Abstract
Several nucleoside analogs are under clinical development for use against hepatitis B virus (HBV). Lamivudine (3TC), a nucleoside analog, and adefovir dipivoxil (ADV), an acyclonucleotide analog, are clinically approved. However, long-term treatment can induce viral resistance, and following the cessation of therapy, viral rebound is frequently observed. There continues to be a need for new antiviral agents with novel mechanisms of action. A library of more than 600 di- and trinucleotide compounds synthesized by parallel synthesis using a combinatorial strategy was screened for potential inhibitors of HBV replication using the chronically HBV-producing cell line 2.2.15. Through an iterative process of synthesis, lead optimization, and screening, three analogs were identified as potent inhibitors of HBV replication: dinucleotides ORI-7246 (drug concentration at which a 10-fold reduction of HBV DNA was observed [EC90], 1.4 μM) and ORI-9020 (EC90, 1.2 μM) and trinucleotide ORI-7170 (EC90, 7.2 μM). These analogs inhibited the replication of both strands of HBV DNA. No suppression of HBV protein synthesis or intracellular core particle formation by these analogs was observed. No inhibition of HBV DNA strand elongation by the analogs or their 5′-triphosphate versions was apparent in in vitro polymerase assays. Although the exact mechanism of action is not yet identified, present data are consistent with an inhibition of the HBV reverse transcriptase-directed priming step prior to elongation of the first viral DNA strand. In transient-transfection assays, these analogs inhibited the replication of 3TC-resistant HBV. Synergistic interactions in combination treatments between the analogs and either 3TC or ADV were observed. These compounds represent a novel class of anti-HBV molecules and warrant further investigation as potential therapeutic agents.
The discovery of safe and effective antiviral drugs continues to present considerable challenges. The rapid emergence of resistance to antiviral drugs is a major problem, and combinations of a limited repertoire of antiviral drugs often need to be employed as a therapeutic strategy. The major stumbling block in antiviral drug development is the limited structural, mechanistic, and functional information on many virus-specific molecular targets. Consequently, the design of target-specific compounds that interfere with viral life cycles is a difficult challenge. There continues to be a substantial unmet clinical need for antiviral drugs with different structures and unique mechanisms of action, other than those conferred by conventional nucleoside analogs (2).
Given a lack of sufficient structural information on new targets, an approach that seems appropriate for antiviral drug discovery is the screening of structurally diverse compounds, generated using combinatorial approaches that would modulate biological pathways without regard to specific molecular targets. In theory, this approach would allow simultaneous functional validation of a target, as well as the discovery of a lead structure that modulates the function of the target. This strategy has been variously referred to as “diversity-oriented organic synthesis for therapeutic target validation” or “combinatorial target-guided ligand assembly” (6, 20).
We describe here the application of this concept for the discovery of anti-hepatitis B virus (HBV) agents. In recent publications, we have reported methods of assembling various classes of di- and trinucleotide libraries (7, 8, 18). Through screening, as well as lead optimization work, and in conjunction with cell-based assays, we have discovered that certain molecules show very promising anti-HBV activity. Furthermore, some of these molecules display synergistic activity when used in combination with lamivudine (3TC) and adefovir dipivoxil (ADV), the two nucleoside analogs currently licensed for the treatment of chronic HBV infection. This work summarizes the in vitro analysis and preliminary toxicity evaluation of these compounds.
(A preliminary report of this work was presented at the 14th International Conference for Antiviral Research, Seattle, Washington, 8 to 12 April 2001.)
MATERIALS AND METHODS
Test compounds.
Nucleotide libraries were synthesized and purified as previously described (7, 8, 18). All test compounds were solubilized in sterile, distilled water. Daily aliquots of test compounds were made in individual tubes and stored at −20°C. On each day of treatment, daily aliquots of the test compounds were suspended in culture medium at room temperature and immediately added to the cell cultures, thereby subjecting each aliquot of test compound to only one freeze-thaw cycle.
Antiviral analysis.
For the antiviral analyses, confluent cultures of 2.2.15 (21) cells were maintained on 96-well flat-bottom tissue culture plates in RPMI 1640 medium with 2% fetal bovine serum (11). Cultures (six per each test concentration on two replicate plates) were treated with nine consecutive daily doses of the test compounds. Medium was changed daily with fresh test compounds. HBV nucleic acid and protein levels were measured 24 h after the last treatment. Extracellular (virion) HBV DNA levels were assessed by quantitative blot hybridization (11). Intracellular HBV DNA levels were measured by quantitative Southern blot hybridization (11).
