Background: Mortalin/mtHsp70 is an essential stress chaperone frequently enriched in cancers.
Results: Mortalin is present in the nucleus of cancer cells where it causes strong inactivation of tumor suppressor protein p53 and activation of telomerase and heterogeneous ribonucleoprotein K (hnRNP-K) proteins.
Conclusion: Nuclear mortalin promotes carcinogenesis.
Significance: This study is important for the development of mortalin-based anticancer treatments.
Keywords: Cancer Biology, Heat Shock Protein (HSP), p53, Telomerase, Tumor Promoter, hnRNP-K, Mortalin, Nuclear Localization, p53 Inactivation, Telomerase Activation
Abstract
The Hsp70 family protein mortalin is an essential chaperone that is frequently enriched in cancer cells and exists in various subcellular sites, including the mitochondrion, plasma membrane, endoplasmic reticulum, and cytosol. Although the molecular mechanisms underlying its multiple subcellular localizations are not yet clear, their functional significance has been revealed by several studies. In this study, we examined the nuclear fractions of human cells and found that the malignantly transformed cells have more mortalin than the normal cells. We then generated a mortalin mutant that lacked a mitochondrial targeting signal peptide. It was largely localized in the nucleus, and, hence, is called nuclear mortalin (mot-N). Functional characterization of mot-N revealed that it efficiently protects cancer cells against endogenous and exogenous oxidative stress. Furthermore, compared with the full-length mortalin overexpressing cancer cells, mot-N derivatives showed increased malignant properties, including higher proliferation rate, colony forming efficacy, motility, and tumor forming capacity both in in vitro and in vivo assays. We demonstrate that mot-N promotes carcinogenesis and cancer cell metastasis by inactivation of tumor suppressor protein p53 functions and by interaction and functional activation of telomerase and heterogeneous ribonucleoprotein K (hnRNP-K) proteins.
Introduction
Mortalin/mtHsp70/Grp75 is a member of the Hsp70 family of proteins (1) that plays an essential role in mitochondrial import, oxidative stress response, regulation of mitochondrial membrane potential, energy generation, intracellular transport, chaperonization, p53 functions, immune response, and protection against apoptosis and tumorigenesis (2–8). Mortalin and p53 colocalize and interact in transformed, but not in normal, human cells, resulting in the nuclear exclusion and transcriptional inactivation of p53 (7, 9–11). Malignant transformation of NIH 3T3 cells, life span extension of MRC-5 cells, and attenuation of differentiation of HL-60 cells by overexpression of mortalin (mot-2) has been attributed, at least in part, to transcriptional inactivation of p53 (12–14). Furthermore, it has been shown to inhibit the p53-dependent suppression of centrosome duplication, leading to aneuploidy (an established hallmark of cancer cells) (6, 15). Walker et al. (11) have reported that, although mortalin and p53 proteins formed complexes in the cytoplasm of leukemic clam hemocytes, normal hemocytes lacked this interaction. Treatment of leukemic clam hemocytes with MKT-077, a cationic mitochondriotropic dye that has been shown to target the mortalin-p53 interaction (16, 17), resulted in the translocation and reactivation of p53 in clam cells (11). These data imply that mortalin-mediated inactivation of p53 is an evolutionarily conserved feature of cancer.
The expression profile of mortalin in normal and a variety of immortal and tumorigenic cell lines revealed its biphasic behavior: an initial elevation during immortalization (relative to a down-regulation during replicative senescence of human fibroblasts), followed by an up-regulation at a later stage that coincides with the acquisition of an invasive phenotype (18–20). In proteomic analyses of cancer tissue arrays, mortalin has been identified as a prognostic marker of colorectal cancers (21, 22). Associated with its phosphorylation, mortalin is known to show enhanced binding with FGF-1 and to be involved in the regulation of its mitogenic activity (23). It has been shown that, although cancers are frequently associated with a higher level of mortalin expression, Alzheimer and Parkinson pathologies involve the loss of mortalin and an imbalance in mitochondrial homeostasis (3, 24–27). Overexpression of mortalin in experimental models of these diseases resulted in the improvement of disease phenotypes and protection against oxidative stress, a hallmark of these dementias (24–26, 28, 29).
In line with the role of mortalin in carcinogenesis, anti-mortalin molecules, such as antisense, ribozyme, siRNA, p53-antagonist polypeptides, and chemicals that abrogated mortalin-p53 interaction and caused the relocation of p53 to the cell nucleus, resulted in growth arrest/apoptosis of cancer cells (2, 4, 6, 30). Mortalin targeting adeno-oncolytic viruses caused tumor suppression in vivo by activation of p53, induction of apoptosis, and inhibition of angiogenesis (31). Furthermore, the up-regulation of mortalin correlated with an early recurrence of hepatocarcinoma in postoperative patients and liver cancer metastasis (32), suggesting that anti-mortalin molecules not only serve as anticancer agents but could also be potentially very important in the prevention of cancer recurrence. Together, these reports have necessitated investigations of the molecular mechanisms of the roles of mortalin in human tumorigenesis.
Mortalin has been reported to exist in multiple subcellular localizations, including the mitochondrion, endoplasmic reticulum, plasma membrane, cytosol, and centrosomes (6, 15, 27, 33, 34). Recently, Rozenberg et al. (22) have reported circulating mortalin in the serum of colorectal cancer patients, and its elevated levels (>60 ng/ml) were assigned as a risk factor for shorter survival. On the other hand, Shih et al. (35) reported that the nuclear translocation of mortalin is critically involved in neuronal cell differentiation. In light of these reports, we examined whether mortalin exists in the nucleus of human normal and transformed cells. We demonstrate that mortalin is present in the nucleus of cancer cells, where it promotes tumor aggressiveness by mechanisms involving inactivation of p53 functions and activation of telomerase, heterogeneous ribonucleoprotein K (hnRNP-K),4 and MMPs.
EXPERIMENTAL PROCEDURES
Cell Culture and Fractionation
Normal human fibroblasts (MRC5, TIG-1, and WI-38), breast carcinoma cells (MCF7, MDA-MB-231, and T47D), osteosarcoma cells (U2OS and Saos-2), fibrosarcoma cells (HT1080), cervical carcinoma cells (HeLa), lung adenocarcinoma cells (A549), colon carcinoma cells (HCT116), and prostate carcinoma cells (DU145) were maintained in DMEM (Invitrogen) as described previously (4). Cells were procured from the Japanese Collection of Research Bioresources, Cell Bank, National Institute of Biomedical Innovation, Japan (MRC5, TIG-1, WI-38, HT1080, HeLa, and A549 cells); from the DS Pharma Biomedical Co. Ltd, Japan (MCF7, T47D, and U2OS cells), and from the Cell Resource Center for Biomedical Research, Institute of Development, Aging, and Cancer, Tohoku University, Japan (DU145 cells). Nuclear and mitochondrial fractions were prepared using the Qproteome cell compartment kit and mitochondrial isolation kit, respectively (Qiagen, Hilden, Germany).
Cell Proliferation Rate and Oxidative Stress Response
Cell proliferation was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Invitrogen) using a 96-well plate (103 cells/well). For oxidative stress, cells were treated with 300 μm (MCF7 cells) or 1.0 mm (U2OS cells) hydrogen peroxide (2 h), followed by recovery in fresh medium (48–72 h). All assays were performed independently and at least three times.
Detection of Reactive Oxygen Species
Cells were cultured on glass coverslips placed in 12-well plates and stained for ROS by fluorescent staining using the Image-iTTM LIVE green ROS detection kit (Molecular Probes, Eugene, OR). Images were captured on a Zeiss Axiovert 200 M microscope and analyzed by AxioVision 4.6 software (Carl Zeiss Microimaging, Thornwood, NY).
Western Blotting
Cells were lysed with radioimmune precipitation assay buffer (Thermo Scientific, Rockford, IL) containing complete protease inhibitor mixture (Roche Applied Science). Cell lysates (or the conditioned medium from cells for MMP-2) were separated on an SDS-polyacrylamide gel and transferred onto a PVDF membrane that was incubated with antibodies against mortalin (17), Myc tag, lamin A/C (Cell Signaling Technology, Danvers, MA), p53, MMP-2, MMP-3, MMP-9 (Santa Cruz Biotechnology, Santa Cruz, CA), α-tubulin (Sigma), hTERT, actin (Abcam, Cambridge, UK), hnRNP-K/J (ImmuQuest), and V5 (Invitrogen) as indicated. Protein bands were detected by chemiluminescence (GE Healthcare) using LAS3000-mini (Fuji Film, Tokyo, Japan) through a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology).
