Background: Correct assembly of coactivators with nuclear receptor (NR) heterodimers is critical for transactivation.
Results: The stoichiometry of SRC1 on the CAR:RXR heterodimer varies with the liganded state.
Conclusion: When both subunits of the heterodimer are in agonist-bound conformation, each subunit independently binds a coactivator molecule.
Significance: A novel mechanism whereby distinct stoichiometric NR·coactivator complexes affect transcriptional levels.
Keywords: Analytical Ultracentrifugation, Nuclear Receptor, Protein Assembly, Small Angle X-ray Scattering (SAXS), Transcription Coactivator, Heterodimer, Stoichiometry, Transactivation
Abstract
The constitutive androstane (CAR) and retinoid X receptors (RXR) are ligand-mediated transcription factors of the nuclear receptor protein superfamily. Functional CAR:RXR heterodimers recruit coactivator proteins, such as the steroid receptor coactivator-1 (SRC1). Here, we show that agonist ligands can potentiate transactivation through both coactivator binding sites on CAR:RXR, which distinctly bind two SRC1 molecules. We also observe that SRC1 transitions from a structurally plastic to a compact form upon binding CAR:RXR. Using small angle x-ray scattering (SAXS) we show that the CAR(tcp):RXR(9c)·SRC1 complex can encompass two SRC1 molecules compared with the CAR(tcp):RXR·SRC1, which binds only a single SRC1. Moreover, sedimentation coefficients and molecular weights determined by analytical ultracentrifugation confirm the SAXS model. Cell-based transcription assays show that disrupting the SRC1 binding site on RXR alters the transactivation by CAR:RXR. These data suggest a broader role for RXR within heterodimers, whereas offering multiple strategies for the assembly of the transcription complex.
Introduction
Nuclear hormone receptors (NR)3 relay cellular signals through distinct multiprotein assemblies (1). At the basic level, small molecule signals produce structural changes within NRs and these changes determine the composition of the interacting protein complex. These changes are essential for transcriptional activity and appear to be conserved among all ligand-activated receptors that have been studied to date. NRs are characteristically modular proteins with distinct functional domains (2). At the N terminus is the DNA binding domain (DBD), which determines target gene selectivity. The ligand binding domain (LBD) is a multifunctional module that contains the ligand binding pocket, a dimerization interface that associates with the retinoid X receptor (RXR) and a C-terminal ligand-dependent transactivation domain (AF2). Multiple biochemical and structural studies on nuclear receptors have demonstrated that ligand binding results in the specific conformational changes that are associated with a transcriptionally active state (3). In this active state, the conformation of the AF2 domains typically rearrange along the receptor surface, thereby creating a new docking site for transcriptional coactivator proteins (4).
Because both receptors within NR heterodimers can bind small molecule agonist ligands, in the simplest model for transactivation, agonist binding to either receptor can generate comparable transcriptional levels of downstream genes. Furthermore, this model would predicate that the presence of agonists to both receptors at once would yield proportionately higher levels of transcription. Such model systems are exemplified by the CAR:RXR (Fig. 1A), PPARα:RXR, and LXR:RXR heterodimers (5). Yet, there are NR heterodimers that exhibit transcriptional responses that are distinct from this model (5, 6). For instance, transactivation by RAR:RXR, VDR:RXR, and TR:RXR only occurs in the presence of the RAR, VDR, and TR agonists, and when used in combination with the RXR agonist, transactivation levels are enhanced, unaffected, or are repressed, respectively (5, 7). Through structural and biophysical studies, the mechanism of transactivation has been recognized to occur through conformational changes that restrict recruitment to a single coactivator protein to RAR:RXR (8) or decrease T3 agonist binding affinity to TR:RXR (7).
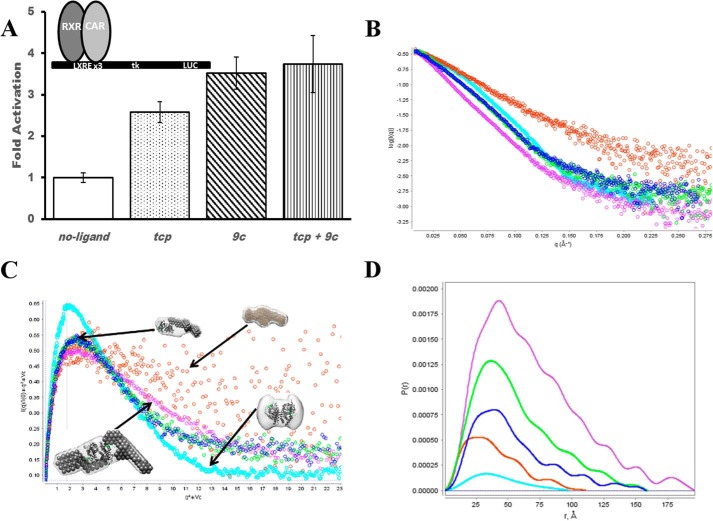
FIGURE 1.
Activity and assembly of CAR:RXR. A, transactivation of CAR:RXR measured in CV-1 cells on 4 copies of liver X receptor response element. Significance in differences in activity between samples was measured by a two-tailed Welch test, and all differences are highly significant (p < 0.05), except tcp versus tcp+9c (p = 0.11) and 9c versus tcp+9c (p = 0.67). B, scattering curve normalized to I(0) to show differences in size and deviation from globular shape. CAR:RXR, cyan; SRC, orange; CAR:RXR·SRC, blue; CAR(tcp):RXR·SRC, green; CAR(tcp):RXR(9c)·SRC, purple. C, Vc-based Kratky plot for visualization of flexibility and surface area to volume ratio. Molecular shapes are generated by MONSA, whereas the individual heterodimer and coactivator envelopes are generated by DAMMIN. D, P(r) distribution. The pairwise distribution of atoms within the complexes gives overall complex size. Greater deviation from a Gaussian distribution indicates an extended structure. CAR:RXR, cyan; SRC, orange; CAR:RXR·SRC, blue; CAR(tcp):RXR·SRC, green; CAR(tcp):RXR(9c)·SRC, purple.