Uptake of neutral red dye was used to determine the relative level of toxicity 24 h following the last treatment (11). The A510 of internalized dye was used for the semiquantitative analysis. Values are presented as a percentage of the average A510 values (± standard deviations) in nine separate cultures of untreated cells maintained on the 96-well plates seeded at the same time with the identical pool of stock cells used for the antiviral analyses and maintained in an identical manner. A total of three cultures were treated with each concentration of test compound.
Combination treatments.
Combination treatments were conducted as previously described (9). Briefly, two agents were mixed together at a predetermined concentration ratio. The relative ratios of the individual agents were based on the monotherapy values of each compound (drug concentrations at which a 10-fold reduction of HBV DNA was observed [EC90s]). For each combination of agents, three concentration ratios, centered upon the use of the compounds at equipotent antiviral concentrations, were used. A dilution series (six threefold-concentration steps, beginning at the approximate EC90s) was then generated with the concentration ratio of the two agents remaining the same in each dilution step. Separate dilution series of monotherapy with each individual antiviral agent at the same concentrations were also used to treat cultures in the same experiment. Toxicity analyses were performed as described above for the monotherapies.
Analysis of drug interactions in the combination studies was determined by the use of the CALCUSYN program (Biosoft, Inc., Cambridge, United Kingdom). This program evaluates synergy, additivity, or antagonism by use of several methodologies, including that of Chou and Talalay with a statistical analysis employing the Monte Carlo technique (3) to provide confidence limits, fraction-affected-confidence interval (FA-CI) plots, isobolograms, and median-effect plots.
Analysis of HBV polymerase activity and HBV protein expression.
An endogenous DNA polymerase activity assay was performed as previously described (14). Briefly, Huh7 cells transfected with HBV DNA were treated with 10 or 50 μM concentrations of test compounds for 72 h. HBV cores were then immunoprecipitated from cell lysates and endogenous polymerase activity was quantified by phosphorimager analysis of band intensities, normalized to that of core proteins from untreated controls.
Intracellular HBV RNA levels in 2.2.15 cells were measured by quantitative Northern blot hybridization (13). Extracellular HBV surface (HBsAg) and e (HBeAg) antigen levels produced from 2.2.15 cells were evaluated by semiquantitative enzyme immunoassay (EIA) methods using commercial kits (HBsAg, Abbott Laboratories; HBeAg, Diasorin, Inc.) as previously described (12). Intracellular HBV core antigen (HBcAg) levels in 2.2.15 cells were assessed using a semiquantitative EIA as previously described (12).
3TC-resistant HBV mutants.
Analysis of antiviral activity against clinically relevant HBV mutants associated with resistance to 3TC and famciclovir was performed using a previously described transfection-based assay (25). For these studies, Huh7 cell cultures were treated with four consecutive daily doses of the test compounds beginning 4 days posttransfection. HBV constructs were those described by Allen et al. (1), and the nomenclature for the drug-resistant mutants is that described by Stuyver et al. (23).
Additional analyses.
Additional cytotoxicity analyses were conducted in the following cell lines as previously described (4): MDBK cells, human foreskin fibroblasts, Vero cells, and human peripheral blood mononuclear cells. In brief, solutions of the compounds were prepared at concentrations of 100, 300, and 1,000 μM in Dulbecco's modified Eagle medium containing 2% fetal bovine serum. The cells were plated in 96-well plates. After 24 h, the compounds were added and the cells were incubated at 37°C in 5% CO2 for 4 days. Relative viability was assessed by the addition of the MTT dye solution (G401B; Promega, Inc.) to the wells. The A570 was recorded.
An in vitro antiviral selectivity assay was conducted against human immunodeficiency virus (HIV) as follows (17). Cord blood mononuclear cells were infected with HIV type 1 IIIb (laboratory strain) for 2 h. The free virus was washed off, and the cells were transferred to plates. The compounds were added as a solution in water. Following 7 days of incubation in culture, the reverse transcriptase (RT) activity of the supernatant was measured to determine antiviral activity as previously described (17). Selectivity assays against other viruses were performed using standard cytopathic effect assays as previously described (7).
Liver microsomal assays were conducted as previously described (5). Briefly, incubations were performed in triplicate at 37°C using 1 and 10 μM concentrations of the compounds in human and mouse liver microsomes, and results were analyzed by reversed-phase high-performance liquid chromatography after 1 h (8).
RESULTS
Anti-HBV activity of di- and trinucleotides.