Immunostaining
Cells were plated on coverslips placed in a 12-well culture plate. Control or etoposide-treated (10 μm for 24 h) cells were incubated with primary antibodies (p53 and myc, Santa Cruz Biotechnology; Aurora A, Sigma; or mortalin (4)) at 4 °C overnight, followed by extensive washing in TPBS (PBS with 0.2% Triton X-100) and incubation with the fluorochrome-conjugated secondary antibodies (Alexa Fluor 488-conjugated goat anti-rabbit or anti-mouse or Alexa Fluor 594-conjugated goat anti-rabbit or anti-mouse (Molecular Probes)) as described previously (4). Stained cells were examined on a Zeiss Axiovert 200 M microscope and analyzed by AxioVision 4.6 software (Carl Zeiss). To examine the presence of mortalin in the nucleus, images were acquired with a confocal laser scanning microscope (Zeiss LSM 700) confocal microscope. The files were transferred to a graphic work station and analyzed with Imaris software (Bitplane, Zurich, Switzerland).
Immunohistochemistry
Paraffin-embedded cancer and normal ovarian tissue slides were purchased from BioChain Institute, Inc. (Newark, CA). Slides were deparaffinized in xylene and incubated with anti-mortalin antibody. For detection of antibody binding, the REAL-HRP system (Dako REALTM EnVisionTM detection system, peroxidase/3,3′-diaminobenzidine+, and rabbit/mouse (Dako, Carpinteria, CA) were used. The color was developed using the liquid 3,3′-diaminobenzidine+ substrate chromogen system (Dako) followed by counterstaining with Mayer's hematoxylin (Wako, Tokyo, Japan). Slides were examined using a fluorescence microscope (BZ-9000, Keyence, Tokyo, Japan), and images were obtained using BZ-II Analyzer software (Keyence).
Construction of Mortalin Mutants and Cell Lines
Full-length mortalin and its three deletion mutants (full-length mortalin (mot-F); mot-N, amino acid residues 42–679; mot-A, N-terminal amino acid residues 1–180; and mot-B, amino acid residues 180–300) were generated by PCR and cloned into the HindIII site of the vector pCX4neo. Retroviruses were produced in Plat-E cells as described previously (18). The stably infected cells were maintained in 100 μg/ml G418-supplemented medium. V5-tagged full-length and a deletion mutant lacking signal peptide were generated using the pcDNA3.1 plasmid vector. Transfections were performed using X-tremeGENE 9 DNA transfection reagent (Roche) following the instructions of the manufacturer.
Colony Forming Assay
A cell transformation assay was performed using a cell transformation detection kit (Chemicon). Colony forming efficacy is expressed as the mean number of colonies from three independent experiments. For colony forming assays, 1 × 103 cells were seeded on 6-well plate. Cells were maintained until the appearance of colonies with regular change of medium. Colonies were fixed in methanol, stained with 0.5% crystal violet, photographed, and counted.
Chemotaxis and Cell Invasion Assay
Cells at 60–70% confluency were trypsinized and resuspended in DMEM. 2 × 104 cells were plated in 12 mm-pored Transwell inserts for the chemotaxis assay (Costar, MA). Fibronectin from human plasma (Sigma) was used as a chemoattractant. A cell invasion assay was performed using a QCMTM 24-well cell invasion assay kit (Millipore). The fluorescence of invaded cells was read using a microplate reader (Infinite 200 PRO, Tecan Group Ltd.) with a 480/520 nm filter set.
Nude Mice Tumor Assay
BALB/c nude mice (4 weeks old, female; Nihon Clea, Japan) were given subcutaneous injections of cells (1 × 106 suspended in 0.2 ml of growth medium). Tumor formation was monitored for a month. The assay was regarded as positive if tumors appeared and grew progressively. For the metastasis assay, cells were injected intravenously into the lateral tail vein of Balb/c nude mice. Two weeks after injection, lungs were harvested and screened for tumor nodules. Five mice were used for each group, and the experiment was repeated twice.
p53-dependent Reporter Assays
Control, mot-F, and mot-N cells (1 × 105/well) were plated in a 6-well plate and transfected with 1 μg of pGL13-luc plasmid vector containing a firefly luciferase gene and p53-responsive elements. The pRL-TK (Promega, Madison, WI) vector was used for transfection efficiency control. Cells were lysed, and luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega) and microplate reader (Infinite 200 PRO, Tecan Group Ltd.).
Telomere Repeat Amplification Protocol Assay
A telomere repeat amplification protocol assay (for semiquantitative detection of telomerase activity) was performed using the TeloTAGGG telomerase PCR ELISA kit (Roche Applied Science). Cell lysates were prepared in lysis reagent from cultured (2 × 105) cells. The supernatant collected after centrifugation at 16,000 × g for 20 min at 4 °C was used for the telomere repeat amplification protocol assay. Telomerase activity in the sample was calculated as units of activity relative to the positive control (MRC5 cells transfected with hTERT). U2OS cells that lacked telomerase activity were used as a negative control. All assays were performed in triplicate.
Coimmunoprecipitation and Immunodepletion Assays
For coimmunoprecipitation, cell lysates (300 μg of protein) in 300 μl of radioimmune precipitation assay buffer were incubated at 4 °C for 1–2 h with anti-myc, anti-hTERT, or anti-mortalin antibody (as indicated). Immunocomplexes were separated by incubation with protein-A/G-Sepharose (Santa Cruz Biotechnology), and Western blotting was performed with the indicated antibodies using the procedure described above. For immunodepletion of hTERT, cell lysates were incubated with anti-telomerase antibody (a gift from Dr. Reddel, Children's Medical Research Institute (CMRI), Australia). Immunoprecipitated hTERT was separated by binding of IgG with the protein-A/G-Sepharose beads. The supernatant was again incubated with the anti-hTERT antibody, and the process was repeated four times. An aliquot of the supernatant after each round of immunoprecipitation was resolved on an SDS gel and was processed for Western blotting with anti-mortalin antibody. Control immunodepletion was performed with isotype-matched, irrelevant preimmune IgG.
Statistical Analysis
Data are expressed as mean ± S.E., and the significance of differences between groups was determined by Mann-Whitney test (nonparametric rank sum test) using Stat View software (Abacus Concepts, Inc., Berkeley, CA). Differences were considered significant when p < 0.05.
RESULTS AND DISCUSSION
Mortalin Is Present in the Nuclei of Cancer Cells
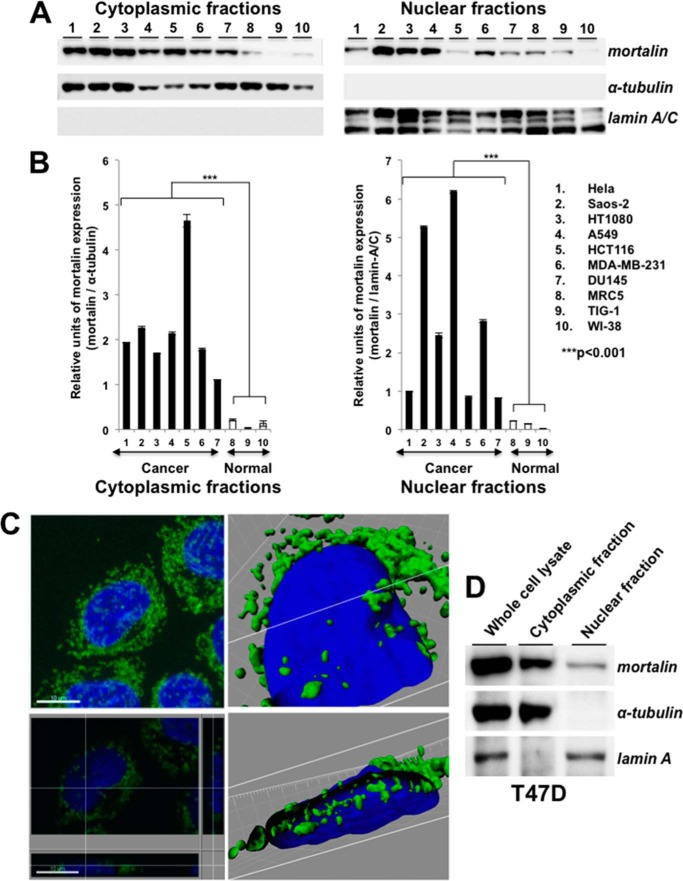
Cytoplasmic and nuclear fractions of several human cancer cell lines were subjected to Western blotting with anti-mortalin antibody. As shown in Fig. 1A, mortalin was enriched in cancer cells and was detected in their nuclear fractions. To rule out the cross-contamination of subcellular fractions, the blots were probed with antibodies to α-tubulin (for cytoplasmic fractions) and lamin A/C (for nuclear fractions) (Fig. 1, A and B). Furthermore, we performed a high-resolution single-cell image analysis that confirmed the presence of mortalin in the nuclei of cancer cells (Fig. 1, C and D) and raised the proposition that nuclear mortalin may contribute functionally to the cancer properties.
