood samples were collected. Active surveillance was conducted as previously described.43,45 Two cohorts of individuals 3 years of age or older totaling over 13,000 individuals and covering 54 city blocks distributed between 10 different Ministry of Health zones were monitored three times a week for febrile illness. One of the cohorts has been previously described and included 38 blocks distributed over two zones, whereas the second cohort included eight zones, with two blocks per zone.46,47 If a participant was found to have a febrile illness, a standard questionnaire was used to obtain demographic and clinical information, and acute and convalescent blood samples were collected.
Viral isolation.
Viral isolation was performed from acute-phase serum using both mosquito (C6/36) and African green monkey kidney (Vero) cells as previously described.40 Briefly, serum was first diluted 1:10 in Eagle's minimal essential medium (Quality Biological Inc., Gaithersburg, MD) supplemented with 2% heat-inactivated fetal bovine serum (FBS; Sigma Aldrich, St. Louis, MO), 200 U/mL penicillin, and 200 μg streptomycin (Sigma Aldrich, St. Louis, MO). For C6/36 cell inoculations, 0.1 mL diluted serum was inoculated into duplicate wells of a 24-well plate. For Vero cell inoculations, 0.2 mL diluted serum was used to inoculate a 12.5-cm2 flask. On evidence of cytopathic effect or 10 days post-inoculation, cultures were harvested. A portion of the harvested culture was processed for an immunofluorescence assay, and the remainder was stored at −80°C. For immunofluorescence assays, cells were first stained using a pool of antiflavivirus hyperimmune ascitic fluid (HMAF) that included anti-DENV, anti-yellow fever virus, and anti-St. Louis encephalitis virus HMAFs. Positive samples were then stained individually with monoclonal antibodies specific for each serotype of DENV, yellow fever virus, and St. Louis encephalitis virus.
Genome sequencing.
All virus sequences were obtained using Illumina sequencing (Illumina, San Diego, CA). Viral RNA (0.1–0.2 μg) was fragmented by incubation at 94°C for 8 minutes in 19.5 μL fragmentation buffer (15016648; Illumina, San Diego, CA). First- and second-strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq RNA Sample Preparation Kit according to the manufacturer's instructions (Illumina, San Diego, CA). Samples were tracked using the index tags incorporated into the adapters as defined by the manufacturer. Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit, version 3 (Illumina, San Diego, CA) and the Illumina cBot Workstation (Illumina, San Diego, CA) using conditions recommended by the manufacturer. Paired-end 50-base sequencing by synthesis was performed using the TruSeq SBS Kit, version 3 (Illumina, San Diego, CA) on an Illumina HiSeq 1000 (Illumina, San Diego, CA) using protocols provided by the manufacturer. Cluster density per lane was 645,000–980,000/mm2, and post-filter reads ranged from 148 million to 178 million per lane. Base call conversion to sequence reads was performed using CASAVA-1.8.2. Virus sequences were edited and assembled using the SeqMan and NextGen modules of the DNAStar Lasergene 7 program (Bioinformatics Pioneer DNAStar, Inc., Madison, WI). In certain cases, pre-filtering of reads to remove host sequence enhanced the assembly process.
Nucleotide sequence accession numbers.
The genomic sequences of the 24 newly obtained DENV-2 sequences that were included in the phylogenetic analyses are KC294200–KC294223.
Phylogenetic and selection pressure analysis.
All available complete genomes of DENV-2 virus were downloaded from GenBank. These sequences, together with sequences from the 24 isolates sequenced in this study, were combined and aligned manually using the Se-Al application based on amino acid sequence alignments. A neighbor-joining tree was built based on this alignment using PAUP* v4.0b Package (Sinauer Associates, Sunderland, MA) as a guide tree. Overly sampled sequences from a single location and year were deleted to accelerate the analysis without sacrificing overall genetic diversity, which led to a final dataset of 137 samples of complete coding sequences (length = 10,173 nucleotides), including all the Peruvian strains under analysis belonging to the southeast Asian/American genotype. A maximum clade credibility (MCC) tree along with evolutionary rate and the lineage divergence times were inferred using BEAST package v1.6.2 based on the GTR nucleotide substitution model, relaxed clock model (uncorrelated lognormal), and Coalescent: GMRF Bayesian Skyride tree prior model.48 Based on the resultant MCC tree, we also traced the amino acid substitutions along the branches by MacClade 4.08 OS X (Sinauer Associates, Sunderland, MA). Finally, to determine the selective pressure on southeast Asian/America genotype, the overall and sitewise dN/dS values were estimated using the internal fixed effects likelihood (IFEL) method in the HyPhy package.49
Viral replication kinetics.
Comparative multistep replication curves of select DENV-2 strains were generated in triplicate on human hepatoma (Huh-7) and C6/36 cells as described previously.38,50 Huh-7 cells (clone JTC-39) were obtained from the Japanese Health Sciences Foundation (Osaka, Japan). Briefly, both cell lines were plated into 12-well plates at 3.6 × 105 (Huh-7) and 7.5 × 105 (C6/36) cells per well and infected at a multiplicity of infection (MOI) of 0.01 focus-forming units (FFUs) in triplicate. Infected dishes containing Huh-7 cells were incubated at 37°C for 1 hour with periodic gentle rocking to facilitate virus adsorption, whereas dishes containing mosquito C6/36 cells were incubated at 28°C. Viral inocula were removed, and cell monolayers were washed three times with phosphate buffered saline (PBS) to remove unadsorbed virus; 2 mL minimal essential medium (MEM) supplemented with 5% FBS, 2 mM L-glutamine, 1% non-essential amino acids, and 50 mg/mL penicillin/streptomycin were then added, and dishes were incubated at 28°C or 37°C for the mosquito or mammalian cell lines, respectively. Virus from individual dishes was harvested daily through day 7 post-inoculation, clarified by low-speed centrifugation, and assayed in C6/36 cells to determine virus titer by focus-forming immunoassay. Virus yield at each time point was recorded as FFUs per cell and represented as the ratio of the total amount of virus present in the sample by the number of cells originally infected.
Isolation, stimulation of peripheral blood mononuclear cells, and DENV infections of mo-DCs.