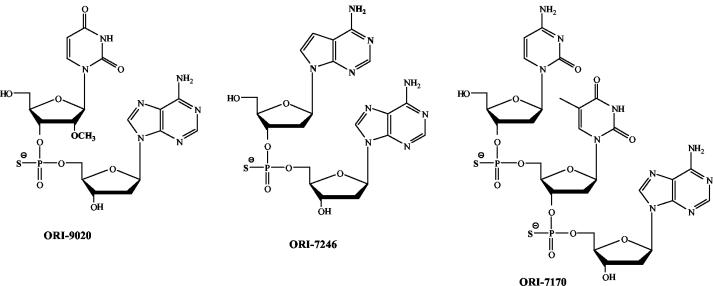
A library of over 600 compounds having a molecular mass range of 550 to 700 kDa (7, 8, 18) was initially screened in 2.2.15 cells at 10 μM (data not shown). The initial library of compounds that were synthesized and evaluated consisted mainly of phosphorothioate analogs as the internucleotidic phosphorothioate linkage is known to provide significant resistance to nuclease-mediated degradation of di- and polynucleotides (8). Those compounds that demonstrated at least 70% inhibition were then resynthesized and subjected to repeat screening. A number of structural analogs of these initial leads were also synthesized and evaluated. This process of iterative synthesis and screening were carried out to select three compounds, ORI-7170, ORI-7246, and ORI-9020, which demonstrated consistent, and potent, anti-HBV activity (Fig. 1; Table 1).
FIG. 1.
Structures of active compounds from this study.
TABLE 1.
Relative potencies of analogs against HBV replication in 2.2.15 cellsa
| Compound | CC50 (μM) (mean ± SD) | Extracellular virion DNA (mean ± SD)
|
Intracellular RI (mean ± SD)
|
SI (CC50/EC90)
|
|||
|---|---|---|---|---|---|---|---|
| EC50 (μM) | EC90 (μM) | EC50 (μM) | EC90 (μM) | Virion | RI | ||
| 3TC | 2,265 ± 86 | 0.061 ± 0.007 | 0.178 ± 0.021 | 0.162 ± 0.024 | 0.473 ± 0.061 | 12,724 | 4,789 |
| ORI-7170 | >300$ | 2.4 ± 0.3 | 7.5 ± 0.8 | 8.4 ± 1.0 | 14 ± 1.2 | >40 | >21 |
| ORI-7246 | >300$ | 0.325 ± 0.044 | 1.7 ± 0.5 | 1.5 ± 0.3 | 5.8 ± 0.5 | >588 | >172 |
| ORI-9020 | >1,000$ | 0.258 ± 0.038 | 1.5 ± 0.2 | 1.4 ± 0.3 | 5.3 ± 0.6 | >667 | >189 |
Values presented were calculated by linear regression analysis (two to four experiments; four to six replicates per concentration). EC50, drug concentration at which a 2-fold depression of HBV DNA (relative to the average levels in untreated cultures), respectively, was observed. The EC90s were used for the calculation of the selectivity index (SI) since at least a threefold depression of HBV RI levels is typically required to achieve statistical significance in this assay system (11). RI, HBV DNA RIs; “$”, no significant cytotoxic effect was observed at the highest (listed) concentration tested.
Analogs ORI-7170, ORI-7246, and ORI-9020 induced significant reductions in extracellular (virion) HBV DNA levels produced by 2.2.15 cells (Table 1). The relative potencies varied severalfold, and two compounds, ORI-7246 and ORI-9020, exhibited anti-HBV potencies that were approximately 10-fold less than that observed for 3TC and approximately equal to that observed for ADV in this assay system (Tables 1 and 2). Compound ORI-7170 was less potent than ORI-7246 and ORI-9020. No apparent cytotoxicity was observed for these compounds at concentrations up to 300 μM under the conditions used for the antiviral analyses (Tables 1 and 2).
TABLE 2.
