Background: rRNA methylation is required for ribosome assembly and/or function.
Results: Knockdown of MRM1, MRM2, and RNMTL1 increases susceptibility of mitochondrial 16 S rRNA to site-specific cleavage by DNAzymes.
Conclusion: The enzymes responsible for all known 2′-O-ribose methylations of 16 S rRNA are now identified.
Significance: Assignment of rRNA modifications to nucleoid-associated proteins implies that mitochondrial ribosome biogenesis begins at the nucleoid.
Keywords: Mitochondria, Ribosomal RNA Processing (rRNA Processing), Ribosome, RNA Methyltransferase, RNA Modification
Abstract
Advances in proteomics and large scale studies of potential mitochondrial proteins have led to the identification of many novel mitochondrial proteins in need of further characterization. Among these novel proteins are three mammalian rRNA methyltransferase family members RNMTL1, MRM1, and MRM2. MRM1 and MRM2 have bacterial and yeast homologs, whereas RNMTL1 appears to have evolved later in higher eukaryotes. We recently confirmed the localization of the three proteins to mitochondria, specifically in the vicinity of mtDNA nucleoids. In this study, we took advantage of the ability of 2′-O-ribose modification to block site-specific cleavage of RNA by DNAzymes to show that MRM1, MRM2, and RNMTL1 are responsible for modification of human large subunit rRNA at residues G1145, U1369, and G1370, respectively.
Introduction
Mitochondria are known for their roles in generating energy and programmed cell death. Although mammalian mitochondrial proteins are mostly encoded by the nuclear genome and imported into the organelle, mitochondria maintain a compact genome encoding 13 mRNAs, 2 rRNAs, and 22 tRNAs completely dedicated to the synthesis of a minor fraction of the subunits of the electron transport chain. Mitochondrial dysfunction is implicated in aging, as well as a broad range of diseases including diabetes, cancer, Parkinson disease, Alzheimer disease, Leigh syndrome, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), myoclonic epilepsy with ragged red fibers (MERRF), Leber's hereditary optic neuropathy (LHON), and progressive external ophthalmoplegia (PEO) (1, 2). Many of the disease mutations are found in tRNA or mRNA sequences; however, some have been found in the 12 S and 16 S rRNAs (3–6) and nuclear-encoded mitochondrial ribosomal proteins (MRPs)2 (7–9). Mutations in AFG3L2 or paraplegin, subunits of an AAA protease that processes mitochondrial ribosomal protein MRPL32 and functions in the assembly of the mitochondrial ribosome (mitoribosome), are associated with hereditary spastic paraplegia, spinocerebellar ataxia type 28, and spastic ataxia-neuropathy syndrome, highlighting the importance of proper mitochondrial protein synthesis in mammals (10, 11).
The mammalian mitoribosome (55 S) consists of a small subunit (SSU; 28 S) composed of 12 S rRNA and 31 MRPs, and a large subunit (LSU; 39 S) composed of 16 S rRNA and 51 MRPs. The mitoribosome has many similarities to its ancestral prokaryotic ribosome, although only 14 of the SSU MRPs and 28 of the LSU MRPs are conserved in bacteria. The reduced lengths of mitochondrial rRNAs are accomplished by multiple small deletions that leave a core structure similar to that of the bacterial rRNA (12, 13), conserving the regions participating in the peptidyl transferase center. Bacterial and mitochondrial ribosomes also share susceptibility to certain antibiotics including chloramphenicol, tetracyclines, and linezolid.
Proteomics has identified a nearly complete set of mitochondrial ribosomal proteins (14–21), some of which appear to be multifunctional (DAP3, AURKAIP, CRIF1, ICT1) (18, 22, 23). Our understanding of the mitoribosome structure is improving with advancements in cryo-EM technology (12, 24–28), but the assembly process has received little attention. We have recently used microscopy and stable isotope labeling by/with amino acids in cell culture (SILAC) proteomic analysis to show that a subset of newly synthesized mitochondrial ribosomal proteins participates in the early stages of ribosome assembly at the nucleoid (29).