Approximately 1 U blood (approximately 500 mL) was obtained from two dengue-naïve, healthy volunteers at different times. Peripheral blood mononuclear cells were isolated from buffy coats by centrifugation over an Accuprep gradient according to the manufacturer's protocol (Accurate Chemical Corp., Westbury, NY). CD14+ monocytes were positively selected using a magnetic cell sorting (MACS) isolation column (Miltenyi Biotec, Auburn, MA). Cells were counted and seeded in six-well plates at a density of 1–2 × 106 cells/well in RPMI 1640 culture medium supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine containing 1,000 U/mL recombinant human interleukin 4 (IL-4; R&D Systems, Minneapolis, MN) and 1,400 U/mL granulocyte–monocyte colony-stimulating factor (GM-CSF; Immunex, Thousand Oaks, CA). On alternate days, one-half of the volume was removed and replaced with medium containing double (2×) the concentration of fresh cytokines. Cultures were maintained under these conditions for a total of 6 days; then, cells were collected with gentle pipetting and washed, and the mo-DC phenotype (purity and maturation markers of mo-DCs) was confirmed by flow cytometry analysis. Subsequently, 1 × 105 cells were infected with the selected DENV at an MOI = 2 for 2 hours at 37°C. The virus inoculum was removed, and the cells were washed three times with PBS to ensure removal of unadsorbed virus. Cells were resuspended in 2 mL RPMI 1640 culture medium supplemented with cytokines and incubated at 37°C. Cell-free supernatant aliquots were removed immediately before and after infection and on days 1 and 2 post-infection, and they were assayed on C6/36 cells to obtain infection and progeny titers.
Flow cytometry.
Human mo-DCs were stained for the confirmation of their immature phenotype with a panel of fluorochrome-conjugated human monoclonal antibodies against CD11c, CD83, CD86, and DC-SIGN (CD209; BD Biosciences, San Jose, CA) and to assess viability using live-dead fixable blue dye (Invitrogen, Carlsbad, CA). As controls, the appropriate isotype and fluorescence intensity minus one (FMO) were used. Cells were allowed to incubate on ice for 30 minutes with the respective antibodies and 20 minutes with the viability dye followed by washing two times with staining buffer (BD Biosciences, San Jose, CA) to remove unbound antibody. Cells were then fixed in buffered 2% paraformaldehyde (pH 9.5) and analyzed using an LSRII Fortessa cytometer (BD Biosciences, San Jose, CA) within 24 hours of staining. The data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Focus-forming assays and immunostaining.
Tenfold serial dilutions of virus in MEM supplemented with 2% FBS and antibiotics (Invitrogen, Carlsbad, CA) were added in duplicate to confluent C6/36 cell monolayers attached to 24-well plates (Costar, Corning, NY) and incubated for 1 hour with periodic gentle rocking to facilitate virus adsorption at 28°C. Wells were then overlaid with 1 mL 0.8% methylcellulose (Sigma-Aldrich, St. Louis, MO) diluted in warm Optimem (Invitrogen, Carlsbad, CA) supplemented with 2% FBS, antibiotics, and 1% (wt/vol) L-glutamine and incubated undisturbed for 4 days at 28°C. Methylcellulose overlay was aspirated, and cell monolayers were rinsed one time with PBS (pH 7.4; Invitrogen, Carlsbad, CA), fixed with a mixture of ice-cold acetone and methanol (1:1), and allowed to incubate for 30 minutes at room temperature. Fixation solution was aspirated, and plates were allowed to air dry. Plates were washed three times with PBS supplemented with 3% FBS followed by a 1-hour-long incubation with DENV-specific HMAF. Plates were washed three times followed by a 1-hour-long incubation with a secondary antibody conjugated to horseradish peroxidase (KPL, Gaithersburg, MD). Detection proceeded with the addition of aminoethylcarbazole substrate (ENZO Life Sciences, Farmingdale, MA) prepared according to the vendor's instructions.
Experimental infection of mosquitoes with DENV.
Ae. aegypti of the National Institutes of Health colony strain were fed for 20 minutes on blood meals composed of 2 mL washed rabbit red blood cells (Hemostat, Dixon, CA) in 10% sucrose combined with 1 mL virus offered in water-jacketed membrane feeders (Lillie Glass, Smyrna, GA) covered with a parafilm seal.51 Virus titer in the blood meals ranged from 5.48 to 6.48 log10 FFUs/mL. Engorged mosquitoes were separated, incubated at 28°C and 80% relative humidity on a 12-hour:12-hour light:dark cycle with ad lib access to 10% sucrose solution for 14 days, and then, frozen at −20°C overnight. To assess infection and dissemination, bodies, heads, and legs were separated, and the former two parts were homogenized using mortar and pestle in 250 μL Hanks Balanced Salt Solution (Life Technologies, Grand Island, NY) supplemented with 10% FBS, 250 μg/mL amphotericin B (Life Technologies, Grand Island, NY), 1% ciprofloxacin, and 150 μg/mL clindamycin and stored at −80°C until assayed. Samples were centrifuged for 10 minutes at 5,000 rpm, and 100 μL each sample supernatant was inoculated into 96-well plates containing C6/36 cells. Cultures were maintained with 100 μL media (DMEM hi-glucose supplemented with 10% FBS, 250 μg/mL amphotericin B, 1% ciprofloxacin, and 150 μg/mL clindamycin) at 28°C and 5% CO2 for 4 days, and then, they were fixed with a mixture of ice-cold acetone and methanol (1:1) solution and immunostained essentially as described above.
Statistical analysis.
The replication kinetics in Huh-7 and C6/36 cells of the two lineages and the individual viruses were compared in separate repeated measures analysis of variance (rmANOVA) using JMP (SAS Institute Inc., Cary, NC). Replication kinetics of the two lineages in mo-DCs were compared with an rmANOVA. All percentile data for mosquito infection and dissemination were arcsin-square root-transformed to render them normal; all such transformations were successful. The association between blood meal titer and transformed infection values was assessed using a parametric correlation; blood meal titers and transformed infection and dissemination were compared between lineages using a Student's t test.
Results
Phylogenetic analysis.