FIGURE 1.
Mortalin is expressed in the nuclei of cancer cells. A, Western blotting of the cytoplasmic and nuclear cell fractions with anti-mortalin antibody showed its presence in the nuclear fractions. Cytoplasmic protein, α-tubulin, and the nuclear protein lamin A/C were used as respective controls for the cytoplasmic and nuclear fractions. B, quantitation of mortalin expression in cytoplasmic and nuclear fractions, S.D. and statistical significance from three independent experiments. C, high-resolution spectral imaging showing mortalin in the cross-section of the nucleus (bottom right panel) of T47D cells. Top left panel, immunostaining of mortalin (green). Nuclei were stained with Hoechst (blue). The white lines in the bottom left, top right, and bottom right panels show the cross-section points. D, Western blot analysis of cytoplasmic and nuclear cell fractions of T47D cells with anti-mortalin antibody showing the presence of mortalin predominantly in the nuclear fraction of cancer cells.
Nuclear Mortalin Has a Proproliferative Function and Protects Cells against Oxidative Stress
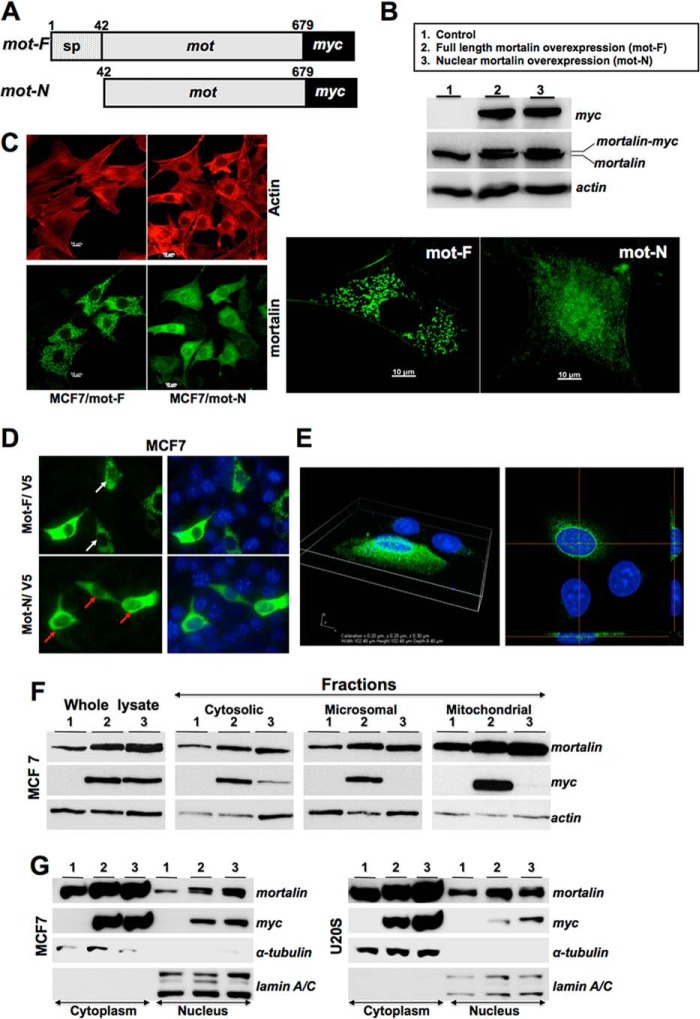
To delineate the functional significance of nuclear mortalin in cancer cells, we generated a mutant that lacked the N-terminal 41 amino acids, the mitochondrial targeting signal peptide sequence. Retroviral expression constructs encoding mot-F (full-length protein, 1–679 residues) and mot-N (amino acid residues 42–679) tagged with a myc epitope were transduced into non-malignant MCF7 breast cancer cells. Expression of mot-F and mot-N proteins was detected by Western blotting with anti-myc and anti-mortalin antibodies. As shown in Fig. 2, A–C, Western blotting and immunostaining of the control and infected cells revealed that the mot-F and mot-N proteins were expressed equally and could be distinguished from the endogenous mortalin (Fig. 2B) and that mot-N was translocated efficiently to the nucleus (Fig. 2C). Similar results were obtained when V5-tagged mot-F and mot-N proteins were expressed in MCF7 and U2OS cells (Fig. 2, D and E). High-resolution three-dimensional imaging confirmed the presence of mot-F and mot-N in the nucleus. There was more mot-N than mot-F. We next performed fractionation of the transduced cells. Western blotting of cell fractions with anti-myc and anti-mortalin antibodies also revealed that the mot-F localized in the cytosol and in the microsomal and mitochondrial fractions (Fig. 2F, lanes 2). On the other hand, only a fraction of the total mot-N protein in the cytosol fractions and none in the microsomal and mitochondrial fractions was detected (Fig. 2F, lanes 3 and data not shown). Examination of the nuclear fractions confirmed the nuclear localization of mot-N (Fig. 2G). Similar results were obtained using different cell lines and in independent experiments. Of note, we found that exogenously expressed full-length mortalin, mot-F, was also detected in the nuclear fractions, confirming that the nuclear translocation of mortalin is an innate phenomenon in cancer cells, as also supported by the data in Fig. 1.
FIGURE 2.
Subcellular localization of native and exogenously expressed normal and mutant mortalin. A, schematic diagram of mortalin constructs. sp, signal peptide. B, expression of exogenously expressed mot-F and mot-N as detected by anti-myc antibody. C, immunostaining of mortalin with anti-mortalin antibody (green) in mot-F- and mot-N-transduced cells. Actin was used as a control. Right panel, enlarged images showing the predominant localization of mot-N in the cell nuclei. D and E, immunostaining of V5-tagged mot-F- and mot-N-transfected cells with anti-V5 antibody (green) in MCF7 (D) and U2OS (E) cells. Nuclei were stained blue with Hoechst 33342. White and red arrows point to the weak and strong nuclear staining in mot-F/V5- and mot-N/V5-transfected cells, respectively. E, three-dimensional image and cut section of a V5-stained U2OS cell showing the presence of V5-tagged mot-N in the nucleus. F and G, Western blot analyses showing mortalin in the subcellular fractions detected by anti-myc and anti-mortalin antibodies. F, Mot-F (lane 2), but not Mot-N (lane 3), was localized in the microsomal and mitochondrial fractions. Mot-N (lane 3) was detected in the cytosol only. G, Western blotting of the cytoplasmic and nuclear fractions of mot-F and mot-N cells revealed that the transfected mot-F protein also localized in the nucleus, although less than mot-N protein. α-tubulin and lamin A/C were used as internal controls for loading and purity of cytoplasmic and nuclear fractions, respectively.