An important step in mitoribosome assembly is the modification of rRNA at conserved regions, often in catalytic domains. The small subunit 12 S rRNA is dimethylated by TFB1M at two adjacent adenosines, 936 and 937 (30). Recently, NSUN4 has been described as a cytosine 5-methyltransferase, acting with MTERF4 at nucleotide C840 in mouse (C841 in human) (31). The importance of these methylations is highlighted by the embryonic-lethal phenotype of mice deficient in these genes (31, 32). 2′-O-Ribose methylation and pseudouridylation are the most common modifications found in eukaryotic and archaeal rRNA (33, 34) and are the only modifications found on mammalian mitochondrial 16 S rRNA (35, 36), as reviewed by Ref. 37. The large subunit 16 S rRNA modifications include three 2′-O-ribose methylations at G1145, U1369, and G1370 and a pseudouridylation at U1397. We have identified RNMTL1 as a novel mitochondrial rRNA methyltransferase localized at nucleoids involved in the 2′-O-methylation of G1370, a unique site that is not methylated in bacterial or yeast mitochondrial rRNA (38). Studies in yeast mitochondria suggest that human homologs of MRM1 and MRM2, both of which we found to co-localize with nucleoids, are involved in 16 S rRNA methylation. These modifications occur at sites that are conserved in bacteria and yeast mitochondria, and contribute to the catalytic domain of the mitoribosome, the peptidyl transferase center, suggesting that they are important for function, yet the exact role of each modification is still unclear (39–42).
There are several established methods of detecting 2′-O-ribose methylation on RNA, including resistance to RNase H when hybridized to a chimeric oligonucleotide (43), splint ligation (44), reverse transcription coupled to PCR (45), mass spectrometry, two-dimensional TLC, boronate affinity chromatography, and other chemical tests (46). One of the most common methods involves inhibition of reverse transcriptase at low deoxynucleotide triphosphate levels (47), as we have recently applied (38). However, a method that has not received much attention is the use of DNAzymes, deoxyoligonucleotides that can anneal to RNA and direct cleavage at a specific unmodified site. DNAzyme sequences were selected in a self-amplifying screen (48) and applied to detecting 2′-O-methylation of yeast nucleo-cytosolic rRNA (34). Here, we combine the use of DNAzymes and Northern blotting for increased sensitivity and specificity to show that MRM1, MRM2, and RNMTL1 are responsible for the 2′-O-ribose methylation of G1145, U1369, and G1370 on 16 S rRNA, respectively.
MATERIALS AND METHODS
Cell Culture and Reagents
HeLa and HEK293 cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Silencer® Select siRNA (Ambion) sequences are as follows: siMRM1-A (5′-CCAGGCCCAUGUUUAUUGA-3′), siMRM1-B (5′-GGAAGUUGAGGGAAAGUUU-3′), siMRM2-A (5′-ACAUCUCAGGGACCCAUUU-3′), and siMRM2-B (5′-GAAUGUAAGGAUCAUCAAA-3′). HeLa cells were reverse-transfected with 3 or 6 nm siRNA and Lipofectamine® RNAiMAX (Life Technologies) for 3 days, as described (38). Antibodies are listed with their suppliers and dilutions: MRM1 (Sigma-Aldrich, 1:3,000), MRM2 (Abcam; 1:1,000), and succinate dehydrogenase subunit A (SDHA, MitoSciences, 1:10,000).
RNA Isolation
HeLa cells treated with siRNA were trypsinized and collected by centrifugation at 500 × g for 5 min at 4 °C. Cells were resuspended in PBS and repelleted. 1 ml of TRIzol® was used to lyse the cells, and RNA was isolated according to the manufacturer's instructions. The RNA was subject to an additional phenol-chloroform extraction and diethyl ether extraction to obtain highly purified RNA. The concentration and purity of RNA was detected by a NanoDrop 1000 spectrophotometer.