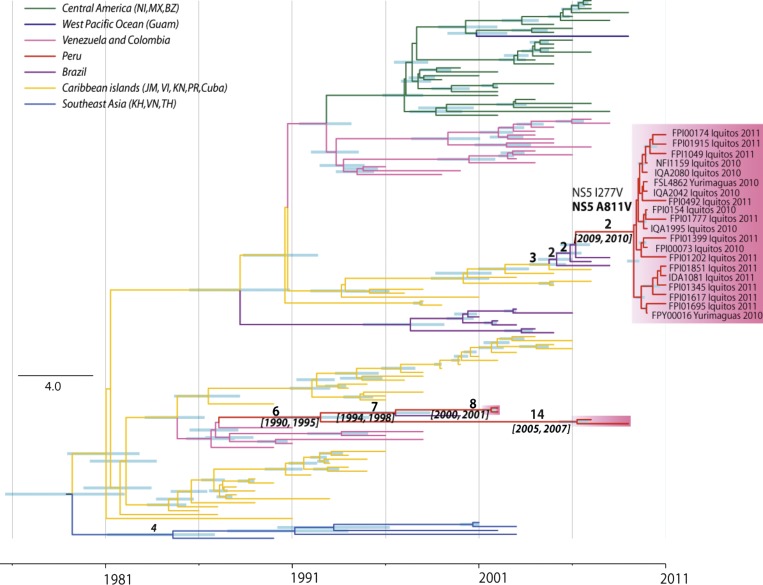
To understand how the DENV-2 that circulated during the 2010 and 2011 outbreak differed genetically from the DENV-2 that had previously circulated in Peru, whole-genome sequencing was conducted on 20 early passage 2010 and 2011 isolates that were collected at time points throughout the outbreak period and four isolates that circulated in Peru before 2010 (Table 1). Although the overall outbreak was noted for its severity (13% of the reported cases were classified as dengue with warning signs), sufficient information to classify dengue disease category in accordance with the World Health Organization (WHO) criteria for each isolate was not available.41,42,52,53 All of the isolates sequenced grouped within the southeast Asian/American genotype (Figure 1). However, within the southeast Asian/American genotype, the strains from the 2010 and 2011 outbreak formed a different lineage from the four strains sampled in Peru from 2002 to 2009. Specifically, the 2010 and 2011 outbreak strains formed a single lineage, which is closely related with strains sampled in Brazil from 2007 to 2008 that were initially introduced from the Caribbean and Central American region. We refer to this group of viruses as lineage II. In contrast, the older Peruvian strains, termed lineage I, were similar to strains that have circulated within South America since 1990. The estimated time to the most recent common ancestor (tMRCA) of 2010 and 2011 outbreak strains was 2009 and 2010 (95% highest posterior density), immediately preceding the outbreak. DENV-2 lineage I seems to have been introduced from Brazil as a result of a recent reintroduction of DENV-2 in South America from the Caribbean, a phenomenon described previously with DENV-2, but the paucity of full-length DENV-2 sequences from South America makes it difficult to determine the exact location of this introduction.54 Within this lineage, the two samples from 2002 and those from 2009 are estimated to have diverged between 1990 and 1995, indicating that they may have been introduced to Peru independently. Furthermore, the observation that both lineage II and the 2002 strains in lineage I are closely related with strains from Brazil in a slightly earlier year indicates that Brazil may have been a regular source for Loreto DENV-2 (Figure 1).
Table 1.
Demographic information for the sequenced DENV-2 virus isolates
| Code | Collection date | Age (years) | Sex | Hospitalization | City | Region |
|---|---|---|---|---|---|---|
| FSL0699 | 02/06/2002 | 19 | F | No | Iquitos | Loreto |
| NFI0052 | 03/22/2002 | 50 | M | No | Iquitos | Loreto |
| FMD1337 | 03/28/2007 | 28 | M | No | Puerto Maldonado | Madre De Dios |
| FMD2303 | 02/13/2009 | 51 | M | No | Puerto Maldonado | Madre De Dios |
| NFI1159 | 10/19/2010 | 37 | F | No | Iquitos | Loreto |
| IQA1995 | 10/21/2010 | 13 | M | No | Iquitos | Loreto |
| IQA2042 | 11/04/2010 | 52 | F | Yes | Iquitos | Loreto |
| IQA2080 | 11/16/2010 | 23 | M | No | Iquitos | Loreto |
| FSL4862 | 11/22/2010 | 36 | F | Yes | Yurimaguas | Loreto |
| FPI00073 | 12/13/2010 | 19 | M | No | Iquitos | Loreto |
| FPY00016 | 12/22/2010 | 26 | F | No | Yurimaguas | Loreto |
| FPI00154 | 12/29/2010 | 34 | M | Yes | Iquitos | Loreto |
| FPI00174 | 01/03/2011 | 28 | F | Yes | Iquitos | Loreto |
| FPI00492 | 01/18/2011 | 17 | F | Yes | Iquitos | Loreto |
| IDA1081 | 01/24/2011 | 14 | M | − | Iquitos | Loreto |
| FPI01049 | 02/07/2011 | 19 | F | Yes | Iquitos | Loreto |
| FPI01202 | 02/11/2011 | 19 | M | Yes | Iquitos | Loreto |
| FPI01345 | 02/23/2011 | 22 | M | Yes | Iquitos | Loreto |
| FPI01399 | 03/01/2011 | 43 | F | Yes | Iquitos | Loreto |
| FPI01617 | 03/23/2011 | 53 | F | Yes | Iquitos | Loreto |
| FPI01695 | 04/04/2011 | 12 | M | − | Iquitos | Loreto |
| FPI01777 | 04/24/2011 | 24 | M | No | Iquitos | Loreto |
| FPI01851 | 04/29/2011 | 20 | M | Yes | Iquitos | Loreto |
| FPI01915 | 05/11/2011 | 14 | M | No | Iquitos | Loreto |
F = female; M = male.
Figure 1.
MCC tree of DENV-2 southeast Asian/American genotype. The 95% highest posterior density (HPD) values of divergence time are shown by the node bar. Divergence events that led to the Peruvian lineages are specifically labeled with numbers of amino acid substitutions (above the branches) and 95% HPD of divergence year (below branches). BZ = Belize; JM = Jamaica; KH = Cambodia; KN = Saint Kitts and Nevis; MX = Mexico; NI = Nicaragua; PR = Puerto Rico; TH = Thailand; VI = Virgin Islands; VN = Vietnam.
Selection pressure analysis.