CAR is most abundantly expressed in the liver and intestine and has been directly linked to the transcription of genes involved in the clearance of both xenobiotics (9–11), and endogenous toxins such as bilirubin (12). These target genes include select P450 family monooxygenases, phase II conjugating enzymes, and xenobiotic transporters. Therefore, CAR serves as a master regulator of xenobiotic clearance and its activation can be considered a form of chemical immunity. Within the nucleus, CAR binds to RXR and forms a functional heterodimer that recognizes its specific target genes. Additionally, the transcriptional activity of CAR is induced simply by association with RXR and with no apparent need for a CAR ligand (13–15) (Fig. 1A). Although ligand is not required for activation, constitutive CAR activity is mediated through the same conserved functional domains as those utilized by ligand-activated receptors; thus the CAR:RXR heterodimer recruits coactivator proteins through the AF2 transactivation domain (16, 17). Transactivation levels mediated by CAR:RXR can be augmented by agonist ligands such as 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (tcp) (18) and 6-(4-cholorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) (19), which are selective for mouse and human CAR, respectively, whereas 9-cis-retinoic acid (9c) can function as an RXR agonist (20). In both cases, these agonists enhance constitutive activity by stabilizing the constitutive AF2-coactivator interaction. Additionally, transactivation by the CAR:RXR heterodimer can be potentiated by the RXR agonist, 9c (Fig. 1A). 9c binds the ligand binding pocket of RXR and evokes the canonical NR conformational changes that result in direct interactions with coactivators. The SRC coactivator proteins function to recruit the cellular transcriptional machinery to activated NRs (21, 22). SRCs also play an essential role as histone acetyltransferases to acetylate histone proteins and consequently enhance transcriptional activity (23). Thus, there is a direct link between agonist ligand binding, coactivator recruitment, and the transcription of downstream genes.
The activity of permissive NR heterodimers such as CAR:RXR that is potentiated by ligands to both CAR and RXR raises interesting questions about the precise structural assembly of nuclear factors that promote such transactivation. In this study, we propose that with permissive NR heterodimers represented by CAR:RXR, the levels of coactivator recruitment are proportional to the liganded state of the heterodimer. Thus, CAR(tcp):RXR·SRC1 is a 1:1 NR:coactivator complex, whereas CAR(tcp):RXR(9c)·SRC1 exists in 1:2 binding stoichiometry. Moreover, these levels of coactivator recruitment are proportional to transcriptional activity (Fig. 1A). These data further suggest that in this subset of NR heterodimers RXR performs a substantial role in regulating transcriptional responses within the cell. Because of the polarity of the liver X receptor response element DNA·CAR:RXR complex used in the transactivation assays (see “Experimental Procedures”), CAR occupies the 3′ half-site, directly upstream from the luciferase gene. Thus, a major role of the coactivator molecule bound to CAR is to recruit the transcriptional machinery needed for luciferase production.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
CAR:RXR was purified as described earlier (24, 25). Briefly, the murine CAR LBD (residues 117–358) was subcloned into the pET15b vector with an N-terminal hexahistidine tag from mCAR cDNA kindly provided by Dr. Barry M. Forman. The human RXRα LBD (residues 225–462) was subcloned into the pACYC184 vector was a kind gift from Dr. Bruce Wisely (Glaxo Smith-Kline, Inc.). Residues 617–769 of SRC1 (accession no. Q15788) encompassing three nuclear receptor interacting motifs (RIDs) were subcloned into the pET-SUMO vector. CAR:RXR and SRC1 were separately isolated by affinity chromatography column using nickel-nitrilotriacetic acid resin (Qiagen Inc.) To prepare various CAR:RXR·SRC complexes, CAR:RXR and SRC(RID1–3) were mixed in a 1:2 molar ratio, after the addition of 2× molar excess of ligands, and loaded onto an S200 Superdex 16/60 column for purification of the resulting complexes (data not shown). Fractions corresponding to the complexes were pooled, measured by Bradford assay, and concentrated for further analysis.
Small Angle X-ray Scattering
Measurements were recorded at the beamlines: SIBLYS at Laurence Berkeley National Laboratory and DND-CAT at Argonne National Laboratory. Scattering for CAR(tcp):RXR(9c) was collected at Argonne National Laboratory at three concentrations ranging from 1.2 to 8 mg/ml. Scattering for all other complexes was collected at SIBYLS with protein concentrations ranging from 0.8 to 5.4 mg/ml. Curves were normalized by concentration to eliminate interparticle effects at higher concentrations. No aggregation was evident within any sample. Data were analyzed using the ATSAS software package (26) and ScÅtter (Table 1). Three-dimensional model building was performed using DAMMIN and MONSA (27) and visualized using Chimera. To generate envelopes, five DAMMIN runs per sample were used and averaged using DAMAVER. χ-Squared values from DAMMIN are in Table 1. Three separate MONSA runs were also performed on the complexes to ensure that the ab initio fitting converged to the relative orientations of the models generated by DAMMIN. Kratky plots were calculated for shape analysis (28, 29). The theoretical values for spherical objects in the Vc-based Kratky plot attain an ordinate maxima of 0.82, and for non-flexible scattering particles, q2 × Vc = √3, where q is the scattering vector (Å) and Vc is defined by the correlation length of the scattering particle as the ratio of the zero angle scattering intensity, I(0), to its total scattered intensity (29). Excluded from these analyses is the CAR:RXR(9c)·SRC1 complex due to severe problems with protein aggregation.