DNAzyme-mediated RNA Cleavage
The 8-17-type DNAzyme sequences used for each site are: G1144 (5′-TCCGAGGTCGCCTATTAGCAATACGAAAACCGAAATTTTT-3′), G1145 (5′-CTCCGAGGTCGCTATTAGCAATACGAACAACCGAAATTTT-3′), and U1369 (5′-TTAATCGTTGAATATTAGCAATACGAAAACGAACCTTTAA-3′), and the 10-23-type DNAzyme sequence for G1370 was (5′-TTTAATCGTTGAAGGCTAGCTACAACGAAAACGAACCTTTA-3′). The underlined sequences correspond to the catalytic region of the DNAzyme that is flanked by targeting sequences to direct the site of cleavage. The reactions were carried out as in Refs. 34 and 48, with minor modifications. 8-17-type reactions were performed by mixing 0.75–4 μg of RNA with 400 pmol of DNAzyme and supplementing with DEPC-treated water to 32 μl. The nucleic acids were boiled in a water bath for 2 min and then cooled to room temperature for 10 min followed by the addition of an equal volume of 2× buffer (200 mm KCl, 800 mm NaCl, 100 mm Hepes, pH 7.5, 15 mm MgCl2, 15 mm MnCl2). The reaction was incubated at 37 °C for 1 h and then stopped by the addition of 3 μl of DEPC-treated 0.25 m EDTA, 1 μl of 10 mg/ml glycogen as carrier, 100 μl of DEPC-treated 0.3 m sodium acetate in TE (10 mm Tris, pH 7.5, 1 mm EDTA) buffer, and 2.5 volumes of 100% ethanol. The RNA was precipitated at −20 °C overnight and spun in a desktop centrifuge at 17,000 × g for 10 min at 4 °C. The supernatant was discarded, and the pellet was washed in 70% ethanol, spun again, and dried in a vacuum centrifuge. The RNA was resuspended in a small volume of TE, typically 6 μl. The 10-23-type reactions were performed by mixing RNA with 400 pmol of DNAzyme supplemented with DEPC-treated water to 12 μl and an equal volume of 2× buffer (20 mm NaCl, 8 mm Tris, pH 8) and boiling for 3 min, chilled on ice for 5 min, and then incubated at room temperature for 10 min. 6 μl of 5× buffer (750 mm NaCl, 200 mm Tris, pH 8) and 2 μl of 300 mm MgCl2 were added. The mixture was incubated at 37 °C for 1 h and stopped as with the 8-17-type reactions. Control reactions used water in place of DNAzyme.
RNA Analysis and Northern Blotting
Denaturing agarose gels were performed as (49) with minor modifications. 9 μl of RNA loading solution (80% deionized formamide, 5% formaldehyde, 20 mm EDTA, 40 μg/ml ethidium bromide, 0.05% bromphenol blue, 0.05% xylene cyanol) were added to 6 μl of the resuspended RNA and heated in a water bath at 65 °C for 5 min and then loaded onto a 2% agarose/MOPS gel with no formaldehyde. The gel was run in 1× MOPS, pH 7.0, for 2.5 h at 83 V with a buffer exchange system. The ethidium bromide-stained gel was photographed and the RNA was then transferred to a Hybond-N+ membrane (GE Healthcare) in 10× SSC (1.5 m NaCl, 150 mm sodium citrate) overnight by capillary action. The membrane was auto-cross-linked by a UV Stratalinker (Stratagene) and baked in a vacuum at 80 °C for 2 h. ULTRAhyb prehybridization buffer (Ambion) was applied according to the manufacturer's instructions, and a biotinylated RNA probe was hybridized overnight at 68 °C. The membrane was then washed two times in 2× SSC, 0.5% SDS and then two times in 0.2× SSC, 0.5% SDS for 10 min each at 65 °C. RNA was detected by streptavidin linked to alkaline phosphatase (BrightStar® BioDetect kit; Ambion) according to the manufacturer's instructions. The biotinylated probe was generated by in vitro T7 RNA polymerase transcription of a sequence complementary to residues 142–458 of 16 S rRNA that was cloned into pBS− plasmid in the presence of biotinylated CTP. In vitro-synthesized full-length 16 S RNA was transcribed from a pBSKII+ plasmid with a 16 S RNA insert as described (38).