The overall and site-specific dN/dS values for the southeast Asian/American genotype were estimated through HyPhy package using the internal fixed effect likelihood (IFEL) method. IFEL calculates the dN/dS value based on internal branches only, therefore reducing the bias caused by transient mutations that will not be fixed in the viral population. The overall dN/dS value for the DENV-2 southeast Asian/American genotype is estimated to be 0.054 (95% confidence interval = 0.051–0.056), suggesting that these viruses were under strong purifying selection; 102 positive selected sites were detected throughout the coding region, with 67 found to be highly positive (dN/dS > 1 × 1010) (Table 2). The sites under selection located within the capsid (C) and NS2A genes showed elevated dN/dS values (0.151 and 0.109, respectively) compared with other genes (0.044–0.074). Amino acid substitutions have been observed along the evolution of both Peruvian lineages. Two amino acid substitutions in the NS5 gene, NS5 I277V and NS5 A811V, occurred during the introduction of lineage II from Brazil. Only the NS5 A811V was the result of positive selection by dN/dS analyses.
Table 2.
Positively and negatively selected sites of the Loreto outbreak DENV-2 viruses
| Gene | Length | dN/dS (95% confidence interval) | Number of positively selected sites | Number of negatively selected sites | |
|---|---|---|---|---|---|
| Strong positive (dN/dS) > 1 × 1010 | Slightly positive (dN/dS) < 10 | ||||
| Total | 3,391 | 0.066 (0.061–0.072) | 67 | 35 | 1,920 |
| C* | 114 | 0.151 (0.102–0.214) | 2 | 3 | 42 (37%) |
| prM | 166 | 0.074 (0.051–0.101) | 2 | 3 | 93 (56%) |
| Envelope | 495 | 0.066 (0.053–0.082) | 8 | 7 | 274 (55%) |
| NS1 | 352 | 0.074 (0.057–0.095) | 5 | 2 | 201 (57%) |
| NS2a* | 218 | 0.109 (0.086–0.135) | 7 | 1 | 136 (62%) |
| NS2b | 130 | 0.061 (0.039–0.090) | 7 | 0 | 67 (52%) |
| NS3 | 618 | 0.040 (0.031–0.051) | 10 | 4 | 366 (59%) |
| NS4a | 149 | 0.074 (0.051–0.102) | 2 | 4 | 90 (60%) |
| NS4b | 249 | 0.044 (0.031–0.061) | 6 | 0 | 143 (57%) |
| NS5 | 900 | 0.068 (0.058–0.080) | 18 | 11 | 508 (56%) |
Elevated dN/dS values.
Replication kinetics.
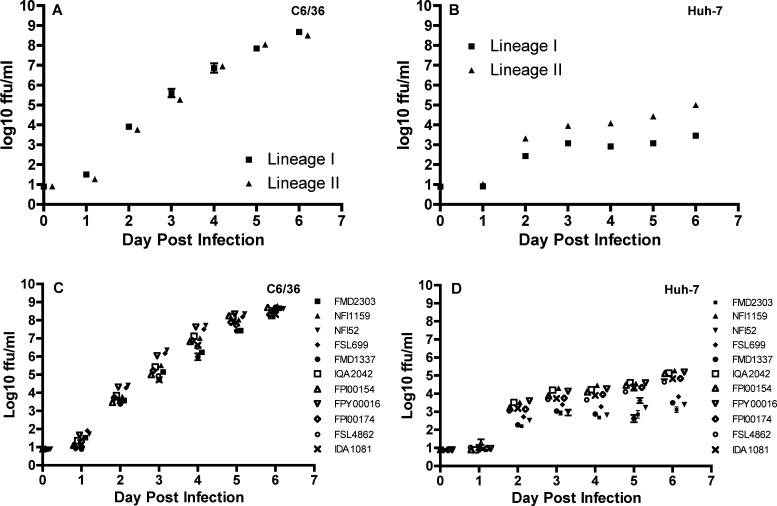
To determine whether the DENV-2 viruses that had circulated in Peru before 2010 (lineage I) and the viruses that circulated during the 2010 and 2011 outbreak in Loreto (lineage II) replicated at different rates in mammalian or mosquito cells, replication of four lineage I isolates and seven lineage II isolates was monitored daily in human hepatoma (Huh-7) or Ae. albopictus (C6/36) cells for 7 days post-infection. Lineages I and II viruses in both Huh-7 and C6/36 cells showed, as expected, a significant increase in titer over the sampling period (rmANOVA, degrees of freedom [df] = 6, 4; P < 0.0001 in both cell lines). The two lineages showed similar patterns of increase over time in C6/36 cells (lineage × sampling day interaction: df = 6, 4; F = 2.83, P = 0.17; lineage: df = 1, 9; F = 0.2, P = 0.65) (Figure 2A). In Huh-7 cells, in contrast, there was a significant interaction between lineage and sampling day (F = 21.0, P = 0.006) (Figure 2B), and lineage II reached a significantly higher titer than lineage I (F = 60.8, P < 0.0001), indicating that both pattern and magnitude of replication differed. Comparison of individual viruses (rmANOVA, df = 10, 60) revealed a significant interaction between virus and sampling day in both C6/36 and Huh-7 cells (P < 0.0001 for all comparisons) (Figure 2C and D) but no overall effect of virus (P > 0.99 for all comparisons).
Figure 2.
Comparative replication curves of lineages I and II DENV-2 strains in vitro. (A and B) Mean virus output of lineage I (triangles) and lineage II (squares) DENV-2 strains in mosquito epithelial (C6/36) and human hepatoma (Huh-7) cell lines; means ± SEs derived from four lineage I (FMD2303, NFI52, FSL699, and FMD1337) and seven lineage II (NFI1159, IQA2042, FPI00154, FPI00174, FPY00016, FSL4862, and IDA1081) DENV-2 strains are shown. (C and D) Daily virus output from days 0–6 after infection at an MOI of 0.01 by lineage I (FMD2303, NFI52, FSL699, and FMD1337) and lineage II (NFI1159, IQA2042, FPI00154, FPI00174, FPY00016, FSL4862, and IDA1081) DENV-2 strains in C6/36 and Huh-7 cell lines.