TABLE 1.
SAXS data
The following abbreviation are used: I(0), interpolated intensity at 0 angle; Rg, radius of gyration; Vc, volume of correlation; Mp, molecular weight of protein; V, volume; Dmax, maximum pairwise distance; P(r), pair-distance distribution function; Rc, cross-sectional radius of gyration; Px, Porod exponent. A noticeably larger Rg, Dmax, and a translated P(r) peak in CAR(tcp):RXR(9c)·SRC suggest a significantly larger species than CAR(tcp):RXR·SRC. Close agreement of real and Guinier-derived values confirm that the data is internally consistent. We use the convention of 4π × sinθ/λ. χ 2 values are for DAMMIN envelope generation.
| Molecular species | I(0) | I(0)_R | Rg | Rg_R | Vc | Mp | V | Dmax | P(r) peak | Rc | Px | χ2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAR(tcp):RXR(9c) | 0.05 | 0.046 | 35.20 | 32.01 | N/A | N/A | 1.43E + 005 | 97 | 32.25 | 17.88 | 3.8 | 0.755 |
| SRC1 | 0.20 | 0.170 | 33.96 | 31.63 | 364 | 31.5 | 1.06E + 005 | 112 | 28–30 | 11.32 | 2.4 | 0.880 |
| CAR:RXR·SRC | 0.37 | 0.330 | 45.63 | 45.15 | 729 | 84.0 | 2.05E + 005 | 159 | 37.00 | 17.07 | 3.9 | 0.971 |
| CAR(tcp):RXR·SRC | 0.56 | 0.520 | 44.46 | 44.89 | 744 | 100 | 2.15E + 005 | 158 | 36.25 | 17.45 | 3.9 | 0.969 |
| CAR(tcp):RXR(9c)·SRC | 1.05 | 0.930 | 55.00 | 54.41 | 1031 | 155 | 3.17E + 005 | 197 | 43.00 | 18.66 | 3.9 | 0.872 |
Analytical Ultracentrifugation
Aliquots of CAR:RXR and SRC(RID1–3) were thawed and mixed in various ratios (1:3, 1:1, and 3:1), incubated briefly with ligands, and analyzed on a Beckman XL ultracentrifuge. Both sedimentation velocity and sedimentation equilibrium experiments were performed. Sedimentation equilibrium was performed at 8–10 °C and 3 rotor speeds, whereas sedimentation velocity was run at 20 °C and 55,000 rpm. Data were analyzed using SEDFIT and SEDPHAT (30). We calculate the parameters, f/f0, frictional ratio; sw(20,w), sedimentation coefficient under standard conditions; and r.m.s. deviations, which reports the quality fit of the solutions of the Lamm equations (30) to the data (Fig. 2A and Table 2).
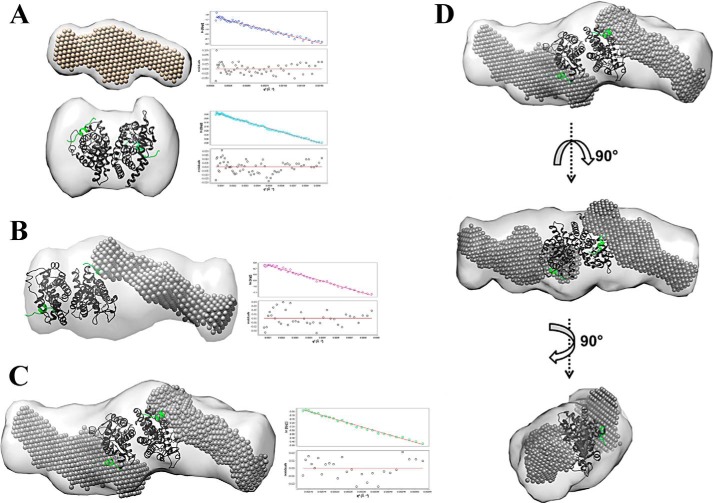
FIGURE 2.
Molecular shapes of scattering species. A, top, molecular shape of free SRC1(RID1–3); bottom, molecular shape of CAR(tcp)·RXR(9c). B and C, molecular shapes of CAR(tcp):RXR·SRC and CAR(tcp):RXR(9c)·SRC (light gray), respectively. Superimposed on this is the SRC (dark gray spheres) envelope from A and the CAR:RXR·SRC(peptide) (green) structure (PDB code 1XLS). Guinier plots (q2(Å−2) versus ln[I(q)]) showing linearity of scattering at low q are shown alongside the corresponding molecular complex. D, CAR(tcp):RXR(9c)·SRC in three different orientations.
TABLE 2.
AUC velocity data
The abbreviations use are: f/f0, frictional ratio; sw(20,w), sedimentation coefficient under standard conditions; root meant square deviations reports the quality fit to the data. A higher sedimentation coefficient for 9c and tcp+9c complexes indicates a larger species, whereas a slightly higher frictional coefficient indicates a more extended structure.