Relative potencies of combination treatments against HBV replication in 2.2.15 cellsa
| Treatment | CC50 (μM) | EC50 (μM) | EC90 (μM) | SI (CC50/EC50) | Type of interaction |
|---|---|---|---|---|---|
| 3TC | 2,304 ± 94 | 0.056 ± 0.007 | 0.170 ± 0.022 | 13,553 | |
| ADV | 425 ± 22 | 0.465 ± 0.051 | 2.4 ± 0.3 | 177 | |
| ORI-7170 | >300 | 2.2 ± 0.3 | 7.2 ± 0.8 | >42 | |
| ORI-7246 | >300 | 0.300 ± 0.041 | 1.4 ± 0.2 | >214 | |
| ORI-9020 | >300 | 0.213 ± 0.030 | 1.2 ± 0.2 | >250 | |
| ORI-7170+3TC@1:1 | >300 | 0.066 ± 0.007 | 0.238 ± 0.014 | >1,261 | Synergistic |
| ORI-7170+3TC@3:1 | >300 | 0.068 ± 0.008 | 0.258 ± 0.028 | >1,161 | Synergistic |
| ORI-7170+3TC@10:1 | >300 | 0.227 ± 0.024 | 0.831 ± 0.067 | >361 | Synergistic |
| ORI-7170+ADV@3:1 | >300 | 0.206 ± 0.016 | 0.656 ± 0.044 | >457 | Synergistic |
| ORI-7170+ADV@1:1 | >300 | 0.272 ± 0.037 | 0.998 ± 0.083 | >301 | Additive, synergistic |
| ORI-7170+ADV@1:3 | >300 | 0.264 ± 0.028 | 0.901 ± 0.039 | >333 | Additive, antagonistic |
| ORI-7246+3TC@1:1 | >300 | 0.015 ± 0.002 | 0.281 ± 0.038 | >1,068 | Synergistic |
| ORI-7246+3TC@3:1 | >300 | 0.103 ± 0.015 | 0.315 ± 0.045 | >953 | Synergistic |
| ORI-7246+3TC@10:1 | >300 | 0.428 ± 0.065 | 2.3 ± 0.2 | >131 | Antagonistic |
| ORI-7246+ADV@3:1 | >300 | 1.3 ± 0.2 | 3.0 ± 0.3 | >100 | Antagonistic |
| ORI-7246+ADV@1:1 | >300 | 0.239 ± 0.030 | 0.903 ± 0.062 | >332 | Additive, synergistic |
| ORI-7246+ADV@1:3 | >300 | 0.083 ± 0.012 | 0.283 ± 0.029 | >1,060 | Synergistic |
| ORI-9020+3TC@1:1 | >300 | 0.026 ± 0.004 | 0.138 ± 0.011 | >2,174 | Synergistic |
| ORI-9020+3TC@3:1 | >300 | 0.090 ± 0.011 | 0.292 ± 0.032 | >1,027 | Synergistic |
| ORI-9020+3TC@10:1 | >300 | 0.207 ± 0.021 | 1.0 ± 0.1 | >300 | Additive |
| ORI-9020+ADV@3:1 | >300 | 2.1 ± 0.3 | 4.5 ± 0.5 | >67 | Antagonistic |
| ORI-9020+ADV@1:1 | >300 | 0.903 ± 0.102 | 1.7 ± 0.2 | >176 | Antagonistic |
| ORI-9020+ADV@1:3 | >300 | 0.187 ± 0.028 | 0.456 ± 0.049 | >658 | Additive |
Values presented (means ± standard deviations) were calculated by linear regression analysis (two experiments; eight replicates per concentration). For the combinations, values are expressed as a concentration of the first compound listed in the mixture. Corresponding concentrations of the second compound can be calculated from the indicated molar ratios. Analysis of the interaction between the compounds in the combination treatments was determined by analysis using Combostat (Biosoft, Inc.).
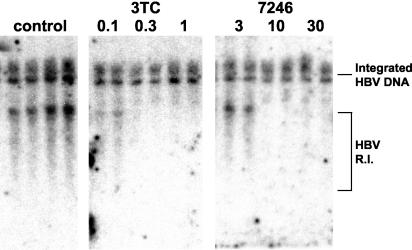
These compounds were also effective inhibitors of intracellular HBV replication, with anti-HBV potencies paralleling those observed against HBV virion production (Table 1). The compounds appeared to inhibit both strands of HBV DNA, similar to that observed for 3TC (Fig. 2, top panel).
FIG. 2.
Effect of analog ORI-7246 on intracellular HBV replication in 2.2.15 cells. Cultures were exposed to the indicated concentrations of agents for 9 consecutive days. DNA was extracted 24 h following the final addition of compounds and analyzed by Southern blot hybridization for HBV DNA.
In order to ascertain if the analogs or their triphosphate forms had activity as chain terminators of elongation by HBV polymerase, the 5′-triphosphate analogs of ORI-7170, ORI-7246, and ORI-9020 were synthesized and used in an HBV DNA polymerase chain elongation assay (14). When used in concentrations up to 50 μM, none of the analogs or their 5′-triphosphate forms produced any significant inhibition of the HBV activity in chain elongation (data not shown).
None of the analogs tested induced significant reductions in the levels of intracellular HBV RNA or in the levels of extracellular HBsAg, extracellular HBeAg, or intracellular HBcAg produced by 2.2.15 cells in this assay as assessed by semiquantitative EIA (12), and none of them affected the levels of intracellular core particles as assessed by an immune blot assay (14) (data not shown).
Combination treatments.