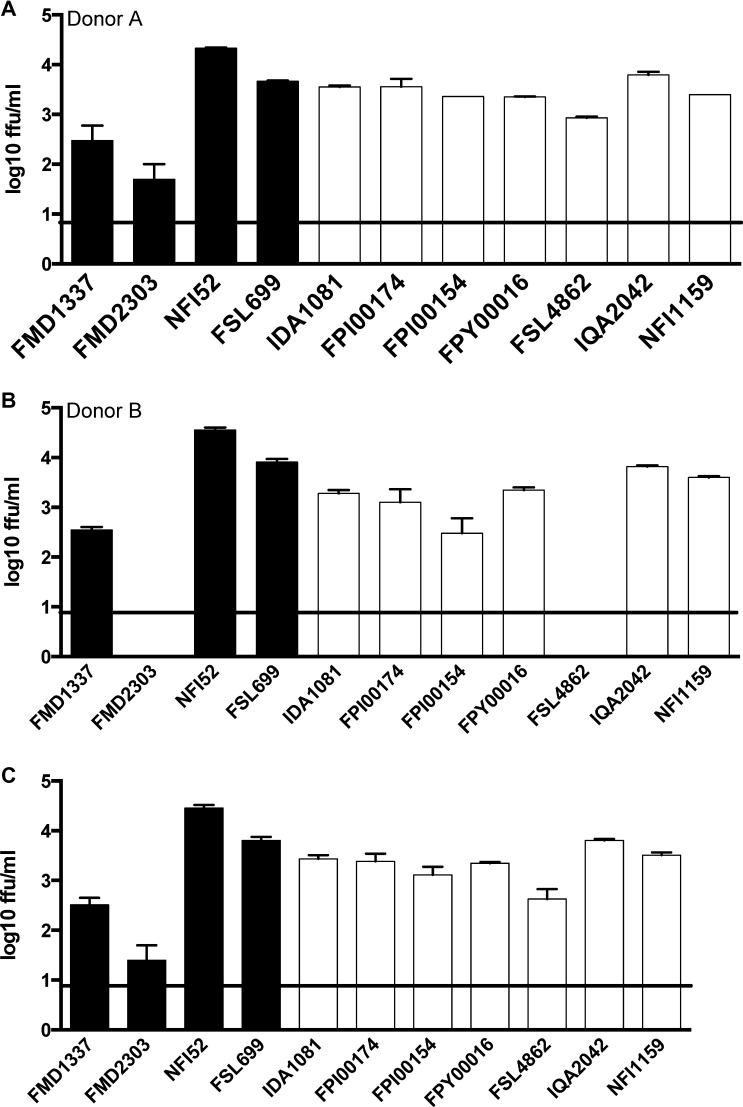
To assess viral replication kinetics in a cell model that more closely reflects replication in vivo, we also monitored the replication of the two lineages daily over 2 days in primary human mo-DCs, which are considered primary targets for DENV infection in humans.55–57 Because two different donors contributed mo-DCs, we initially tested for an effect of donor on viral replication, but we detected neither an interaction between donor and lineage (rmANOVA, df = 1, 18; P = 0.69) nor an overall effect of donor (rmANOVA, df = 1, 18; P = 0.98) on viral replication kinetics at 48 hours post-infection (Figure 3A and B). We, therefore, combined data from both donors. As expected, virus titers from both lineages increased with time (rmANOVA, df = 1, 19; P < 0.0001), but there was no interaction between lineage and time (rmANOVA, df = 1, 19; P = 0.53) and no overall effect of lineage on viral replication kinetics (rmANOVA, df = 1, 20; P = 0.43) (Figure 3C).
Figure 3.
Comparative replication curves of lineages I and II DENV-2 strains ex vivo. (A and B) Mean virus output of lineage I (black columns) and lineage II (white columns) DENV-2 strains in mo-DCs from two healthy human donors at 48 hours post-infection; means ± SEs derived from four lineage I and seven lineage II DENV-2 strains are shown. (C) Combined mean virus output of lineage I (solid columns) and lineage II (open columns) DENV-2 strains in mo-DCs from both donors (data from two individual experiments pooled together) at 48 hours post-infection. The limit of detection of the assay is 0.9 log10 FFUs/mL (solid line). Although the statistical analysis of viral replication in these cells included days 0–2 post-infection, to allow clear visualization of both donors and all virus isolates, only titers on day 2 post-infection are shown.
Mosquito infection.
To assess potential differences in transmission between the two lineages, Ae. aegypti were fed blood meals containing one of three different isolates from each of the two lineages. There was no difference in mean blood meal titer between the two lineages (Student's t test, df = 4; t = 0.26, P = 0.40) and no correlation between blood meal titer (which only varied by 1 log10 FFU/mL across all virus isolates) and infection (pairwise correlations, df = 4; P = 0.18). The two lineages did not differ in infection, total dissemination, or dissemination from infected midguts (Student's t test, df = 4; P > 0.30 for all comparisons) (Table 3).
Table 3.
Infection and dissemination of Peruvian lineages I and II DENV-2 in the vector Ae. aegypti
| Lineage/strain | Blood meal titer (log10 FFUs/mL) | Number infected/number engorged | Infected* (%) | Number disseminated/ number engorged | Total dissemination† (%) | Number disseminated/ number infected | Disseminated from infected midgut‡ (%) |
|---|---|---|---|---|---|---|---|
| I | |||||||
| FSL699 | 6.48 | 15/28 | 54 | 7/28 | 25 | 7/15 | 47 |
| NFI52 | 6.38 | 17/32 | 53 | 10/32 | 31 | 10/17 | 59 |
| FMD2303 | 5.78 | 22/46 | 48 | 2/46 | 4 | 2/22 | 9 |
| II | |||||||
| FPI01617 | 6.60 | 8/27 | 30 | 2/27 | 7 | 2/8 | 25 |
| FPI00154 | 6.25 | 13/30 | 43 | 2/30 | 7 | 2/13 | 15 |
| NFI1159 | 5.48 | 19/30 | 63 | 4/30 | 13 | 4/19 | 21 |
No significant difference between lineages I and II, Student's t test on arcsin-square root-transformed percentage values (df = 4; P = 0.55).
No significant difference between lineages I and II, Student's t test on arcsin-square root-transformed percentage values (df = 4; P = 0.33).
No significant difference between lineages I and II, Student's t test on arcsin-square root-transformed percentage values (df = 4; P = 0.37).