| Molecular species | f/fo | sw(20,w) | Root mean square deviations |
|---|---|---|---|
| CAR:RXR | 1.28 | 4.12 | 0.005 |
| SRC1 | 1.5 | 2.1 | 0.007 |
| CAR:RXR·SRC | 1.46 | 2.27, 4.7 | 0.007 |
| CAR(tcp):RXR·SRC | 1.46 | 2.4, 4.7 | 0.006 |
| CAR:RXR(9c)·SRC | 1.5 | 2.48, 5.69 | 0.006 |
| CAR(tcp):RXR(9c)·SRC | 1.5 | 2.27, 5.6 | 0.009 |
Reporter Gene Assays
These assays were performed as reported earlier (31). Briefly, CV-1 cells were maintained in DMEM/F-12 media containing 10% fetal bovine serum and 1000 units/ml of penicillin and 1 mg/ml of streptomycin. Immediately prior to the assay, the media was changed to DMEM/F-12 with 10% charcoal-dextran-treated FBS and no antibiotics. Effectene® (Qiagen Inc.) was used to transfect cells with 50 ng/well of pCMX mCAR and 100 ng/well of pCMV-TK-luc containing three copies of the liver X receptor response element and 10 ng/well of pRL CMV expressing Renilla luciferase as an internal control. The cells were dispensed on 24-well plates and ligands were added 24 h post-transfection. The ligand concentrations used were TCPOBOP (1.0 μm), 0.5 μm 9-cis-retinoic acid. After 48 h, cells were lysed. Activity was determined using the dual luciferase assay kit (Promega Inc.) following the manufacturer's instructions. The reported results are the average from three separate experiments. Significance in differences in activity between liganded samples was measured by a two-tailed Welch test (32).
Gal4-DBD Assays
The E456K mutation was made on pCMX-Gal4DBD-hRXRαLBD (40 ng) (gift from Prof. Barry Forman). Transfection and assay were performed as described above with full-length CAR (50 ng), four copies of a Gal4 binding site (pUC8-MH100 × 4-TK-Luc) (100 ng), and pCMV-Renilla luciferase (10 ng). As described above the reported results are an average of three separate experiments. Significance in differences in activity between liganded samples was measured by a one-tailed Welch test (32). Also, liganded values were found to differ significantly from unliganded (p < 0.05) in both CAR:RXR and CAR:RXRE456K.
RESULTS
CAR(tcp):RXR(9c)·SRC1 Assembles as a 1:2 Heterodimer·Coactivator Complex
To develop an understanding of how the transcriptionally active CAR:RXR assembles with coactivators, we prepared CAR:RXR LBD and SRC1(RID1–3) in Escherichia coli and isolated multiple complexes of CAR:RXR·SRC1. Because CAR:RXR can also bind SRC1 in the absence of agonist, for the study here we isolated complexes CAR:RXR·SRC1, CAR(tcp):RXR·SRC1, and CAR(tcp):RXR(9c)·SRC1 representing the unliganded, singly liganded, and doubly liganded CAR·RXR complexes, respectively. SRC1(RID1–3) used in these studies comprises amino acids 617–769, and encompasses the three nuclear RIDs (33, 34). Also, SRC1(RID1–3) has previously been shown to interact in a ligand-dependent manner with CAR:RXR (15, 25). Using small angle x-ray scattering (SAXS), we have determined the global molecular assembly and structural properties of the CAR:RXR·SRC1, CAR(tcp):RXR·SRC1, and CAR(tcp):RXR(9c)·SRC1 complexes (Fig. 2 and Table 1). For comparisons, we also measured scattering from CAR(tcp):RXR(9c) and SRC1 alone and the shapes of the scattering curves are distinct and typical of their molecular size and flexibility. Therefore, the scattering curve of the CAR:RXR heterodimer alone is characteristic of folded protein, whereas the scattering curve of SRC alone is representative of disordered proteins (Fig. 1B). We applied Kratky analyses (28) and shape comparisons to identify any noticeable difference in compactness between CAR(tcp):RXR·SRC1 and CAR(tcp):RXR(9c)·SRC1. From these analyses we could conclude that the CAR(tcp):RXR(9c)·SRC1 complex is more elongated and shows greater flexibility than CAR(tcp):RXR·SRC1, which suggests the presence of a second SRC1 molecule within the CAR(tcp):RXR(9c)·SRC1 particle. We also compare the I(0)-scaled and Vc-based Kratky scattering curves, which emphasize differences in size and geometry of the scattering particles (29). In the I(0)-normalized graphical plot, we compare the linearity and negative slope of the scaled intensity versus scattering angle for the complexes (Fig. 1B). The I(0)-normalized scattering plot of CAR(tcp):RXR(9c)·SRC1 complex is more linear with a sharper slope than the corresponding plots of either CAR(tcp):RXR·SRC1 or CAR:RXR·SRC1 complexes. This clearly suggests that CAR(tcp):RXR(9c)·SRC1 is a relatively more extended molecule of higher molecular weight than the unliganded and singly liganded complexes. From the Vc-Kratky plot using experimental SAXS data we are able to infer that CAR:RXR alone is mostly spherical with no apparent flexibility, whereas free SRC1 shows a hyperbolic plateau that is indicative of a highly flexible structure and with higher surface area to volume ratio (Fig. 1C). When comparing the heterodimer:coactivator complexes we note that there is a decrease in peak height between CAR(tcp):RXR·SRC1 and CAR(tcp):RXR(9c)·SRC1, which further indicates that CAR(tcp):RXR(9c)·SRC1 is a relatively extended particle (Fig. 1C). Overall, the observed scattering pattern is consistent with the presence of a second SRC molecule in the CAR(tcp):RXR(9c)·SRC complex. Furthermore, we note from a comparison of molecular envelopes of free and bound SRC1 that this molecule adopts a relatively more compact structure upon binding CAR:RXR, a feature that has been observed previously in the RAR:RXR·SRC1 complex (8).