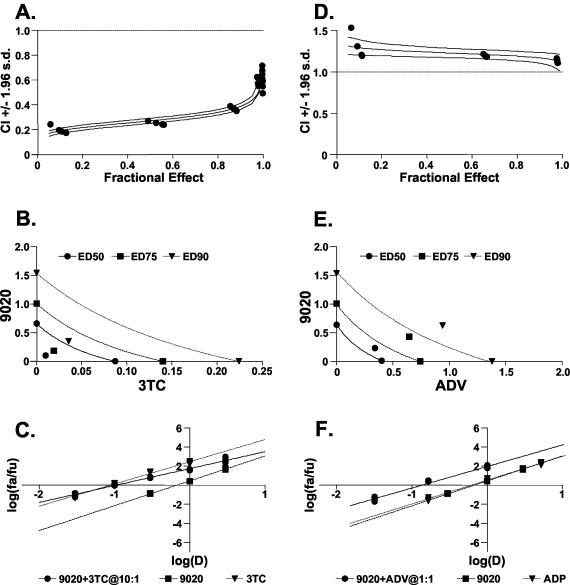
All three analogs appeared to work cooperatively overall in combination with 3TC (Table 2; Fig. 3). In essentially all mixtures examined, these compounds acted in a generally synergistic manner against HBV production in 2.2.15 cells. Combinations of the compounds and ADV generally exhibited less favorable interactions (Table 2; Fig. 3). In several instances, antagonistic interactions with ADV were observed, and only the trinucleotide, ORI-7170, exhibited synergistic interactions with this nucleotide analog. The most favorable interactions in the combination treatments were generally observed at lower relative concentrations of 3TC, and at higher relative concentrations of ADV, to the analogs. The cytotoxicity profiles for these compounds were not changed by the addition of either 3TC or ADV in these combination treatments (Table 2).
FIG. 3.
Examples of the analysis of compound interactions during combination treatments. The interactions between ORI-9020 and either 3TC (A to C) or ADV (D to F) by the CALCUSYN software program (Biosoft, Inc., Cambridge, United Kingdom) are displayed. Three types of drug interaction analyses are displayed: drug-effect plots (A and D), FA-CI plots (with Monte Carlo analysis) (B and E), and conservative isobolograms (C and F). For the FA-CI plot analysis, a combination index (CI) greater than 1.0 indicates antagonism and a CI less than 1.0 indicates synergism. Evaluations of synergy, additivity (summation), or antagonism at different levels of virus inhibition (e.g., 5% [FA = 0.5] to 99% [FA = 0.99]) are provided. The triple lines on these plots, i.e., median values ±1.96 standard deviations, were calculated by Monte Carlo analysis (3). For the isobolograms, ED50, ED75, and ED90 values (ED, effective antiviral dose) for the combination treatments are displayed as single points. Values that fall to the left of the lines (representing values for additive interactions based on the monotherapies) indicate synergy, and values to the right indicate antagonism. For the dose-effect and median-effect plots, plotted values for the combination treatments that fall above both sets of lines for the monotherapies generally indicate synergism.
In an effort to further enhance the potency of the compounds, a number of additional structural analogs of ORI-7170, ORI-7246, and ORI-9020 were also synthesized and evaluated. In all instances, replacement of the phosphorothioate linkages by nonionic N-alkyl phosphoramidate linkages (8) reduced or eliminated the anti-HBV potencies of these compounds (data not shown). In the case of ORI-7170, analogs with chimeric phosphorothioate and phosphoramidate linkages (8) also had reduced potencies compared to ORI-7170. Compounds with other structural variations—such as substitution of the purines by pyrimidines or those resulting from switching the nucleobase positions within the di- and trinucleotide structures—had severely reduced potencies compared to those of the corresponding parent molecules (data not shown). In addition, the individual deoxy- and ribonucleosides such as 2′-OMe U, dA, dC, and T were devoid of anti-HBV activity in these assays (data not shown).
Activity against 3TC-resistant HBV.
In a standard DNA-transfection assay (25), analogs ORI-7246 and ORI-9020 were active against 3TC-resistant HBV variants (Table 3). Essentially identical potencies were observed against wild-type virus and constructs carrying either C domain (M204V/I) or B domain (L180M) mutations (1) that are clinically known to confer differing levels of resistance to 3TC (but not ADV).
TABLE 3.
Relative potencies of analogs against HBV drug-resistant HBV mutantsa
| HBV construct | Mean EC90 ± SD (μM)
|
|||
|---|---|---|---|---|
| 3TC | ADV | ORI-7246 | ORI-9020 | |
| Wild type | 0.6 ± 0.1 | 7.8 ± 1.0 | 9.9 ± 1.0 | 9.0 ± 1.2 |
| M204V | >100 | 9.5 ± 1.2 | 12 ± 1.6 | 9.8 ± 1.0 |
| M204I | >100 | 9.0 ± 1.1 | 11 ± 1.5 | 10 ± 1.6 |
| L180M | 18 ± 2.5 | 10 ± 1.5 | 13 ± 1.8 | 12 ± 1.7 |
Cultures were treated for 3 days with different doses of the indicated compounds, beginning 72 h posttransfection (25). Values presented were calculated by linear regression analysis (four replicates per concentration). Nomenclature for HBV drug resistance mutations has been previously described (23).
bEC90s for the indicated intracellular RIs are given.