Discussion
Using whole-genome sequences, we have shown that the DENV-2 responsible for a severe DF outbreak in Loreto, Peru during 2010 and 2011 belongs to the southeast Asian/American lineage II and was genetically different from DENV-2 southeast Asian/American lineage I, which had previously circulated in the region. Two amino acid substitutions occurred during the emergence of the 2010 and 2011 outbreak lineage, but only one (NS5 A81V) was the result of positive selection. Because continuous substitutions and adaptations were observed along the branches of this lineage, it is not clear what role these particular substitutions played in the emergence of the 2010 and 2011 DENV-2 outbreak. We detected a replication advantage for lineage II over lineage I viruses in Huh-7 mammalian cells but not in mo-DCs or C6/36 mosquito cells. While acknowledging the many limitations of cell culture as a model for replication in vivo, these results suggest that southeast Asian/American DENV-2 lineage II viruses do not differ from lineage I viruses with respect to their replication kinetics in human mo-DCs or mosquito cells. Additionally, we did not detect a difference in infection or dissemination rates between the two lineages when Ae. aegypti mosquitoes were infected with these viruses.
Our results suggest that the emergence of lineage II during the severe 2010 and 2011 Loreto outbreak was not attributable to an advantage in replication in humans or mosquitoes. This finding contrasts with other studies of DENV lineage invasions that showed such an advantage.36,51,58 Notably, although other lineage/genotype replacement events associated with increased disease severity involved a period of lineage/genotype cocirculation, the southeast Asian/American lineage II viruses from our study were introduced into Loreto in the absence of any DENV-2 competitors.59,60
It is clear that both host and viral factors can play a role in determining dengue disease severity. Our results indicate that the large number of severe dengue cases observed during the 2010 and 2011 Loreto outbreak was not associated with increased viral replication in human mo-DCs or dissemination in mosquitoes. Instead, the severity of the 2010 and 2011 Loreto outbreak may have been caused by the introduction of southeast Asian/American lineage II DENV-2 at just the right (or wrong, depending on perspective) time in the context of the population's pre-existing immunity to other DENVs. It has previously been shown that particular sequences of dengue infection can affect disease severity.27,61 Even more specifically, it has been shown that disease severity in the context of a given immunological background can be affected by the genotype/clade of the DENV-2 infecting virus.59,62 In Iquitos, a single DENV serotype typically dominates, and historically, the introduction of new DENV serotypes has resulted in epidemics of DF. These epidemics in Iquitos have been well-documented and were marked by the introduction of American DENV-2 in 1995, DENV-3 in 2001, and DENV-4 in 2008. Additional studies addressing the role that pre-existing immunity played in this outbreak will provide valuable insights about this outbreak and determinants of dengue severity in general.
ACKNOWLEDGMENTS
The authors thank Robert Tesh and Hilda Guzman for providing DENV antisera and the Japanese Health Sciences Foundation, Osaka, for providing the Huh-7 cells (clone JTC-39).
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the US Government. Some of the authors are military service members or employees of the US Government. This work was prepared as part of their official duties. Title 17 USC §105 provides that “[c]opyright protection under this title is not available for any work of the United States Government.” Title 17 USC §101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person's official duties.
Footnotes
Financial support: This work was supported by the Global Emerging Infections Surveillance and Response System, a Division of the Armed Forces Health Surveillance Center (Work Unit Number 800000.82000.25GB.B0016) and the Military Infectious Diseases Research Program (Work Unit Number 6000 RAD1.S.B0302). W.L.J. and K.A.H. were supported by National Center for Research Resources Grant 5P20RR016480-12 and National Institute of General Medical Sciences Grant 8 P20 GM103451-12. N.V. was supported by start-up funds provided by the Department of Pathology, University of Texas Medical Branch and NIH contract HHSN272201000040I/HHSN27200004/D04.
Authors' addresses: Maya Williams, Viral and Rickettsial Diseases Department, Naval Medical Research Center, Silver Spring, MD, E-mail: maya.williams@med.navy.mil. Sandra V. Mayer and Rubing Chen, Department of Pathology and Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX, E-mails: samayer@utmb.edu and ruchen@utmb.edu. William L. Johnson, Colorado School of Public Health, Fort Collins, CO, E-mail: w.johnson@rams.colostate.edu. Kathryn A. Hanley, Department of Biology, New Mexico State University, Las Cruces, NM, E-mail: khanley@nmsu.edu. Evgeniya Volkova, Laboratory of Emerging Pathogens, Food and Drug Administration Center for Biologics Evaluation and Research, Division of Emerging and Transfusion Transmitted Diseases, Bethesda, MD, E-mail: Evgeniya.volkova@fda.hhs.gov. Stalin Vilcarromero, Department of Virology, US Naval Medical Research Unit No. 6, Iquitos, Peru, E-mail: stalinf@yahoo.com. Steven G. Widen and Thomas G. Wood, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, E-mails: sgwiden@utmb.edu and tgwood@utmb.edu. Luis Suarez-Ognio, Faculty of Health Sciences, Universidad de Ciencias Aplicadas, Lima, Peru, E-mail: luis.suarez@upc.edu.pe. Kanya C. Long, Department of Biology, Andrews University, Berrien Springs, MI, E-mail: kanya@andrews.edu. Amy C. Morrison, Department of Virology, US Naval Medical Research Unit No. 6, Iquitos, Peru, and Entomology Department, University of California, Davis, CA, E-mail: amy.aegypti@gmail.com. Nikos Vasilakis, Department of Pathology, Center for Biodefense and Emerging Infectious Diseases and Institute for Human Infections and Immunity and Center for Tropical Diseases, University of Texas Medical Branch, Galveston, TX, E-mail: nivasila@utmb.edu. Eric S. Halsey, Malaria Branch, US Centers for Disease Control and Prevention, Atlanta, GA, E-mail: ehalsey@cdc.gov.