RESULTS
DNAzymes Can Target the Cleavage of Unmodified RNA at Specific Sites
Baer and Dubin (35) and Dubin et al. (50) used radioactive labeling and RNA fingerprinting to identify the methylation sites on mammalian (hamster) mitochondrial large and small subunit rRNA. The large subunit rRNA methylation sites all occur on the ribose 2′-OH, whereas the small subunit rRNA methylation sites all occur at the nucleotide base. The location of the 2′-O-methylations corresponds to homologous sites in bacteria that participate in the peptidyl transferase center of the ribosome. These residues are identified within 16 S rRNA structures similar to the well described A-loop and P-loop secondary structures of Escherichia coli 23 S rRNA (Fig. 1A). An NMR study suggested that methylation of U2552 alters the ribose pucker in such a way as to promote pairing with a cytidine residue and increased exposure of the adjacent G2553 (51). These residues are conserved in the mitochondrial A-loop with the additional feature that the corresponding G1370 in 16 S rRNA is also methylated.
FIGURE 1.
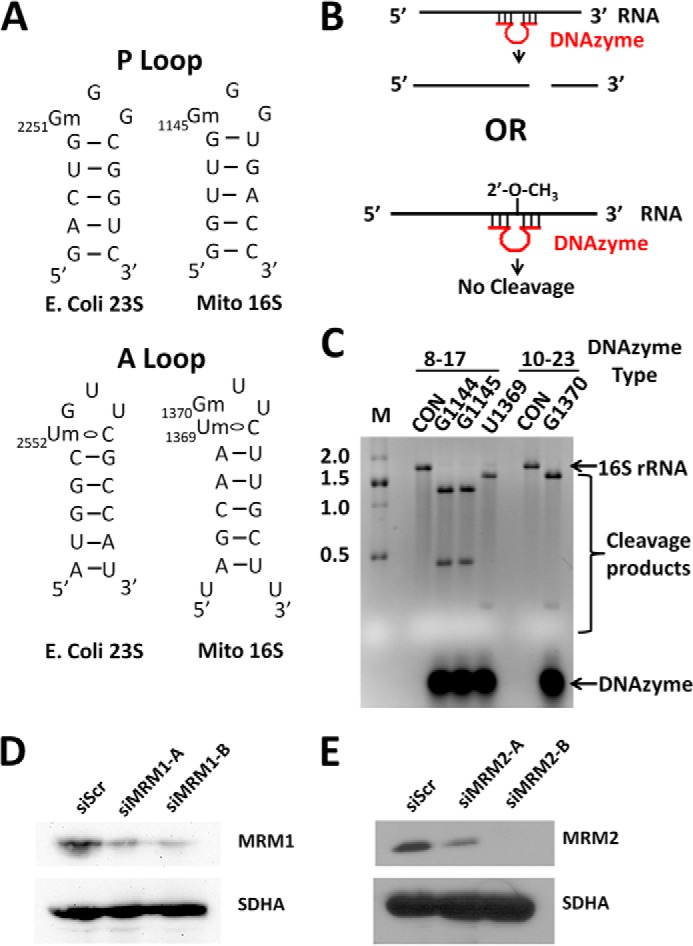
DNAzyme cleavage of 16 S rRNA. A, the secondary structures of the mammalian mitochondrial large ribosomal subunit 16 S rRNA A- and P-loop regions are shown with those of E. coli rRNA for comparison. B, DNAzymes target RNA cleavage at specific sites employing the 2′-OH group to attack the phosphodiester linkage. C, denaturing gel analysis of in vitro-transcribed 16 S rRNA shows that DNAzymes completely cleave unmodified RNA. 0.75 μg of RNA were loaded per lane and detected by ethidium bromide fluorescence. CON, control reaction without DNAzyme; M, RNA ladder with sizes in kB. D and E, HeLa cells were treated with siRNA targeting a scrambled sequence (siScr) or one of two sequences targeting either MRM1 (siMRM1-A and siMRM1-B) or MRM2 (siMRM2-A and siMRM2-B). 5.3 μg (D) or 4 μg (E) of mitochondrial protein were loaded per lane onto 12% SDS-PAGE and blotted for MRM1, MRM2, or succinate dehydrogenase subunit A (SDHA).