Additional characterization of anti-HBV dinucleotides.
A more extended analysis of compounds ORI-7246 and ORI-9020 against a number of cell lines displayed a favorable cytotoxicity profile with high values for the drug concentration at which a twofold reduction of neutral red dye uptake (relative to the average levels in untreated cultures) was observed (CC50): MDBK (800 μM), human foreskin fibroblasts (300 μM), and human peripheral blood mononuclear cells (>300 μM). Compounds ORI-7246 and ORI-9020 were found to be specific and selective inhibitors of HBV (Table 4). All three analogs were metabolically stable in liver microsomes for up to 1 h, remaining essentially undegraded (data not shown).
TABLE 4.
Activities of analogs ORI-7246 and ORI-9020 against different viruses
| Virusa | EC50 (μM)
|
|
|---|---|---|
| ORI-7246 | ORI-9020 | |
| HBV (2.2.15) | 0.2 | 0.3 |
| HCMV (AD169) | >20 | >20 |
| YFV | >50 | >50 |
| HSV (KOS) | >20 | >20 |
| HIV type 1 (IIIb) | >2.0 | >2.0 |
Abbreviations: HCMV, human cytomegalovirus; YFV, yellow fever virus; HSV, herpes simplex virus.
DISCUSSION
In this report, the anti-HBV activities of two phosphorothioate dinucleotides (ORI-7246 and ORI-9020) and a phosphorothioate trinucleotide (ORI-7170) have been described. These compounds represent a new, unique class of antiviral agents with reasonable potencies compared to the two anti-HBV nucleosides, 3TC and ADV. It is significant that these compounds inhibited intracellular HBV DNA replication, confirming that these compounds are directly inhibiting virus replication. The compounds described in this report appear to act by mechanisms different from those previously reported for dinucleotides used to inhibit HIV integrase (16) (integration of HBV is not part of the virus replication cycle [24]).
Our antiviral drug discovery strategy was to design a library of small molecules that mimic the repertoire of interactions that exist among nucleic acids and proteins. Many structural and functional proteins are known to contain nucleotide-binding domains within protein α-helices and β-sheets (19). The binding pockets can accommodate nucleotides as substrates or ligands. Presumably, the exquisite specificity of such binding interactions is due to a network of hydrogen bonding, hydrophobic, ionic, and van der Waals interactions. The nucleic acid-based (NAB) scaffold (7, 8, 18) used as a basis for the construction of the compounds discussed in this report appears to be a logical template to incorporate diversity attributes to potentially target protein-protein and protein-nucleic acid interactions. Both rigid and flexible NAB scaffolds can be used to create variable spatial display of hydrogen bonding, hydrophobic, charge transfer, electrostatic, and other noncovalent interactions. Also, by linking the individual scaffolds together, certain shape-defining motifs such as circles, pseudoknots, bulges, and stem-loops can be incorporated into libraries (7, 8, 18). Furthermore, from a synthetic perspective, the NAB libraries can be assembled using well-developed solid-phase or solution-phase methods (7, 8, 18).
The library members screened in this study are all d-nucleoside analogs and possess most of the key diversity attributes enunciated above. Synthesis of these compounds and the design rationale have been described in detail elsewhere (7, 8, 18). A key structural feature of the members of the library is that the 3′-5′ di- or trinucleotides carry phosphorothioate and phosphoramidate internucleotidic modifications which are expected to provide desirable metabolic stability to the compounds when used in cell-based assays. The phosphorothioates could also potentially participate in electrostatic interactions, while the nonionic phosphoramidates could facilitate hydrophobic and hydrogen-bonding interactions with the target receptor and also facilitate delivery into cells. Our results suggest that replacement of the phosphorothioate by phosphoramidate resulted in much-reduced activity of the compounds. The dominant furanose modification was the substitution of a 2′-OMe group in place of a 2′-hydrogen in the deoxyribofuranoside ring. The 2′ substituent can act as a conformational switch that transforms the furanose ring pucker from 2′-endo to 3′-endo, thereby affecting the global conformation of the individual library members (e.g., compound ORI-9020). The nucleobase modifications included both the replacement of the parent heterocyclic moiety and substitution on the heterocycle that provided additional hydrophobicity to the library members (compound ORI-7246 has a deaza-adenosine moiety).