To visualize the assembly of these complexes we generated molecular envelopes of CAR:RXR·SRC1, CAR(tcp):RXR·SRC1, and CAR(tcp):RXR(9c)·SRC1 (Fig. 2, B and C). The molecular shapes of CAR:RXR·SRC1 and CAR(tcp):RXR·SRC1 are of an elongated species and are nearly identical at SAXS resolution (Fig. 2B). The molecular shape of the CAR(tcp):RXR(9c)·SRC1 complex is also elongated and is ∼1.4-fold larger than the CAR:RXR·SRC1 and CAR(tcp):RXR·SRC1 complexes (Fig. 2, C and D). To depict the assembly of each complex, the molecular envelopes of the heterodimer and coactivator were superimposed upon the envelope of each CAR:RXR·SRC1 complex. Both manual and automated fitting (see “Experimental Procedures”) suggest that both the CAR:RXR·SRC1 and CAR(tcp):RXR·SRC1 envelopes can encompass the CAR:RXR heterodimer bound to a single SRC1 molecule (Fig. 2B). On the other hand, the molecular shape of CAR(tcp):RXR(9c)·SRC1 readily corresponds to a single CAR:RXR heterodimer that is bound to two SRC1(RID1–3) molecules (Fig. 2, C and D). Thus, these SAXS analyses suggest molecular complexes with stoichiometries of 1:1 heterodimer:coactivator for the singly liganded CAR(tcp):RXR·SRC1 and 1:2 heterodimer:coactivator and CAR(tcp):RXR(9c)·SRC complexes.
Analytical Ultracentrifugation for Size Determination
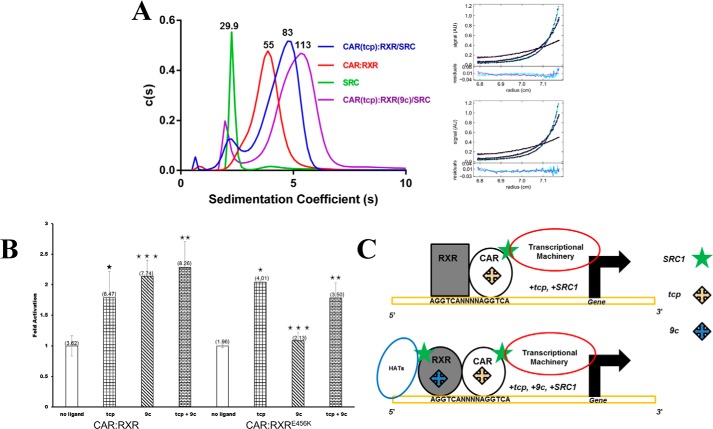
To establish if the shapes of CAR:RXR·SRC1, CAR(tcp):RXR·SRC1, and CAR(tcp):RXR(9c)·SRC1 correspond to their relative sizes, these complexes were independently analyzed through sedimentation velocity and equilibrium studies by analytical ultracentrifugation (AUC). The sedimentation velocity data consistently shows a species of higher sedimentation coefficient with CAR(tcp):RXR(9c)·SRC1 than CAR(tcp):RXR·SRC1 (Fig. 3A and Table 2). From sedimentation equilibrium analyses we confirmed the molecular masses of these species to be 84.0 kDa (CAR:RXR·SRC1 and CAR(tcp):RXR·SRC1), which corresponds to the size of one CAR:RXR heterodimer (55 kDa) bound to a single SRC1 (30 kDa) and 113.0 kDa for the CAR(tcp):RXR(9c)·SRC1 complex, which corresponds to one CAR:RXR heterodimer bound to two SRC1 molecules (Fig. 3A). The equilibrium data therefore suggests a strong preference for 2:1 complex formation in the doubly liganded state. Taken together, the ultracentrifugation data confirm that the molecular envelopes determined by SAXS correspond to the molecular weights of these complexes as determined by AUC. Therefore, we hypothesize that transactivation by CAR(tcp):RXR(9c)·SRC1 relies on coactivator binding to both CAR and RXR.
FIGURE 3.
Hydrodynamic analysis of CAR:RXR and transactivation by CAR:RXRE456K. A, sedimentation coefficients and stoichiometry from sedimentation velocity and equilibrium experiments using AUC. Molecular masses shown above each peak are in kDa (also Table 2). Fitting of CAR(tcp):RXR(9c)·SRC to heterodimer:coactivator stoichiometry of 1:1 (r.m.s. deviations = 0.010, top) and 1:2 models (r.m.s. deviations = 0.0068, bottom). The better fit has a lower overall r.m.s. deviation, and this discrepancy is consistent across three data sets. B, transactivation by CAR:Gal4 DBD-RXR LBD and CAR:Gal4 DBD-RXRE456K LBD measured in CV-1 cells on four copies of a Gal4 binding site (mh100 × 4-tk-luc) response element. Data are normalized to no-ligand activity = 1.0 and absolute luciferase activity is in parentheses above each column. Statistical significance was measured by a one-tailed Welch test and differences are listed as highly significant (***, p < 0.05), moderately significant (**, p = 0.198), and not significant (*, p = 0.5). C, models of the NR transactivation complexes. Top, CAR:RXR·SRC1 and/or CAR(tcp):RXR·SRC1. Bottom, CAR(tcp):RXR(9c)·SRC1.