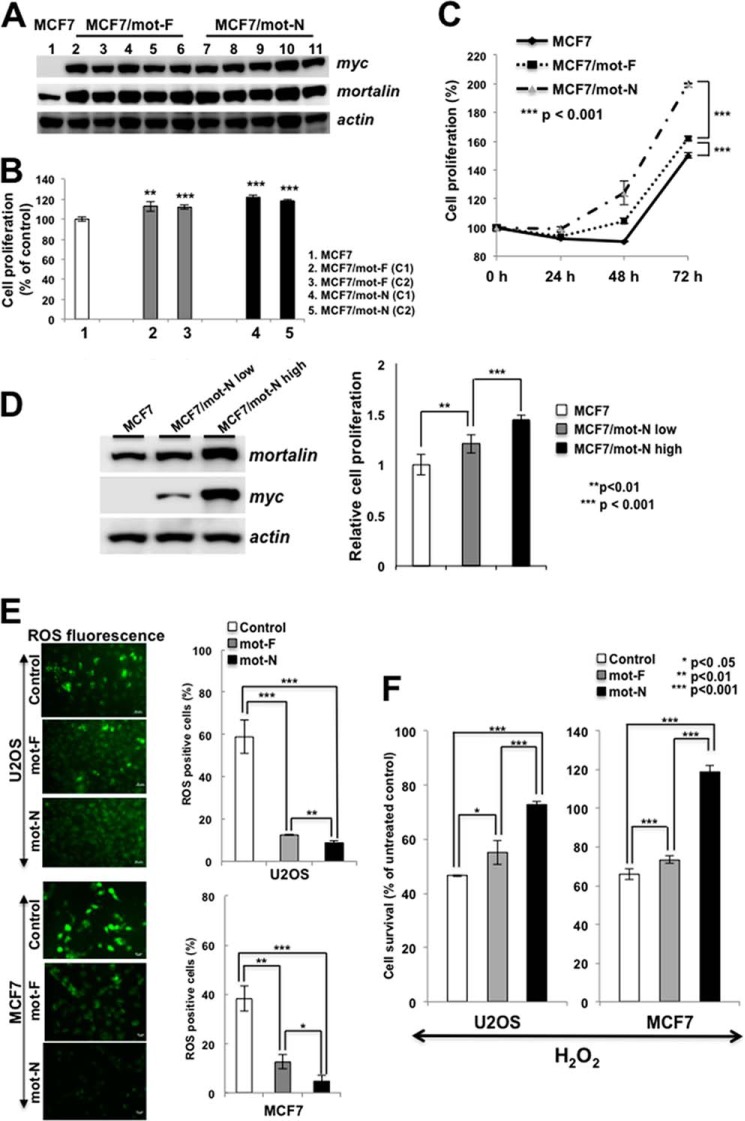
We next examined the effect of mot-F (which mimics the endogenous mortalin localized at multiple subcellular sites) and mot-N (which is nuclear-enriched) on cancer cell properties. Multiple clones with stable expression of mot-F and mot-N were isolated and first examined the level of mortalin expression for two to three passages (Fig. 3A). The clones were then subjected to cell proliferation, oxidative stress, and malignant transformation assays. As shown in Fig. 3B, mortalin-overexpressing cells showed a higher proliferation rate compared with the untransfected/vector-transfected (control) cells. Furthermore, mot-N derivatives showed higher proliferation compared with the mot-F derivatives (Fig. 3, B and C, and data not shown). To further resolve the effect of mot-N on cell proliferation, we isolated low and high mot-N expression clones. As shown in Fig. 3, C and D, a dose-dependent increase in cell proliferation was detected. Cells were examined for their endogenous and exogenous oxidative stress tolerance by quantitation of ROS. As shown in Fig. 3E, mot-F- and mot-N-overexpressing cells showed significantly lower ROS levels (3-fold), suggesting a decrease in the level of oxidative stress in these cells compared with the untransfected as well as vector-transfected (control) cells. Furthermore, mot-F- and mot-N-overexpressing cells, when exposed to oxidative stress by incubation in H2O2-supplemented medium, showed better survival than the control cells, suggesting their increased tolerance to exogenous oxidative stress (Fig. 3F). In both assays, mot-N cells were more tolerant than mot-F cells. Similar data were obtained with U2OS and MCF7 cells (Fig. 3, E and F).
FIGURE 3.
Nuclear mortalin contributes to proproliferative and oxidative stress tolerance in cancer cells. A, expression level of transduced mot-F and mot-N protein in several independent clones as examined by Western blotting. All isolated clones (five each) showed comparable levels of expression. B, comparison of the growth rate of control of mot-F and mot-N derivatives as determined by the time-dependent increase in cell number (48 h post-plating). C, average proliferation capacity of the control, mot-F and mot-N derivatives showed increased proliferation capacity of mot-N cells compared with control and mot-F cells. D, proliferation rate of cells corresponding to low and high levels of mot-N expression. E, endogenous level of oxidative stress as detected by ROS levels, showing a low level of oxidative stress in mot-N cells. Quantitation of the data from three independent experiments in U2OS and MCF7 cells is shown. Scale bars = 20 μm. F, response to the exogenous oxidative stress as evaluated by exposure of cells to hydrogen peroxide showed better stress tolerance in mot-N cells compared with control and mot-F cells.
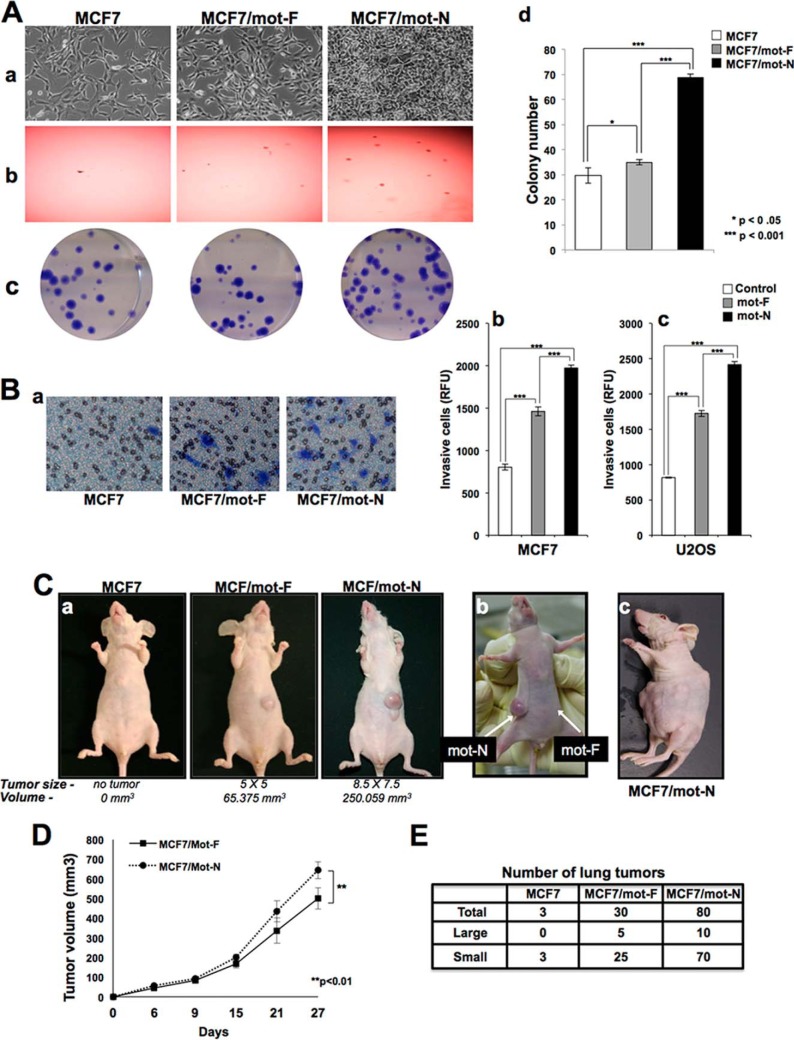
In vitro and in vivo cell transformation assays revealed the malignant transformation capacity of mot-F- and mot-N-overexpressing cells (Fig. 4, A–C). Overexpression of mot-F caused a malignant transformation of cancer cells as detected by subcutaneous bone and breast xenograft models of nude mice in a tumor formation assay (12, 18 and data not shown). Furthermore, we found that the mot-N derivatives were more aggressively transformed than mot-F cells (Fig. 4). The mot-N derivatives showed a rounded morphology (Fig. 4A, a), higher anchorage-independent growth capability (Fig. 4A, b), and higher colony forming efficiency (Fig. 4A, c and d) compared with the mot-F derivatives. Cell migration assays revealed higher migration (Fig. 4B, a) and invasion capacity (Fig. 4B, b and c) of mot-N cells. An in vivo tumor-forming capacity assay in nude mice revealed that mot-N derivatives were transformed aggressively. They formed tumors rapidly and aggressively compared with the mot-F derivatives (Fig. 4C, a, and D). Furthermore, when MCF7/mot-F and MCF7/mot-N cells were injected into the opposite flank of the same mouse, MCF7/mot-N tumors appeared earlier and increased rapidly in size compared with the MCF7/mot-F tumors (Fig. 4C, b). Some mice injected with mot-N derivatives showed multiple tumors all over the injected side of the body (Fig. 4C, c). Furthermore, consistent with the in vitro migration assay data, MCF7/mot-N cells showed aggressive lung metastasis in in vivo (tail vein injection assays) models (Figs. 4E). Taken together, the data demonstrate that mot-N has a proproliferative function and that it contributes to the malignant transformation of cancer cells.
FIGURE 4.
Nuclear mortalin contributes to proproliferative and malignant characteristics of cancer cells. A, effect of exogenous expression of mot-F and mot-N on cell morphology (a), anchorage independent growth of cells (b), and colony forming efficacy (c and d). B, cell migration assay (a) and cell invasion assay (b and c) showing increased migration of MCF7/mot-N compared with MCF7/mot-F cells. C, nude mice tumor formation for MCF7, MCF7/mot-F, and MCF7/mot-N cells (a) and aggressiveness of mot-N compared with the mot-F tumors (b and c). D, average tumor volume of mot-F and mot-N derivatives of MCF7. MCF7 untransduced cells did not form tumors. E, in vivo lung metastasis of MCF7, MCF7/mot-F and MCF7/mot-N cells showing significantly higher number of tumors in case of MCF7/mot-N cells.