References
- 1.Armstrong PM, Rico-Hesse R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am J Trop Med Hyg. 2003;68:539–544. doi: 10.4269/ajtmh.2003.68.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 4.Anderson KB, Chunsuttiwat S, Nisalak A, Mammen MP, Libraty DH, Rothman AL, Green S, Vaughn DW, Ennis FA, Endy TP. Burden of symptomatic dengue infection in children at primary school in Thailand: a prospective study. Lancet. 2007;369:1452–1459. doi: 10.1016/S0140-6736(07)60671-0. [DOI] [PubMed] [Google Scholar]
- 5.Beaute J, Vong S. Cost and disease burden of dengue in Cambodia. BMC Public Health. 2010;10:521. doi: 10.1186/1471-2458-10-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Min N. Assessment of dengue hemorrhagic fever in Myanmar. Southeast Asian J Trop Med Public Health. 2000;31:636–641. [PubMed] [Google Scholar]
- 7.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luz PM, Grinsztejn B, Galvani AP. Disability adjusted life years lost to dengue in Brazil. Trop Med Int Health. 2009;14:237–246. doi: 10.1111/j.1365-3156.2008.02203.x. [DOI] [PubMed] [Google Scholar]
- 9.Meltzer MI, Rigau-Perez JG, Clark GG, Reiter P, Gubler DJ. Using disability-adjusted life years to assess the economic impact of dengue in Puerto Rico: 1984–1994. Am J Trop Med Hyg. 1998;59:265–271. doi: 10.4269/ajtmh.1998.59.265. [DOI] [PubMed] [Google Scholar]
- 10.Anez G, Balza R, Valero N, Larreal Y. Economic impact of dengue and dengue hemorrhagic fever in the State of Zulia, Venezuela, 1997–2003. Rev Panam Salud Publica. 2006;19:314–320. doi: 10.1590/s1020-49892006000500004. [DOI] [PubMed] [Google Scholar]
- 11.Armien B, Suaya JA, Quiroz E, Sah BK, Bayard V, Marchena L, Campos C, Shepard DS. Clinical characteristics and national economic cost of the 2005 dengue epidemic in Panama. Am J Trop Med Hyg. 2008;79:364–371. [PubMed] [Google Scholar]
- 12.Clark DV, Mammen MP, Jr, Nisalak A, Puthimethee V, Endy TP. Economic impact of dengue fever/dengue hemorrhagic fever in Thailand at the family and population levels. Am J Trop Med Hyg. 2005;72:786–791. [PubMed] [Google Scholar]
- 13.Garg P, Nagpal J, Khairnar P, Seneviratne SL. Economic burden of dengue infections in India. Trans R Soc Trop Med Hyg. 2008;102:570–577. doi: 10.1016/j.trstmh.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Harving ML, Ronsholt FF. The economic impact of dengue hemorrhagic fever on family level in Southern Vietnam. Dan Med Bull. 2007;54:170–172. [PubMed] [Google Scholar]
- 15.Huy R, Wichmann O, Beatty M, Ngan C, Duong S, Margolis HS, Vong S. Cost of dengue and other febrile illnesses to households in rural Cambodia: a prospective community-based case-control study. BMC Public Health. 2009;9:155. doi: 10.1186/1471-2458-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepard DS, Coudeville L, Halasa YA, Zambrano B, Dayan GH. Economic impact of dengue illness in the Americas. Am J Trop Med Hyg. 2011;84:200–207. doi: 10.4269/ajtmh.2011.10-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suaya JA, Shepard DS, Siqueira JB, Martelli CT, Lum LC, Tan LH, Kongsin S, Jiamton S, Garrido F, Montoya R, Armien B, Huy R, Castillo L, Caram M, Sah BK, Sughayyar R, Tyo KR, Halstead SB. Cost of dengue cases in eight countries in the Americas and Asia: a prospective study. Am J Trop Med Hyg. 2009;80:846–855. [PubMed] [Google Scholar]
- 18.Valdes LG, Mizhrahi JV, Guzman MG. Economic impact of dengue 2 epidemic in Santiago de Cuba, 1997. Rev Cubana Med Trop. 2002;54:220–227. [PubMed] [Google Scholar]
- 19.San Martin JL, Brathwaite O, Zambrano B, Solorzano JO, Bouckenooghe A, Dayan GH, Guzman MG. The epidemiology of dengue in the Americas over the last three decades: a worrisome reality. Am J Trop Med Hyg. 2010;82:128–135. doi: 10.4269/ajtmh.2010.09-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, Vazques S, Delgado I, Halstead SB. Epidemiologic studies on dengue in Santiago de Cuba, 1997. Am J Epidemiol. 2000;152:793–799. doi: 10.1093/aje/152.9.793. [DOI] [PubMed] [Google Scholar]
- 21.Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 22.Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de Chacon, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halstead SB. Dengue in the Americas and Southeast Asia: do they differ? Rev Panam Salud Publica. 2006;20:407–415. doi: 10.1590/s1020-49892006001100007. [DOI] [PubMed] [Google Scholar]
- 24.Spence L, Jonkers AH, Casals J. Dengue type 3 virus isolated from an antiguan patient during the 1963–64 Caribbean epidemic. Am J Trop Med Hyg. 1969;18:584–587. doi: 10.4269/ajtmh.1969.18.584. [DOI] [PubMed] [Google Scholar]
- 25.Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 26.Kouri GP, Guzman MG, Bravo JR, Triana C. Dengue haemorrhagic fever/dengue shock syndrome: lessons from the Cuban epidemic, 1981. Bull World Health Organ. 1989;67:375–380. [PMC free article] [PubMed] [Google Scholar]
- 27.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 28.Bennett SN, Drummond AJ, Kapan DD, Suchard MA, Munoz-Jordan JL, Pybus OG, Holmes EC, Gubler DJ. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27:811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–809. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steel A, Gubler DJ, Bennett SN. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology. 2010;405:505–512. doi: 10.1016/j.virol.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnes WJ, Rosen L. Fatal hemorrhagic disease and shock associated with primary dengue infection on a Pacific island. Am J Trop Med Hyg. 1974;23:495–506. doi: 10.4269/ajtmh.1974.23.495. [DOI] [PubMed] [Google Scholar]
- 32.Loison G, Rosen L, Papillaud J, Tomasini J, Vaujany J, Chanalet G. La dengue en Nouvelle-Caledonie (1971–1972) Bull Soc Pathol Exot. 1973;66:511–519. [Google Scholar]
- 33.Lopez-Correa RH, Cline BL, Ramirez-Ronda C, Bermudez R, Sather GE, Kuno G. Dengue fever with hemorrhagic manifestations: a report of three cases from Puerto Rico. Am J Trop Med Hyg. 1978;27:1216–1224. doi: 10.4269/ajtmh.1978.27.1216. [DOI] [PubMed] [Google Scholar]
- 34.Moreau JP, Rosen L, Saugrain J, Lagraulet J. An epidemic of dengue on Tahiti associated with hemorrhagic manifestations. Am J Trop Med Hyg. 1973;22:237–241. doi: 10.4269/ajtmh.1973.22.237. [DOI] [PubMed] [Google Scholar]