Transactivation by CAR:RXRE456K Is Distorted from the Native CAR·RXR Complex
There are multiple factors within the cell that function to regulate transactivation by NRs (35, 36). Of these factors, the SRC family of coactivator proteins are recruited specifically to the agonist-bound conformation of the NR LBD (37). In this agonist-bound conformation the AF2 domain is realigned along the receptor surface, and in doing so creates a new interface that can bind SRC proteins (38). The amino acid Glu456 is within the AF2 domain of RXR and interacts with SRC1 (16). To better understand the role of the RXR coactivator binding site within CAR:RXR, we compare the transactivation of CAR:RXRE456K with the native protein complex in a cell-based reporter system. This Glu → Lys amino acid substitution has previously been shown to disrupt SRC1 recruitment by RXR (39–42). Therefore, we predicted that the native and CAR:RXRE456K would have distinct transcriptional responses to RXR-specific agonists. When tested in CV1 cells, we observe that the transcriptional response of CAR:RXR and CAR:RXRE456K to tcp (relative to no exogenous ligand) is similar. However, transactivational levels in response to exogenously applied 9c alone (Fig. 3B) and to the combination of 9c+tcp by the native CAR:RXR and mutant CAR:RXRE456K receptor complexes are markedly different. Specifically, 1) transactivation of CAR(tcp):RXRE456K is greater than CAR:RXRE456K, which is analogous to the trend observed with the native protein where the increased activity is potentiated by tcp (Figs. 1A and 3B); 2) transactivation of CAR:RXR(9c)E456K is at the same level as the unliganded CAR:RXRE456K, which suggests that in the mutant complex 9c is unable to increase transactivation above the basal level of unliganded CAR:RXRE456K, unlike within the native protein; and 3) transactivation by CAR(tcp):RXR(9c)E456K is at the same level as CAR(tcp):RXRE456K, but lower than by the native CAR(tcp):RXR(9c) (Figs. 1A and 3B). The measurements of SAXS and AUC above propose a distinct role for RXR within the CAR:RXR complex. Together, these results suggest that in CAR:RXR the SRC-binding site within RXR is essential for the heterodimer to achieve maximum transcriptional activity. Thus, relative to other NR heterodimers such as RAR:RXR and VDR:RXR (8, 43), RXR can undertake a more significant role in transactivation by CAR:RXR.
DISCUSSION
Nuclear receptors are a superfamily of structurally and functionally conserved proteins that have evolved to regulate transcription in response to small molecule ligands through multiprotein assemblies. Hormonal agonist molecules evoke the correct structural changes within NRs to interact directly with coactivators, such as SRC1. Normal transactivation is dependent on the precise assembly of the component molecules. However, there is only a superficial mechanistic understanding of how this multiprotein assembly takes place, how it can be modulated, and how it relates to transactivation. The study here illustrates the role of the agonist ligand in defining the molecular assembly of the NR·coactivator complex (Fig. 3C).
Both CAR and RXR in the CAR:RXR heterodimer can independently bind their respective agonists (24). Also, crystallographic studies have shown that CAR(agonist):RXR(agonist) can bind two 13-mer LXXLL coactivator-derived peptides through binding sites on both CAR and RXR (16, 17). This structural assembly has also been observed in other permissive NR heterodimers such as LXR:RXR (44) and PPARγ:RXR (45). It is now clear from our data that these LXXLL motifs that are bound to permissive NR heterodimers are derived from two independent SRC1 molecules, although the intact SRC1 molecule has three distinct LXXLL-containing RIDs (46).
This agonist-mediated heterodimer:coactivator stoichiometry has important mechanistic implications for transactivation and in pharmacology. First, among the several functions ascribed to SRCs are the recruitment of the cellular transcriptional machinery to activated NRs (21, 22, 47) and as histone acetyltransferases (23). The polarity of heterodimers such as CAR:RXR on the direct repeat response element places CAR toward the 3′ end of the promoter (48–50). Therefore, the most likely function of the coactivator molecule bound directly to CAR is to assemble the transcriptional machinery through interactions with p300/cAMP-response element-binding protein (22). The addition of 9c to permissive NR heterodimers allows for the recruitment of a second SRC coactivator molecule directly to RXR on the 5′ end of the promoter. From this location, the second SRC can function as and recruit other histone acetyltransferases thereby enhancing transactivational levels (Fig. 3C), also observed with the progesterone and glucocorticoid receptors (51). Second, this mechanism of transactivation is a distinct alternative to that proposed for the RAR:RXR heterodimer (8, 43, 52). In RAR:RXR, a single coactivator molecule is recruited directly to RAR upon activation by agonist in a conformation similar to CAR(tcp):RXR·SRC. However, the recruitment of a second coactivator to RAR(agonist):RXR(agonist) is restricted through long-range, agonist-induced conformational changes that disrupt the RXR coactivator-binding site (53). Third, targeting coactivators for therapy is of growing interest (54, 55), thus requiring a detailed knowledge of such binding events. These coactivators display different binding specificities for the receptors both independently (56) and within the heterodimer (15), thus, it is likely that the specific interactions between each SRC1 molecule and the two binding sites on CAR:RXR are distinct. The activation of CAR:RXR is not always beneficial as hepatic metabolism can convert certain therapeutic drugs to potent toxins. For instance, CAR:RXR-mediated metabolism of acetaminophen results in a reactive quinone metabolite (N-acetyl-p-benzoquinone imine). This metabolic by-product promotes acute liver failure by binding to cellular macromolecules and generating reactive oxygen species (10, 57). The hepatotoxic effects of cocaine are also mediated via a CAR:RXR-dependent pathway (9). As a result, the activity of CAR:RXR can have either protective or deleterious consequences to the organism depending on the particular chemical challenges faced. Also, the discoveries of endogenous RXR ligands such as polyunsaturated fatty acids (58–60) increase the likelihood of two agonists binding the CAR:RXR heterodimer at once. Consequently, this novel assembly has important implications for the design of small molecules directed at regulating transactivation by modulating the formation and composition of the NR-coactivator assembly.
Acknowledgments
We thank Drs. Cynthia Peterson, Ed Wright, Chris Stanley, and Richard Gillilan for expert advice on AUC and SAXS, Jeremy Vincent for the transactivation assays, and Robert Fletterick, Bruce McKee, Bert O'Malley, Mariano Labrador, Kumar Putcha, and Martin Privalsky for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK097337-01 (to E. J. F.).