Nuclear Mortalin Contributes to Malignant Transformation by Strong Inactivation of p53
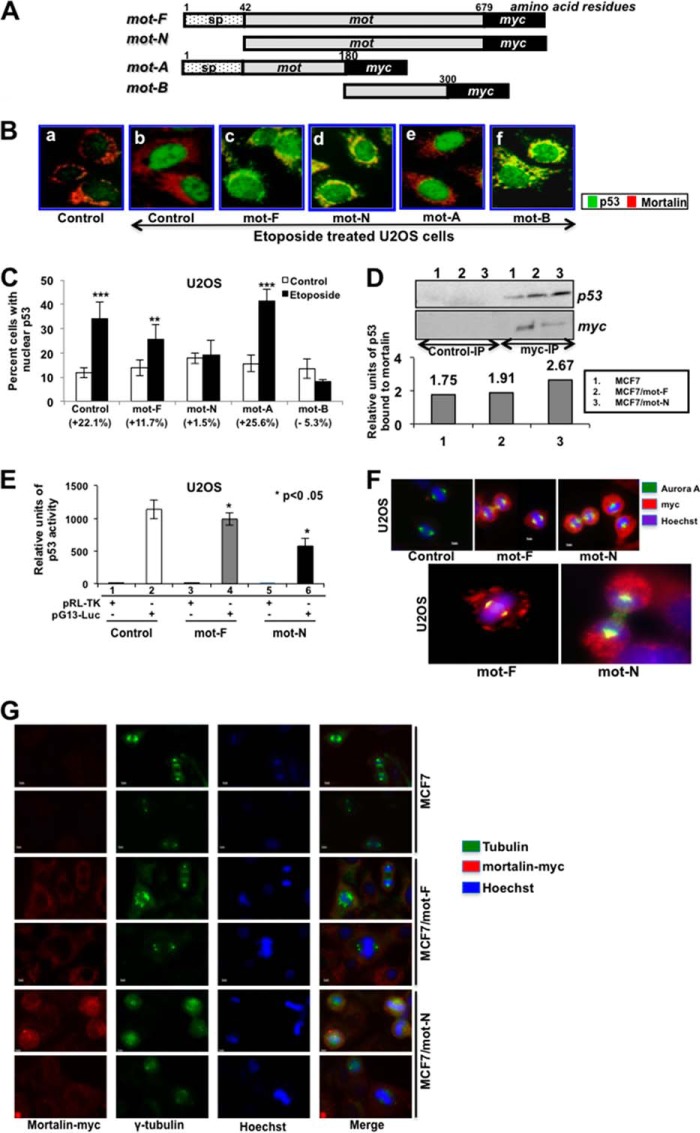
Mortalin has been shown to inactivate p53 by sequestering it in the cytoplasm and also by binding it in the nucleus at the G1 stage of the cell cycle, where it inactivates its control of the centrosome duplication function (4, 6, 9, 10, 15). In light of the finding that mortalin exists in the nucleus of malignantly transformed cancer cells, we investigated the impact of mot-F and mot-N on p53 function by binding and activity assays. For this study, two more deletion mutants (mot-A (N-terminal amino acid residues 1–180 that lack the p53 binding region) and mot-B (amino acid residues 180–300 containing the p53 binding region)) were generated. Similar to mot-F and mot-N, these constructs were designed to have a myc tag at the carboxyl terminus (Fig. 5A). The cells infected with mot-F, mot-N, mot-A, and mot-B were examined for nuclear translocation of p53 in response to DNA damage stress in U2OS cells that harbor wild-type p53 function. As shown in Fig. 5B, control cells showed that the etoposide caused nuclear translocation of p53 (green) as seen by its intense nuclear staining and absence of p53 (green) in the cytoplasm (Fig. 5B, compare a with b). Cells infected with mot-F, mot-N, and mot-B strongly inhibited the nuclear translocation of p53 (Fig. 5B, c, d, and f). The presence of p53 in the cytoplasm was marked by an overlay of p53 and mortalin (yellow) (Fig. 5B, c, d, and f). Mutant mot-A-infected cells (which lacked the p53 binding region of mortalin) did not show any difference from the control etoposide-treated cells, and there was no colocalization of mortalin and p53 in the cytoplasm (Fig. 5B, b and e). To quantitate this effect, we counted the cells with very intense p53 staining (Fig. 5C) and found that mot-F, mot-N, and mot-B caused an approximately 2- to 20-fold decrease in the etoposide-induced nuclear translocation of p53. Of note, we found that the inactivation of p53 by mot-N was significantly higher than that of mot-F. Similar results were obtained in etoposide-treated MCF7 cells overexpressing either mot-F or mot-N (data not shown).
FIGURE 5.
Nuclear mortalin causes strong inactivation of p53. A, schematic diagram of mortalin constructs. sp, signal peptide. B, overexpression of mortalin and its p53-binding mutants inhibited etoposide-induced nuclear translocation of p53. Double immunostaining with anti-p53 (green) and anti-mortalin (red) antibodies showed nuclear translocation of p53 in control, mot-A mutant-transfected etoposide-treated cells. Mot-F-, mot-N-, and mot-B-transduced cells showed cytoplasmic retention of p53, as seen by cytoplasmic staining that is colocalized with mortalin and, hence, seen as yellow. C, quantitation of cells with etoposide-induced nuclear translocation of p53. D, coimmunoprecipitation (IP) of mot-F and mot-N with p53. Immunoprecipitated mortalin-myc by anti-myc antibody (Myc-IP) was examined for the presence of p53. Control immunoprecipitation was performed with isotype-matched IgG. Quantitation of p53 precipitated with mortalin-myc is shown. E, a p53-dependent reporter assay showed a stronger inactivation of p53 activity in mot-N-transduced cells. Luc, luciferase. F, coimmunostaining of mortalin and Aurora A revealed a stronger intensity of coimmunostaining (yellow signal) of the two proteins in MCF7/mot-N than MCF/mot-F cells. G, coimmunostaining of mortalin-myc and γ-tubulin showing the presence of aneuploid cells in mot-F and mot-N cells. The cell aneuploidy and anomalies score was higher in mot-N cells.
We anticipated that the stronger inactivation of p53 by mot-N may explain the increased malignant characteristics of cancer cells. To investigate this proposition, we examined the binding of p53 to mot-F and mot-N derivatives by coimmunoprecipitation assays. As shown in Fig. 5D, the amount of p53 precipitated with mot-N was more than with mot-F, suggesting higher complex formation between p53 and mot-N (Fig. 5D). Furthermore, p53-dependent reporter assays on mot-F and mot-N derivatives of U2OS cells revealed stronger inactivation in the latter (Fig. 5E). Control of centrosome duplication by p53 has been shown to be regulated by a p53-mortalin interaction at the centrosome and by the mitotic kinase Aurora A (6, 15). Hence, we examined the localization of mot-F and mot-N at the centrosome by coimmunostaining of mortalin and Aurora A. As shown in Fig. 5F, mot-N cells showed enhanced colocalization with Aurora A. Furthermore, inactivation of the control of the centrosome duplication function of p53, resulting in aneuploid chromosomes as examined by γ-tubulin staining, revealed higher aneuploidy in mot-F and mot-N derivatives compared with control MCF7 cells. The aneuploidy score was higher in cells expressing mot-N compared with the ones expressing mot-F protein (Fig. 5G and data not shown). These data suggested that nuclear mortalin contributes to the malignant properties of cancer cells through the strong inactivation of p53 functions, including transcriptional activation and control of centrosome duplication.