- 35.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- 36.Cologna R, Armstrong PM, Rico-Hesse R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J Virol. 2005;79:853–859. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cologna R, Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J Virol. 77. 2003:3929–3938. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasilakis N, Shell EJ, Fokam EB, Mason PW, Hanley KA, Estes DM, Weaver SC. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007;358:402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watts DM, Porter KR, Putvatana P, Vasquez B, Calampa C, Hayes CG, Halstead SB. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354:1431–1434. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]
- 40.Forshey BM, Guevara C, Laguna-Torres VA, Cespedes M, Vargas J, Gianella A, Vallejo E, Madrid C, Aguayo N, Gotuzzo E, Suarez V, Morales AM, Beingolea L, Reyes N, Perez J, Negrete M, Rocha C, Morrison AC, Russell KL, Blair PJ, Olson JG, Kochel TJ. Group NFSW Arboviral etiologies of acute febrile illnesses in Western South America, 2000–2007. PLoS Negl Trop Dis. 2010;4:e787. doi: 10.1371/journal.pntd.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boletín Epidemiológico . (52) Vol. 20. Dirección General de Epdemiologia MoH; Peru: 2011. http://www.dge.gob.pe/boletin.php Available at. Accessed September 5, 2013. [Google Scholar]
- 42.Durand Velazco S, Fiestas Solorzano V, Sihuincha Maldonado M, Chavez Lencinas C, Vasquez Vela V, Torrejon Flores C, Rodriguez Ferruchi H, Cabezas Sanchez C. Impact of the dengue epidemic due to a new lineage of DENV-2 American/ Asian genotype in the health services demand in hospital “Cesar Garayar Garcia”, Iquitos. Rev Peru Med Exp Salud Publica. 2011;28:157–159. doi: 10.1590/s1726-46342011000100027. [DOI] [PubMed] [Google Scholar]
- 43.Morrison AC, Minnick SL, Rocha C, Forshey BM, Stoddard ST, Getis A, Focks DA, Russell KL, Olson JG, Blair PJ, Watts DM, Sihuincha M, Scott TW, Kochel TJ. Epidemiology of dengue virus in Iquitos, Peru 1999 to 2005: interepidemic and epidemic patterns of transmission. PLoS Negl Trop Dis. 2010;4:e670. doi: 10.1371/journal.pntd.0000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamani E, Alvarez C, Garcia MM, Figueroa D, Gatti M, Guio H, Merino S, Valencia P, Calampa C, Franco L, Cabezas C. Circulation of a different lineage of dengue virus serotype 2 American/Asian genotype in the Peruvian Amazon, 2010. Rev Peru Med Exp Salud Publica. 2011;28:72–77. doi: 10.1590/s1726-46342011000100011. [DOI] [PubMed] [Google Scholar]
- 45.Rocha C, Morrison AC, Forshey BM, Blair PJ, Olson JG, Stancil JD, Sihuincha M, Scott TW, Kochel TJ. Comparison of two active surveillance programs for the detection of clinical dengue cases in Iquitos, Peru. Am J Trop Med Hyg. 2009;80:656–660. [PubMed] [Google Scholar]
- 46.Forshey BM, Laguna-Torres VA, Vilcarromero S, Bazan I, Rocha C, Morrison AC, Stoddard ST, Alegre Y, Gomez J, Scott TW, Kochel TJ. Epidemiology of influenza-like illness in the Amazon Basin of Peru, 2008–2009. Influenza Other Respi Viruses. 2010;4:235–243. doi: 10.1111/j.1750-2659.2010.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoddard ST, Forshey BM, Morrison AC, Paz-Soldan VA, Vazquez-Prokopec GM, Astete H, Reiner RC, Jr, Vilcarromero S, Elder JP, Halsey ES, Kochel TJ, Kitron U, Scott TW. House-to-house human movement drives dengue virus transmission. Proc Natl Acad Sci USA. 2013;110:994–999. doi: 10.1073/pnas.1213349110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 50.Vasilakis N, Fokam EB, Hanson CT, Weinberg E, Sall AA, Whitehead SS, Hanley KA, Weaver SC. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology. 2008;377:296–307. doi: 10.1016/j.virol.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanley KA, Nelson JT, Schirtzinger EE, Whitehead SS, Hanson CT. Superior infectivity for mosquito vectors contributes to competitive displacement among strains of dengue virus. BMC Ecol. 2008;8:1. doi: 10.1186/1472-6785-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.WHO . Dengue Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. Geneva: World Health Organization; 2009. [PubMed] [Google Scholar]
- 53.Fiestas Solorzano V, Sihuincha Maldonado M, Donaires Toscano F, Durand Velazco S, Garcia MM, Mamani E, Gomez de la Torre Pretell J. Clinical characteristics of patients admitted to hospital “Cesar Garayar Garcia”, Iquitos during the dengue epidemic, January–February 2011. Rev Peru Med Exp Salud Publica. 2011;28:78–82. doi: 10.1590/s1726-46342011000100012. [DOI] [PubMed] [Google Scholar]
- 54.Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blackley S, Kou Z, Chen H, Quinn M, Rose RC, Schlesinger JJ, Coppage M, Jin X. Primary human splenic macrophages, but not T or B cells, are the principal target cells for dengue virus infection in vitro. J Virol. 2007;81:13325–13334. doi: 10.1128/JVI.01568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for dengue virus dissemination. PLoS Pathog. 2012;8:e1002465. doi: 10.1371/journal.ppat.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6:816–820. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- 58.Lambrechts L, Fansiri T, Pongsiri A, Thaisomboonsuk B, Klungthong C, Richardson JH, Ponlawat A, Jarman RG, Scott TW. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J Virol. 2012;86:1853–1861. doi: 10.1128/JVI.06458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, Nunez A, Lennon NJ, Birren BW, Gordon A, Henn MR, Harris E. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3003084. 114ra128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anez G, Morales-Betoulle ME, Rios M. Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS ONE. 2011;6:e27459. doi: 10.1371/journal.pone.0027459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guzman MG, Kouri G. Dengue haemorrhagic fever integral hypothesis: confirming observations, 1987–2007. Trans R Soc Trop Med Hyg. 2008;102:522–523. doi: 10.1016/j.trstmh.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Kochel TJ, Watts DM, Halstead SB, Hayes CG, Espinoza A, Felices V, Caceda R, Bautista CT, Montoya Y, Douglas S, Russell KL. Effect of dengue-1 antibodies on American dengue-2 viral infection and dengue haemorrhagic fever. Lancet. 2002;360:310–312. doi: 10.1016/S0140-6736(02)09522-3. [DOI] [PubMed] [Google Scholar]