- NR
- nuclear receptor
- 9c
- 9-cis-retinoic acid
- AF1 (or 2)
- activation function 1 (or 2)
- AUC
- analytical ultracentrifugation
- CAR
- constitutive androstane receptor
- DBD
- DNA binding domain
- LBD
- ligand binding domain
- RID
- receptor interacting domain
- RXR
- retinoid X receptor
- SAXS
- small angle x-ray scattering
- SRC
- steroid receptor coactivator
- TCPOBOP (or tcp)
- 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene.
REFERENCES
- 1. Rosenfeld M. G., Glass C. K. (2001) Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 276, 36865–36868 [DOI] [PubMed] [Google Scholar]
- 2. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) The nuclear receptor superfamily: the second decade. Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Billas I., Moras D. (2013) Allosteric controls of nuclear receptor function in the regulation of transcription. J. Mol. Biol. 425, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 4. Onate S. A., Boonyaratanakornkit V., Spencer T. E., Tsai S. Y., Tsai M. J., Edwards D. P., O'Malley B. W. (1998) The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem. 273, 12101–12108 [DOI] [PubMed] [Google Scholar]
- 5. Shulman A. I., Larson C., Mangelsdorf D. J., Ranganathan R. (2004) Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116, 417–429 [DOI] [PubMed] [Google Scholar]
- 6. Forman B. M., Umesono K., Chen J., Evans R. M. (1995) Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81, 541–550 [DOI] [PubMed] [Google Scholar]
- 7. Putcha B. D., Wright E., Brunzelle J. S., Fernandez E. J. (2012) Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers. Proc. Natl. Acad. Sci. U.S.A. 109, 6084–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rochel N., Ciesielski F., Godet J., Moman E., Roessle M., Peluso-Iltis C., Moulin M., Haertlein M., Callow P., Mély Y., Svergun D. I., Moras D. (2011) Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol. 18, 564–570 [DOI] [PubMed] [Google Scholar]
- 9. Wei P., Zhang J., Egan-Hafley M., Liang S., Moore D. D. (2000) The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407, 920–923 [DOI] [PubMed] [Google Scholar]
- 10. Zhang J., Huang W., Chua S. S., Wei P., Moore D. D. (2002) Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science 298, 422–424 [DOI] [PubMed] [Google Scholar]
- 11. Sonoda J., Rosenfeld J. M., Xu L., Evans R. M., Xie W. (2003) A nuclear receptor-mediated xenobiotic response and its implication in drug metabolism and host protection. Curr. Drug Metab. 4, 59–72 [DOI] [PubMed] [Google Scholar]
- 12. Huang W., Zhang J., Chua S. S., Qatanani M., Han Y., Granata R., Moore D. D. (2003) Induction of bilirubin clearance by the constitutive androstane receptor (CAR). Proc. Natl. Acad. Sci. U.S.A. 100, 4156–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baes M., Gulick T., Choi H. S., Martinoli M. G., Simha D., Moore D. D. (1994) A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol. Cell. Biol. 14, 1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi H. S., Chung M., Tzameli I., Simha D., Lee Y. K., Seol W., Moore D. D. (1997) Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 272, 23565–23571 [DOI] [PubMed] [Google Scholar]
- 15. Dussault I., Lin M., Hollister K., Fan M., Termini J., Sherman M. A., Forman B. M. (2002) A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol. Cell. Biol. 22, 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suino K., Peng L., Reynolds R., Li Y., Cha J. Y., Repa J. J., Kliewer S. A., Xu H. E. (2004) The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol. Cell 16, 893–905 [DOI] [PubMed] [Google Scholar]
- 17. Xu R. X., Lambert M. H., Wisely B. B., Warren E. N., Weinert E. E., Waitt G. M., Williams J. D., Collins J. L., Moore L. B., Willson T. M., Moore J. T. (2004) A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol. Cell 16, 919–928 [DOI] [PubMed] [Google Scholar]
- 18. Tzameli I., Pissios P., Schuetz E. G., Moore D. D. (2000) The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell. Biol. 20, 2951–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maglich J. M., Parks D. J., Moore L. B., Collins J. L., Goodwin B., Billin A. N., Stoltz C. A., Kliewer S. A., Lambert M. H., Willson T. M., Moore J. T. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J. Biol. Chem. 278, 17277–17283 [DOI] [PubMed] [Google Scholar]
- 20. Levin A. A., Sturzenbecker L. J., Kazmer S., Bosakowski T., Huselton C., Allenby G., Speck J., Kratzeisen C., Rosenberger M., Lovey A. (1992) 9-cis-Retinoic acid stereoisomer binds and activates the nuclear receptor RXR α. Nature 355, 359–361 [DOI] [PubMed] [Google Scholar]
- 21. Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85, 403–414 [DOI] [PubMed] [Google Scholar]
- 22. Yao T. P., Ku G., Zhou N., Scully R., Livingston D. M. (1996) The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. U.S.A. 93, 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spencer T. E., Jenster G., Burcin M. M., Allis C. D., Zhou J., Mizzen C. A., McKenna N. J., Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389, 194–198 [DOI] [PubMed] [Google Scholar]
- 24. Wright E., Vincent J., Fernandez E. J. (2007) Thermodynamic characterization of the interaction between CAR-RXR and SRC-1 peptide by isothermal titration calorimetry. Biochemistry 46, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright E., Busby S. A., Wisecarver S., Vincent J., Griffin P. R., Fernandez E. J. (2011) Helix 11 dynamics is critical for constitutive androstane receptor activity. Structure 19, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Receveur-Brechot V., Durand D. (2012) How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rambo R. P., Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shan L., Vincent J., Brunzelle J. S., Dussault I., Lin M., Ianculescu I., Sherman M. A., Forman B. M., Fernandez E. J. (2004) Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol. Cell 16, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Welch B. L. (1947) The generalization of students problem when several different population variances are involved. Biometrika 34, 28–35 [DOI] [PubMed] [Google Scholar]