To probe for 2′-O-methylation of these sites, we first established the efficiency of DNAzymes on unmodified RNA. DNAzymes cause the self-cleavage of unmodified RNA at a specific site by using two arms of complementary nucleotides to anneal to the RNA target promoting nucleophilic attack of the RNA 2′-OH on its own 3′-phosphate (34). However, if the ribose 2′-OH is methylated, then the RNA is resistant to cleavage at that site (Fig. 1B). We designed single-stranded DNA oligonucleotides with the catalytic 15- or 16-nucleotide sequences for the 8-17 or 10-23-type DNAzymes, respectively, flanked by sequences complementary to 16 S rRNA, according to existing guidelines (34, 48). DNAzymes were designed to target the cleavage of a control unmodified site (G1144) and the three suspected methylation sites (G1145, U1369, and G1370). In separate reactions, 750 ng of in vitro-transcribed 16 S RNA, which lacks any modifications, were annealed to 400 pmol of each DNAzyme, such that the DNAzyme is in stoichiometric excess to the RNA, and incubated as described under “Materials and Methods.” The RNA was precipitated from the reactions, denatured in a loading solution, and separated on a 2% agarose/MOPS gel. As expected, with all of the DNAzymes used, the unmodified RNA undergoes site-specific cleavage to near completion, generating RNA fragments of the expected size (Fig. 1C). The buffer and incubation conditions alone do not cause cleavage.
Because DNAzymes can distinguish unmodified and 2′-O-methylated RNA, we then set out to compare the susceptibility of mitochondrial RNA prepared from cells with normal and reduced methyltransferase protein levels to DNAzyme-mediated cleavage. To increase the sensitivity and specificity of mitochondrial RNA detection relative to ethidium bromide staining, we prepared a biotinylated RNA probe complementary to 16 S rRNA, 5′ of the cleavage sites to permit detection of RNA cleavage using nonradioactive Northern blotting. Streptavidin-linked alkaline phosphatase allows chemiluminescent detection of the biotinylated probe.
Assignment of Mitochondrial Methyltransferases to Target Sites
We have previously used siRNA to significantly reduce the levels of RNMTL1 protein in HeLa cells (38). In this analysis, primer extension in the presence of limiting deoxynucleotide triphosphate concentrations was used to conclude that RNMTL1 methylates residue G1370 of 16 S rRNA, but this analysis was complicated somewhat by methylation of the adjacent U1369 residue. In our present work, we treated HeLa and HEK293 cells with siRNA targeting a negative control scrambled sequence (siScr), RNMTL1, MRM1, or MRM2 for 3 days. Two different siRNA sequences were each tested for reduction of MRM1 and MRM2 protein levels. Although both were effective, later experiments were conducted with the siRNAs causing a greater effect, siMRM1-B and siMRM2-B (Fig. 1, D and E). Efficacy of the RNMTL1 siRNA was established previously (38).
Whole cell RNA was isolated from HeLa and HEK293 cells that were transfected with siRNA. 2 μg of each RNA sample were treated with each DNAzyme, and the products were separated by electrophoresis on a 2% agarose/MOPS gel and transferred to Hybond-N+ membrane. Fig. 2 shows results obtained with RNA preparations from HeLa (A) and HEK293 cells (B and C). Ethidium bromide staining in the lower panels reveals loading controls for the cytosolic rRNAs, which are estimated to be ∼100 times more abundant than the mitochondrial rRNAs (52). The membrane was probed with biotinylated RNA complementary to 16 S and developed with chemiluminescent reagents detected with film. Overall levels of 16 S rRNA relative to total cell RNA did not appear to be significantly affected by the siRNA treatments. As a positive control, RNAs were treated with a DNAzyme designed to cleave at G1144, which is known not to be methylated. This control confirmed essentially complete cleavage by the DNAzyme at this site on 16 S rRNA. Knockdown of RNMTL1 significantly increased accessibility of 16 S rRNA to DNAzyme cleavage at G1370, confirming our previous results (38) that RNMTL1 is involved in the 2′-O-methylation of G1370 (Fig. 2, A and B, arrows indicate the 5′ cleavage product). Likewise, knockdown of MRM1 in both HeLa and HEK293 cells significantly increased the cleavage product at G1145, providing evidence that MRM1 is involved in the 2′-O-ribose methylation of that site. We consistently found weak cleavage of 17.3 ± 3% at this site in RNA samples derived from control cells or cells treated with siRNAs targeted to different enzymes. This appears to reflect some steady-state level of unmodified RNA. The amount of cleavage product was increased more than 2-fold following siRNA depletion of MRM1.