The data obtained in this study are consistent with a mechanism of action centered on a direct interference with HBV DNA replication. In theory, this could be accomplished by direct termination of DNA replication by the HBV polymerase (RT and/or DNA polymerase) by incorporation followed by chain termination or by direct interaction with the polymerase to prevent further binding and elongation of DNA (e.g., noncompetitive inhibition with deoxynucleoside triphosphates). However, there is currently no evidence of either intracellular phosphorylation of these compounds (7, 8, 18) or their incorporation into HBV DNA. The patterns of HBV DNA replication intermediates (RIs) resulting from inhibition by the test compound show no accumulation of minus strand HBV DNA (the first strand synthesized by the HBV RT). This pattern is similar to that observed for inhibition of HBV replication induced by the internal assay control compound, 3TC, a documented inhibitor of HBV RT and, to a lesser extent, HBV DNA polymerase (22). This pattern of inhibition is distinctly different from that induced by inhibition of HBV replication by ara-AMP in 2.2.15 cells (a well-characterized inhibitor of DNA polymerase with no documented activity against HBV RT) in which a substantial accumulation of negative-strand DNA is observed (10).
The present data are consistent with an inhibition of the HBV RT-directed priming step prior to elongation of the negative (first) strand of HBV DNA. This could conceivably take place either by direct interaction with the polymerase or by competition for the binding of the four-base primer sequence to the initiation site for elongation of the HBV negative strand of DNA. The lack of a change in HBV RNA levels indicates that the test compounds did not directly interfere with the transcription of HBV RNA, and indicates that the molecules were not acting by mechanisms similar to those associated with antisense oligonucleotides (12). The lack of change in HBV protein levels is consistent with the observation that HBV RNA levels were not changed and makes it unlikely that the compounds were inhibiting HBV production by a direct interference in virus protein production. It is possible that other steps, such as packaging of viral RNA into the HBV core or encapsidation of the HBV nucleocapsid by surface antigen, may be inhibited by the test compounds. Alternatively, secondary processing (e.g., glycosylation or phosphorylation) could also be affected by the test compounds. Such alterations would not change the apparent levels of the HBV proteins as assessed by EIA analysis, but could change their properties. However, alteration of the glycosylation patterns of the HBV surface antigens by the test compounds is unlikely in that one consequence of such changes is a dramatic intracellular accumulation of HBV DNA RIs (15), which was not observed in these experiments.
The compounds identified in this study appear to act cooperatively in combination with 3TC, although less promising interactions with ADV were observed. The mechanisms related to the observed interactions are currently unclear, but the apparent differences in interactions between the di- and trinucleotides and these nucleoside analogs may be an indication of potential differences in intracellular metabolism or mechanisms of activity against HBV. Since these compounds are not phosphorylated and do not appear to be inhibitors of HBV polymerase via incorporation, cooperative effects with potent nucleoside analogs are not unexpected. These observations, along with apparent activity against several known HBV drug-resistant mutants, a promising lack of cytotoxicity in standard cell culture assays, and stability in liver microsomal assays, make these compounds excellent candidates for animal testing and potential clinical studies.
Acknowledgments
This work was partially supported by contract NO1-AI-85349 between Georgetown University and the National Institute of Allergy and Infectious Diseases.
We thank Mark Wainberg, McGill AIDS Center, Lady Davis Institute for Medical Research (Montreal, Quebec, Canada), for the screening of compounds against HIV. We thank Guy Boivin, University of Laval (Laval, Quebec, Canada), for screening of the compounds against herpes simplex virus and human cytomegalovirus. We also thank In Vitro Technologies (Baltimore, Md.) for the in vitro metabolism studies of the compounds. We thank Robert E. Lanford (Southwest Foundation for Biomedical Research, San Antonio, Texas) for the HBV polymerase assays. The HBV mutant constructs were provided by L. Condreay (Glaxo-SmithKline).