Nuclear Mortalin Contributes to Malignant Transformation by Activation of Telomerase and hnRNP-K
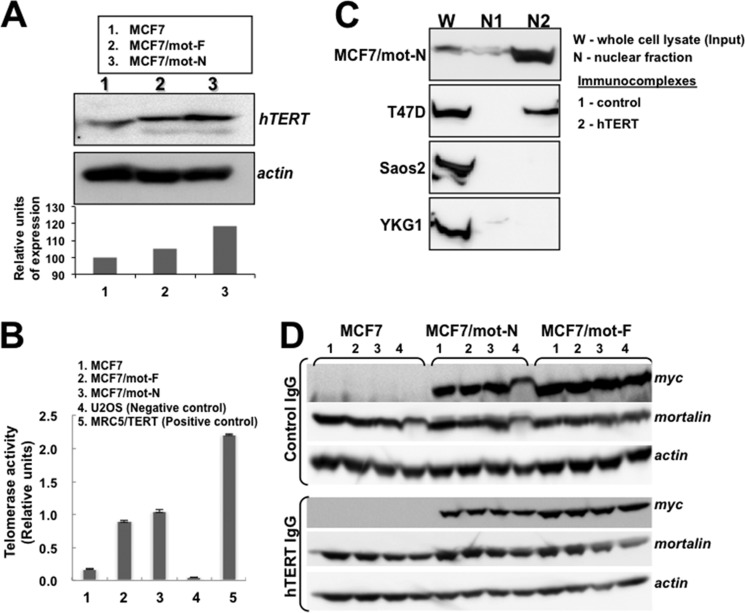
In view of the nuclear localization of mortalin and its impact on cell proliferation, transformation, and metastasis of cancer cells, we next examined its impact on two oncogenes, telomerase and hnRNP-K, that have been shown to act as key regulators of tumorigenesis and metastasis, respectively. As shown in Fig. 6, A and B, mot-F and mot-N cells showed higher levels of telomerase protein (Fig. 6A) and activity (Fig. 6B) compared with the control. Furthermore, telomerase activity was higher in mot-N derivatives than in mot-F cells. We next investigated whether the increase in telomerase activity was due to direct interaction with mortalin in the nucleus. Coimmunoprecipitation of hTERT and mortalin from the nuclear fractions of highly malignant cells, such as MCF7/mot-N and T47D cells, showed the presence of mortalin-hTERT complexes. Non-malignant cells (MCF7, Saos-2, and YKG-1), on the other hand, lacked a mortalin-telomerase interaction (Fig. 6C and data not shown). To quantitate these interactions, we performed serial immunodepletion assays on mot-F and mot-N cell lysates. Serial immunodepletion of hTERT using a hTERT-specific antibody caused parallel immunodepletion of mortalin (detected by anti-myc and anti-mortalin antibodies) in mot-F and mot-N cells (Fig. 6D), suggesting the direct interaction of telomerase and mortalin. These data demonstrate, for the first time, that mortalin interacts with telomerase in the nucleus, causes its stabilization and activation, and contributes to the malignant transformation of cancer cells.
FIGURE 6.
Mortalin interacts with telomerase, leading to activation of its function. A, Western blotting of hTERT showing a higher level of hTERT expression in mot-N cells. B, telomere repeat amplification protocol assay showing activation of telomerase in mot-F and mot-N cells. U2OS (an alternative lengthening of telomeres cell line) and MRC5 cells transfected with hTERT were used as negative and positive controls, respectively. C, coimmunoprecipitation of mortalin and hTERT in which hTERT complexes were examined for the presence of mortalin. The latter was seen to coimmunoprecipitate with hTERT both in the whole cell lysates and the nuclear fractions of malignantly transformed cells. D, immunodepletion of hTERT by repeated immunoprecipitation (bottom panel, lanes 1–4) of hTERT from cell lysates revealed a decrease in mortalin both in mot-F and mot-N cells. Irrelevant IgG immunoprecipitation used as a control did not show a decrease in mortalin in serial immunodepletion rounds (top panel, lanes 1–4).
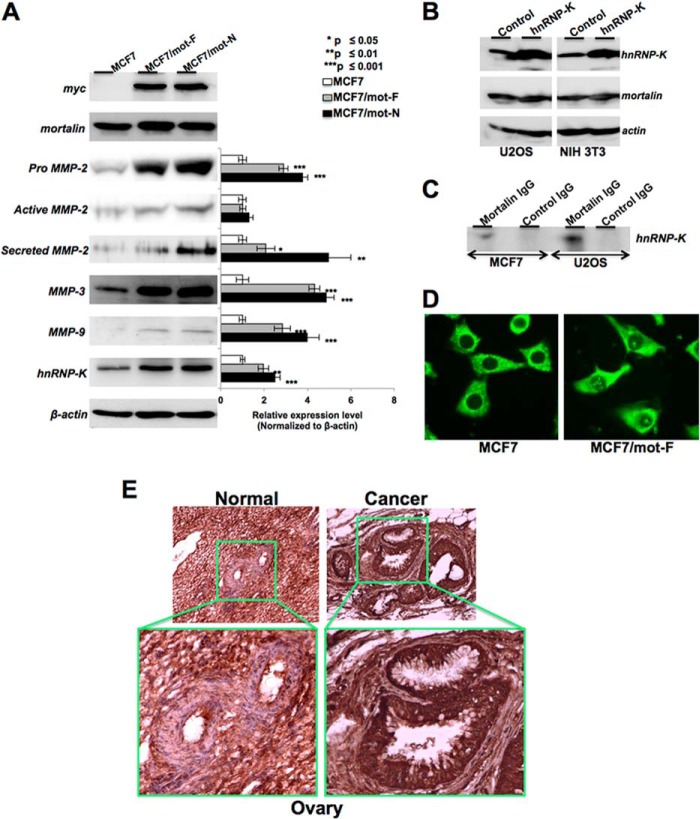
We next investigated the mechanism of increased metastatic properties of mot-F and mot-N cells (Figs. 7, A–D). Analyses of matrix metalloproteases (well established markers of cancer cell metastasis) in MCF7, MCF7/mot-F, and MCF7/mot-N cells revealed a statistically significant increase in the level of MMP-2, MMP-3, and MMP-9 expression (Fig. 7A). Furthermore, we investigated whether the increase in MMPs was due to activation of their upstream regulator, hnRNP-K (36–38), by nuclear mortalin. As shown in Fig. 7A, MCF7/mot-F and mot-N cells showed a higher level of expression of hnRNP-K that matched well with their high migratory characteristics in vitro and in vivo (Fig. 4). To investigate the cross-talk between mortalin and hnRNP-K, we also generated hnRNP-K-overexpressing cells and examined the level of mortalin expression. We found that the hnRNP-K-overexpressing cells possessed highly malignant characteristics (39). However, as shown in Fig. 7B, the level of mortalin expression in these cells remained unchanged compared with the parent cells, suggesting that mortalin may work upstream of hnRNP-K in promoting the cancer metastasis. Coimmunoprecipitation of mortalin and hnRNP-K showed that the two proteins form a complex (Fig. 7C), and an increased level of expression of hnRNP-K in mortalin-overexpressing cells might be the result of its stabilization and protection from HDM2-mediated degradation (40). In support of this, immunostaining revealed that a significantly high proportion of MCF7/mot-F and MCF7/mot-N cells have nuclear hnRNP-K, in contrast with MCF7 cells. Although only 10–20% MCF7 cells showed nuclear hnRNP-K, more than 75% of either the MCF7/mot-F or MCF7/mot-N cells exhibited strong nuclear staining (Fig. 7D).
FIGURE 7.
Mortalin interacts with hnRNP-K and leads to activation of its downstream effectors MMP-2, MMP-3, and MMP-9. A, MCF7/mot-F and mot-N cells showed an increase in MMP-2, MMP-3, MMP-9, and hnRNP-K. B, hnRNP-K overexpression did not affect the level of mortalin. C, coimmunoprecipitation assay showing the presence of hnRNP-K in mortalin immunocomplexes. D, immunostaining of hnRNP-K revealed its enhanced nuclear localization in mot-F and mot-N cells. E, immunohistochemical detection of mortalin in human normal and cancer ovarian tissues. An increased level of mortalin expression and its enrichment in nuclei are seen in cancer tissue.
Telomerase is a ribonucleoprotein complex composed of the catalytic protein subunit human telomerase reverse transcriptase (hTERT) and the RNA subunit human telomerase RNA that serves as the template for the synthesis of telomeric DNA and is expressed constitutively expressed in cells. Telomerase activity is closely associated with hTERT expression. In contrast to normal somatic cells, most human tumor cells possess a high level of hTERT expression. Several studies have demonstrated that telomerase activity is required for the malignant properties of cancer cells, and, hence, that they may serve as a good target for the development of anticancer drugs (41). Studies of the regulation of hTERT expression have revealed that it is regulated by trans-regulatory factors, including Sp1/Sp3, AP-2β, Myc/Max heterodimer, HIF-2α, TAK1, MSH2, GRHL2, p53, HER2, hnRNP-K, and hnRNP-D (42–44), that show physical interactions with the hTERT promoter. It has been shown earlier that mortalin overexpression cooperates with telomerase to extend the in vitro lifespan of normal human fibroblasts (13). Taken together with our data, we conclude that nuclear mortalin interacts with telomerase and activates its function, contributing to proliferation and malignant characteristics of cancer cells.