- 33. Westin S., Rosenfeld M. G., Glass C. K. (2000) Nuclear receptor coactivators. Adv. Pharmacol. 47, 89–112 [DOI] [PubMed] [Google Scholar]
- 34. Leo C., Chen J. D. (2000) The SRC family of nuclear receptor coactivators. Gene 245, 1–11 [DOI] [PubMed] [Google Scholar]
- 35. Lonard D. M., O'Malley B. W. (2012) Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 8, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Millard C. J., Watson P. J., Fairall L., Schwabe J. W. (2013) An evolving understanding of nuclear receptor coregulator proteins. J. Mol. Endocrinol. 51, T23–T36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson A. B., O'Malley B. W. (2012) Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol. Cell Endocrinol. 348, 430–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wurtz J. M., Bourguet W., Renaud J. P., Vivat V., Chambon P., Moras D., Gronemeyer H. (1996) A canonical structure for the ligand-binding domain of nuclear receptors. Nat. Struct. Biol. 3, 87–94 [DOI] [PubMed] [Google Scholar]
- 39. Schulman I. G., Chakravarti D., Juguilon H., Romo A., Evans R. M. (1995) Interactions between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. Proc. Natl. Acad. Sci. U.S.A. 92, 8288–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schulman I. G., Li C., Schwabe J. W., Evans R. M. (1997) The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 11, 299–308 [DOI] [PubMed] [Google Scholar]
- 41. Thompson P. D., Remus L. S., Hsieh J. C., Jurutka P. W., Whitfield G. K., Galligan M. A., Encinas Dominguez C., Haussler C. A., Haussler M. R. (2001) Distinct retinoid X receptor activation function-2 residues mediate transactivation in homodimeric and vitamin D receptor heterodimeric contexts. J. Mol. Endocrinol. 27, 211–227 [DOI] [PubMed] [Google Scholar]
- 42. Ito M., Fukuzawa K., Mochizuki Y., Nakano T., Tanaka S. (2008) Ab initio fragment molecular orbital study of molecular interactions between liganded retinoid X receptor and its coactivator; part II: influence of mutations in transcriptional activation function 2 activating domain core on the molecular interactions. J. Phys. Chem. A 112, 1986–1998 [DOI] [PubMed] [Google Scholar]
- 43. Germain P., Iyer J., Zechel C., Gronemeyer H. (2002) Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415, 187–192 [DOI] [PubMed] [Google Scholar]
- 44. Lou X., Toresson G., Benod C., Suh J. H., Philips K. J., Webb P., Gustafsson J. A. (2014) Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA. Nat. Struct. Mol. Biol. 21, 277–281 [DOI] [PubMed] [Google Scholar]
- 45. Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T. P., Rastinejad F. (2008) Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature 456, 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 47. Kalkhoven E., Valentine J. E., Heery D. M., Parker M. G. (1998) Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perlmann T., Rangarajan P. N., Umesono K., Evans R. M. (1993) Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 7, 1411–1422 [DOI] [PubMed] [Google Scholar]
- 49. Kurokawa R., Yu V. C., Näär A., Kyakumoto S., Han Z., Silverman S., Rosenfeld M. G., Glass C. K. (1993) Differential orientations of the DNA-binding domain and carboxyl-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 7, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 50. Zechel C., Shen X. Q., Chen J. Y., Chen Z. P., Chambon P., Gronemeyer H. (1994) The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J. 13, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2003) Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23, 3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Westin S., Kurokawa R., Nolte R. T., Wisely G. B., McInerney E. M., Rose D. W., Milburn M. V., Rosenfeld M. G., Glass C. K. (1998) Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 395, 199–202 [DOI] [PubMed] [Google Scholar]
- 53. Osz J., Brélivet Y., Peluso-Iltis C., Cura V., Eiler S., Ruff M., Bourguet W., Rochel N., Moras D. (2012) Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proc. Natl. Acad. Sci. U.S.A. 109, E588–E594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu Z., Boss O. (2007) Targeting PGC-1 α to control energy homeostasis. Expert. Opin. Ther. Targets 11, 1329–1338 [DOI] [PubMed] [Google Scholar]
- 55. Hwang J. Y., Attia R. R., Zhu F., Yang L., Lemoff A., Jeffries C., Connelly M. C., Guy R. K. (2012) Synthesis and evaluation of sulfonylnitrophenylthiazoles (SNPTs) as thyroid hormone receptor-coactivator interaction inhibitors. J. Med. Chem. 55, 2301–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. (1998) Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol. Endocrinol. 12, 302–313 [DOI] [PubMed] [Google Scholar]
- 57. Gill R. Q., Sterling R. K. (2001) Acute liver failure. J. Clin. Gastroenterol. 33, 191–198 [DOI] [PubMed] [Google Scholar]
- 58. de Urquiza A. M., Liu S., Sjöberg M., Zetterström R. H., Griffiths W., Sjövall J., Perlmann T. (2000) Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science 290, 2140–2144 [DOI] [PubMed] [Google Scholar]
- 59. Lengqvist J., Mata De Urquiza A., Bergman A. C., Willson T. M., Sjövall J., Perlmann T., Griffiths W. J. (2004) Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor α ligand-binding domain. Mol. Cell Proteomics 3, 692–703 [DOI] [PubMed] [Google Scholar]
- 60. Volakakis N., Joodmardi E., Perlmann T. (2009) NR4A orphan nuclear receptors influence retinoic acid and docosahexaenoic acid signaling via up-regulation of fatty acid binding protein 5. Biochem. Biophys. Res. Commun. 390, 1186–1191 [DOI] [PubMed] [Google Scholar]