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Balfour, H. H., Jr. 1999. Antiviral drugs. N. Engl. J. Med. 340:1255-1268. [DOI] [PubMed] [Google Scholar]
- 3.Belen'kii, M. S., and R. Schinazi. 1994. A method for the analysis of combination therapies with statistical analysis. Antivir. Res. 25:11-18. [Google Scholar]
- 4.Bryant, M. L., E. G. Bridges, L. Placidi, A. Faraj, A. G. Loi, C. Pierra, D. Dukhan, G. Gosselin, J. L. Imbach, B. Hernandez, A. Juodawlkis, B. Tennant, B. Korba, P. Cote, P. Marion, E. Cretton-Scott, R. F. Schinazi, and J. P. Sommadossi. 2001. Antiviral l-nucleosides specific for hepatitis B virus infection. Antimicrob. Agents Chemother. 45:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chauret, N., A. Gauthier, J. Martin, and D. A. Nicoll-Griffith. 1997. In vitro comparison of cytochrome P450-mediated metabolic activities in human, dog, cat and horse. Drug Metab. Dispos. 25:1130-1136. [PubMed] [Google Scholar]
- 6.Ellman, J. A. 1996. Design, synthesis, and evaluation of small-molecule libraries. Acc. Chem. Res. 29:132-143. [Google Scholar]
- 7.Jin, Y., A. Roland, W. Zhou, M. Fauchon, J. Lyaku, and R. P. Iyer. 2000. Synthesis and antiviral evaluation of nucleic acid-based (NABTM) libraries. Bioorg. Med. Chem. Lett. 10:1921-1925. [DOI] [PubMed] [Google Scholar]
- 8.Jin, Y., X. Chen, M.-E. Cote, A. Roland, B. Korba, S. Mounir, and R. P. Iyer. 2001. Parallel solid-phase synthesis of nucleoside phosphoramidate libraries. Bioorg. Med. Chem. Lett. 11:2057-2060. [DOI] [PubMed] [Google Scholar]
- 9.Korba, B. E. 1996. In vitro evaluation of combination therapies against hepatitis B virus replication. Antivir. Res. 29:49-51. [DOI] [PubMed] [Google Scholar]
- 10.Korba, B. E., and G. Milman. 1991. A cell culture assay for compounds which inhibit hepatitis B virus replication. Antivir. Res. 15:217-228. [DOI] [PubMed] [Google Scholar]
- 11.Korba, B. E., and J. L. Gerin. 1992. Use of a standardized cell culture assay to determine activities of nucleoside analogs against hepatitis B virus replication. Antivir. Res. 19:55-70. [DOI] [PubMed] [Google Scholar]
- 12.Korba, B. E., and J. L. Gerin. 1995. Antisense oligonucleotides are effective inhibitors of hepatitis B virus replication in vitro. Antivir. Res. 28:225-242. [DOI] [PubMed] [Google Scholar]
- 13.Korba, B. E., P. J. Cote, W. E. Hornbuckle, B. C. Tennant, and J. L. Gerin. 2000. Treatment of chronic woodchuck hepatitis virus infection in the Eastern woodchuck (Marmota monax) with nucleoside analogues is predictive of therapy for chronic hepatitis B virus infection in humans. Hepatology 31:1165-1175. [DOI] [PubMed] [Google Scholar]
- 14.Lanford, R. E., L. Notvall, and B. Beames. 1995. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J. Virol. 69:4431-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, X., A. Mehta, M. Dadmarz, R. Dwek, B. S. Blumberg, and T. M. Block. 1997. Aberrant trafficking of hepatitis B virus glycoproteins in cells in which N-glycan processing is inhibited. Proc. Natl. Acad. Sci. USA 94:2380-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair, V. 2002. HIV integrase as a target for antiviral chemotherapy. Rev. Med. Virol. 12:179-193. [DOI] [PubMed] [Google Scholar]
- 17.Quan, Y., B. Brenner, G. Oliveira, and M. A. Wainberg. 2003. Lamivudine can exert a modest antiviral effect against human immunodeficiency virus type 1 containing the M184V mutation. Antimicrob. Agents Chemother. 47:747-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roland, A., Y. Xiao, Y. Jin, and R. P. Iyer. 2001. A novel linker for the solid-phase synthesis of a library of 3′-thiophosphorylated dinucleotides. Tetrahedron Lett. 42:3669-3672. [Google Scholar]
- 19.Sanger, W. 1984. Protein-nucleic acid interactions, p. 385-431. In W. Sanger (ed.), Principles of nucleic acid structure. Springer-Verlag, New York, N.Y.
- 20.Schreiber, S. L. 2000. Target-oriented and diversity oriented organic synthesis in drug discovery. Science 287:1964-1969. [DOI] [PubMed] [Google Scholar]
- 21.Sells, M. A., A. Z. Zelent, M. Shvartsman, and G. Acs. 1988. Replication intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 62:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severini, A., X. Y. Liu, J. S. Wilson, and D. L. Tyrrell. 1995. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 39:1430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuyver, L. J., S. A. Locarnini, A. S. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 24.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis-B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 25.Tatti, K. M., B. E. Korba, H. L. Stang, S. Peek, J. L. Gerin, B. C. Tennant, and R. F. Schinazi. 2002. Mutations in the conserved woodchuck hepatitis virus polymerase FLLA and YMDD regions conferring resistance to lamivudine. Antivir. Res. 55:141-150. [DOI] [PubMed] [Google Scholar]