hnRNP-K is a member of the family of about 20 hnRNP proteins that are expressed ubiquitously, complex with heterogeneous nuclear RNA, and influence pre-mRNA processing and transport involved in transcription and posttranscriptional messenger RNA metabolism. hnRNP-K is known to regulate a multitude of gene expressions, including p53 (40), as a transcription factor or by alteration of mRNA stability and translation (45). Kang et al. (44) have shown that hnRNP-K associates physically with the hTERT promoter in vitro and in vivo. Although knockdown of hnRNP-K did not affect telomerase activity, its knockdown, along with GRHL2, resulted in an inhibition of telomerase function in oral squamous cell carcinoma (44). Consistent with its telomerase activity-enhancing function, hnRNP-K was enriched in human oral squamous cell carcinoma cells but not in normal cells (which lack telomerase activity). hnRNP proteins are known to shuttle between the nucleus and the cytoplasm and have a role during cell cycle progression. Cytoplasmic accumulation, which is required for the ability of hnRNP-K to silence mRNA translation, is dependent on phosphorylation mediated by MAPK/ERK (46). Several studies have reported up-regulation of hnRNP-K and its altered subcellular localization in tumors (47). A higher level of nuclear hnRNP-K has been reported in proliferating compared with resting cells. In contrast, the level of cytoplasmic hnRNP-K protein was either the same or lower in dividing compared with quiescent cells. States of enhanced proliferation were also associated with increased phosphorylation of hnRNP-K. On the basis of these findings, it has been suggested that the nuclear hnRNP-K is involved in cell proliferation regulatory signaling (48). It is modified in response to changes in the intracellular and extracellular environment, including cytokines, growth factors, and oxidative stress (49), and bridges signal transduction pathways to chromatin remodeling and nucleic acid-directed processes. hnRNP-K has also been shown to bind to the 3′ UTR of VEGF mRNA and regulate its translation (50) and, in turn, would affect the expression of MMPs. Mot-F and Mot-N cells showed an increase in the level of expression of MMP-2, MMP-3, and MMP-9. MMPs are up-regulated in many tumors. They cause epithelial-mesenchymal transition, malignant transformation, and genomic instability by mechanisms involving articulation of the tumor environment, oxidative stress, and induction of transcription factors.
To obtain further insights into the clinical relevance of nuclear mortalin in cancer, we examined normal and tumor (ovary, kidney, lung, liver, and brain) tissues by mortalin immunohistochemistry. Mortalin was found to be highly expressed and enriched in the nuclei of all tumor tissues examined compared with the controls (Fig. 7E and data not shown). A similar increase in mortalin has also been reported in liver cancers and melanoma biopsy samples (2, 10, 51). In agreement with the role of nuclear mortalin in tumors, human normal lung fibroblasts lacked endogenous nuclear mortalin staining (data not shown). Furthermore, although the exogenous mot-F and mot-N were localized in the nuclei, the cells neither showed an increase in proliferation or transformation nor in malignant properties (data not shown). The unaltered phenotype of normal cells could be due to the lack of mortalin-p53 interaction (9), telomerase, and hnRNP-K proteins (39–41).
Taken together, this study demonstrates, for the first time, a new line of action of the stress chaperone mortalin in human carcinogenesis. In addition to the cytoplasmic sequestration and inactivation of p53 described previously, it localizes in the nucleus, inactivates p53-mediated control of centrosome duplication, causing genomic instability, activates the telomere maintaining enzyme hTERT (a hallmark of most cancers) and a multifunctional chromatin-remodeling protein, hnRNP-K, involved in regulation cell migration, and, therefore, promotes carcinogenesis.
This work was supported by grants-in-aid for scientific research (Kakenhi) from the Japanese Society for the Promotion of Science, Japan (to S. C. K.); by Ministry of Knowledge Economy Grant 10030051 (to C.-O. Y.); and by Korea Science Engineering Foundation Grants 2010-0029220, 2013K1A1A2A02050188, and 2013M3A9D3045879 (to C.-O. Y.).
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- MMP
- matrix metalloprotease
- ROS
- reactive oxygen species
- mot
- mortalin
- mot-F
- full-length mortalin
- mot-N
- nuclear mortalin
- hTERT
- human telomerase reverse transcriptase.
REFERENCES
- 1. Wadhwa R., Kaul S. C., Ikawa Y., Sugimoto Y. (1993) Identification of a novel member of mouse hsp70 family: its association with cellular mortal phenotype. J. Biol. Chem. 268, 6615–6621 [PubMed] [Google Scholar]
- 2. Lu W. J., Lee N. P., Kaul S. C., Lan F., Poon R. T., Wadhwa R., Luk J. M. (2011) Induction of mutant p53-dependent apoptosis in human hepatocellular carcinoma by targeting stress protein mortalin. Int. J. Cancer 129, 1806–1814 [DOI] [PubMed] [Google Scholar]
- 3. Park S. J., Shin J. H., Jeong J. I., Song J. H., Jo Y. K., Kim E. S., Lee E. H., Hwang J. J., Lee E. K., Chung S. J., Koh J. Y., Jo D. G., Cho D. H. (2014) Down-regulation of mortalin exacerbates aβ-mediated mitochondrial fragmentation and dysfunction. J. Biol. Chem. 289, 2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaul S. C., Aida S., Yaguchi T., Kaur K., Wadhwa R. (2005) Activation of wild type p53 function by its mortalin-binding cytoplasmically localizing carboxy-terminus peptides. J. Biol. Chem. 280, 39373–39379 [DOI] [PubMed] [Google Scholar]
- 5. Pilzer D., Saar M., Koya K., Fishelson Z. (2010) Mortalin inhibitors sensitize K562 leukemia cells to complement-dependent cytotoxicity. Int. J. Cancer 126, 1428–1435 [DOI] [PubMed] [Google Scholar]
- 6. Ma Z., Izumi H., Kanai M., Kabuyama Y., Ahn N. G., Fukasawa K. (2006) Mortalin controls centrosome duplication via modulating centrosomal localization of p53. Oncogene 25, 5377–5390 [DOI] [PubMed] [Google Scholar]
- 7. Gestl E. E., Anne Böttger S. (2012) Cytoplasmic sequestration of the tumor suppressor p53 by a heat shock protein 70 family member, mortalin, in human colorectal adenocarcinoma cell lines. Biochem. Biophys. Res. Commun. 423, 411–416 [DOI] [PubMed] [Google Scholar]
- 8. Voloboueva L. A., Emery J. F., Sun X., Giffard R. G. (2013) Inflammatory response of microglial BV-2 cells includes a glycolytic shift and is modulated by mitochondrial glucose-regulated protein 75/mortalin. FEBS Lett. 587, 756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wadhwa R., Takano S., Robert M., Yoshida A., Nomura H., Reddel R. R., Mitsui Y., Kaul S. C. (1998) Inactivation of tumor suppressor p53 by mot-2, a hsp70 family member. J. Biol. Chem. 273, 29586–29591 [DOI] [PubMed] [Google Scholar]
- 10. Lu W. J., Lee N. P., Kaul S. C., Lan F., Poon R. T., Wadhwa R., Luk J. M. (2011) Mortalin-p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy. Cell Death Differ. 18, 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker C., Böttger S., Low B. (2006) Mortalin-based cytoplasmic sequestration of p53 in a nonmammalian cancer model. Am. J. Pathol. 168, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaul S. C., Duncan E. L., Englezou A., Takano S., Reddel R. R., Mitsui Y., Wadhwa R. (1998) Malignant transformation of NIH3T3 cells by overexpression of mot-2 protein. Oncogene 17, 907–911 [DOI] [PubMed] [Google Scholar]
- 13. Kaul S. C., Yaguchi T., Taira K., Reddel R. R., Wadhwa R. (2003) Overexpressed mortalin (mot-2)/mthsp70/GRP75 and hTERT cooperate to extend the in vitro lifespan of human fibroblasts. Exp. Cell Res. 286, 96–101 [DOI] [PubMed] [Google Scholar]
- 14. Xu J., Xiao H. H., Sartorelli A. C. (1999) Attenuation of the induced differentiation of HL-60 leukemia cells by mitochondrial chaperone HSP70. Oncol. Res. 11, 429–435 [PubMed] [Google Scholar]
- 15. Kanai M., Ma Z., Izumi H., Kim S. H., Mattison C. P., Winey M., Fukasawa K. (2007) Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells 12, 797–810 [DOI] [PubMed] [Google Scholar]
- 16. Wadhwa R., Colgin L., Yaguchi T., Taira K., Reddel R. R., Kaul S. C. (2002) Rhodacyanine dye MKT-077 inhibits in vitro telomerase assay but has no detectable effects on telomerase activity in vivo. Cancer Res. 62, 4434–4438 [PubMed] [Google Scholar]
- 17. Wadhwa R., Sugihara T., Yoshida A., Nomura H., Reddel R. R., Simpson R., Maruta H., Kaul S. C. (2000) Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 60, 6818–6821 [PubMed] [Google Scholar]
- 18. Wadhwa R., Takano S., Kaur K., Deocaris C. C., Pereira-Smith O. M., Reddel R. R., Kaul S. C. (2006) Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer 118, 2973–2980 [DOI] [PubMed] [Google Scholar]
- 19. Ando K., Oki E., Zhao Y., Ikawa-Yoshida A., Kitao H., Saeki H., Kimura Y., Ida S., Morita M., Kusumoto T., Maehara Y. (2014) Mortalin is a prognostic factor of gastric cancer with normal p53 function. Gastric Cancer 17, 255–262 [DOI] [PubMed] [Google Scholar]
- 20. Chen J., Liu W. B., Jia W. D., Xu G. L., Ma J. L., Huang M., Deng Y. R., Li J. S. (2014) Overexpression of Mortalin in hepatocellular carcinoma and its relationship with angiogenesis and epithelial to mesenchymal transition. Int. J. Oncol. 44, 247–255 [DOI] [PubMed] [Google Scholar]
- 21. Dundas S. R., Lawrie L. C., Rooney P. H., Murray G. I. (2005) Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J. Pathol. 205, 74–81 [DOI] [PubMed] [Google Scholar]
- 22. Rozenberg P., Kocsis J., Saar M., Prohászka Z., Füst G., Fishelson Z. (2013) Elevated levels of mitochondrial mortalin and cytosolic HSP70 in blood as risk factors in patients with colorectal cancer. Int. J. Cancer 133, 514–518 [DOI] [PubMed] [Google Scholar]
- 23. Mizukoshi E., Suzuki M., Misono T., Loupatov A., Munekata E., Kaul S. C., Wadhwa R., Imamura T. (2001) Cell-cycle dependent tyrosine phosphorylation on mortalin regulates its interaction with fibroblast growth factor-1. Biochem. Biophys. Res. Commun. 280, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 24. Qu M., Zhou Z., Xu S., Chen C., Yu Z., Wang D. (2011) Mortalin overexpression attenuates beta-amyloid-induced neurotoxicity in SH-SY5Y cells. Brain Res. 1368, 336–345 [DOI] [PubMed] [Google Scholar]
- 25. Burbulla L. F., Schelling C., Kato H., Rapaport D., Woitalla D., Schiesling C., Schulte C., Sharma M., Illig T., Bauer P., Jung S., Nordheim A., Schöls L., Riess O., Krüger R. (2010) Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum. Mol. Genet. 19, 4437–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiasserini D., Tozzi A., de Iure A., Tantucci M., Susta F., Orvietani P. L., Koya K., Binaglia L., Calabresi P. (2011) Mortalin inhibition in experimental Parkinson's disease. Mov. Disord. 26, 1639–1647 [DOI] [PubMed] [Google Scholar]
- 27. Flachbartová Z., Kovacech B. (2013) Mortalin: a multipotent chaperone regulating cellular processes ranging from viral infection to neurodegeneration. Acta Virol. 57, 3–15 [DOI] [PubMed] [Google Scholar]
- 28. Li H. Y., Yang L., Liu W., Zuo J. (2011) GRP75 overexpression inhibits apoptosis induced by glucose deprivation via Raf/Mek/Erk1/2 signaling pathway. Sheng Li Xue Bao 63, 69–74 [PubMed] [Google Scholar]
- 29. Yang L., Guo W., Zhang Q., Li H., Liu X., Yang Y., Zuo J., Liu W. (2011) Crosstalk between Raf/MEK/ERK and PI3K/AKT in suppression of Bax conformational change by Grp75 under glucose deprivation conditions. J. Mol. Biol. 414, 654–666 [DOI] [PubMed] [Google Scholar]
- 30. Wadhwa R., Takano S., Taira K., Kaul S. C. (2004) Reduction in mortalin level by its antisense expression causes senescence-like growth arrest in human immortalized cells. J. Gene Med. 6, 439–444 [DOI] [PubMed] [Google Scholar]
- 31. Yoo J. Y., Ryu J., Gao R., Yaguchi T., Kaul S. C., Wadhwa R., Yun C. O. (2010) Tumor suppression by apoptotic and anti-angiogenic effects of mortalin-targeting adeno-oncolytic virus. J. Gene Med. 12, 586–595 [DOI] [PubMed] [Google Scholar]
- 32. Yi X., Luk J. M., Lee N. P., Peng J., Leng X., Guan X. Y., Lau G. K., Beretta L., Fan S. T. (2008) Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell Proteomics 7, 315–325 [DOI] [PubMed] [Google Scholar]
- 33. Ran Q., Wadhwa R., Kawai R., Kaul S. C., Sifers R. N., Bick R. J., Smith J. R., Pereira-Smith O. M. (2000) Extramitochondrial localization of mortalin/mthsp70/PBP74/GRP75. Biochem. Biophys. Res. Commun. 275, 174–179 [DOI] [PubMed] [Google Scholar]
- 34. Kaul S. C., Deocaris C. C., Wadhwa R. (2007) Three faces of mortalin: a housekeeper, guardian and killer. Exp. Gerontol. 42, 263–274 [DOI] [PubMed] [Google Scholar]
- 35. Shih Y. Y., Lee H., Nakagawara A., Juan H. F., Jeng Y. M., Tsay Y. G., Lin D. T., Hsieh F. J., Pan C. Y., Hsu W. M., Liao Y. F. (2011) Nuclear GRP75 binds retinoic acid receptors to promote neuronal differentiation of neuroblastoma. PLoS ONE 6, e26236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Inoue A., Sawata S. Y., Taira K., Wadhwa R. (2007) Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl. Acad. Sci. U.S.A. 104, 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L. P., Lu C. H., Chen Z. P., Ge F., Wang T., Wang W., Xiao C. L., Yin X. F., Liu L., He J. X., He Q. Y. (2011) Subcellular proteomics revealed the epithelial-mesenchymal transition phenotype in lung cancer. Proteomics 11, 429–439 [DOI] [PubMed] [Google Scholar]
- 38. He Z. Y., Wen H., Shi C. B., Wang J. (2010) Up-regulation of hnRNP A1, Ezrin, tubulin β-2C and Annexin A1 in sentinel lymph nodes of colorectal cancer. World J. Gastroenterol. 16, 4670–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao R., Yu Y., Inoue A., Widodo N., Kaul S. C., Wadhwa R. (2013) Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 288, 15046–15056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moumen A., Masterson P., O'Connor M. J., Jackson S. P. (2005) hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 41. Shay J. W., Keith W.N. (2008) Targeting telomerase for cancer therapeutics. Br. J. Cancer 98, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujiki T., Miura T., Maura M., Shiraishi H., Nishimura S., Imada Y., Uehara N., Tashiro K., Shirahata S., Katakura Y. (2007) TAK1 represses transcription of the human telomerase reverse transcriptase gene. Oncogene 26, 5258–5266 [DOI] [PubMed] [Google Scholar]
- 43. Papanikolaou V., Iliopoulos D., Dimou I., Dubos S., Tsougos I., Theodorou K., Kitsiou-Tzeli S., Tsezou A. (2009) The involvement of HER2 and p53 status in the regulation of telomerase in irradiated breast cancer cells. Int. J. Oncol. 35, 1141–1149 [DOI] [PubMed] [Google Scholar]
- 44. Kang X., Chen W., Kim R. H., Kang M. K., Park N. H. (2009) Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene 28, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ostareck-Lederer A., Ostareck D. H. (2004) Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol. Cell 96, 407–411 [DOI] [PubMed] [Google Scholar]
- 46. Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., Hentze M. W., Ronai Z. (2001) ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3, 325–330 [DOI] [PubMed] [Google Scholar]
- 47. Hope N. R., Murray G. I. (2011) The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum. Pathol. 42, 393–402 [DOI] [PubMed] [Google Scholar]
- 48. Ostrowski J., Kawata Y., Schullery D. S., Denisenko O. N., Bomsztyk K. (2003) Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res. 31, 3954–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ostrowski J., Bomsztyk K. (2003) Nuclear shift of hnRNP K protein in neoplasms and other states of enhanced cell proliferation. Br. J. Cancer 89, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sataranatarajan K., Lee M. J., Mariappan M. M., Feliers D. (2008) PKCδ regulates the stimulation of vascular endothelial factor mRNA translation by angiotensin II through hnRNP K. Cell Signal. 20, 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu P. K., Hong S. K., Veeranki S., Karkhanis M., Starenki D., Plaza J. A., Park J. I. (2013) A mortalin/HSPA9-mediated switch in tumor suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol. Cell. Biol. 33, 4051–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]