FIGURE 2.
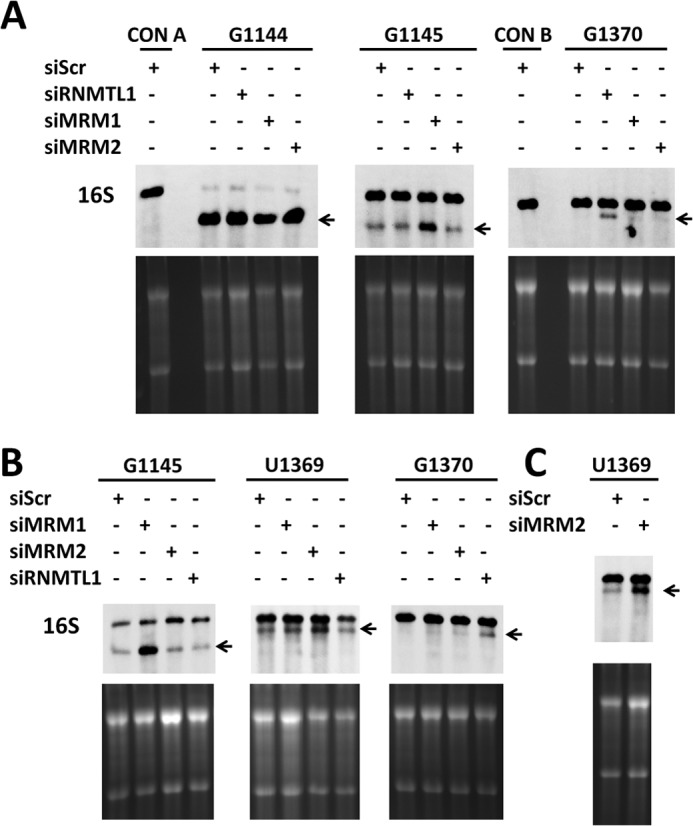
Assignment of 2′-O-methyltransferases to their respective modification sites. A–C, HeLa (A) or HEK293 cells (B and C) were subjected to the indicated siRNA treatments, and isolated RNA was treated with DNAzymes. 2 μg of RNA were loaded per lane and visualized by ethidium bromide staining to provide a loading control (lower panels), then transferred to Hybond-N+ and probed for the 5′ end of 16 S rRNA (upper panels). siScr, siRNA targeting a scrambled sequence. siRNMTL1, siRNA targeting RNMTL1; siMRM1 and siMRM2, siRNA targeting MRM1 and MRM2., A, DNAzymes targeting the cleavage of G1144, G1145, and G1370 show that MRM1 and RNMTL1 are involved in RNA methylation in HeLa cells. B, HEK293 cells were treated with siRNA targeting MRM1, MRM2, or RNMTL1 for 3 days prior to RNA isolation and analysis. C, HEK293 cells were treated with 6 nm siRNA targeting MRM2 prior to RNA isolation and analysis. Arrows indicate the 16 S rRNA 5′ cleavage products. CON A and CON B refer to buffer controls using 8-17 and 10-23 type DNAzyme reaction conditions, without DNAzyme.
We also observed appreciable DNAzyme cleavage of U1369 in 16 S rRNA from either HeLa or HEK293 cells, averaging 30 ± 7% of total RNA, when cells were either not treated with siRNA or treated with the scrambled control. This most likely represents rRNA that is not modified in vivo because 2′-O-methylation chemically blocks RNA strand scission catalyzed by the DNAzyme. This high background of unmodified RNA complicated efforts to document an increased accessibility following siRNA-directed depletion of MRM2, as shown by the modest effect on DNAzyme susceptibility after 3 days of standard siRNA treatment of HEK293 cells in Fig. 2B (HeLa results not shown). We observed a more dramatic effect when the dose of siRNA was increased to 6 nm (Fig. 2C). Under these conditions the cleavage at residue U1369 was increased more than 2-fold, clearly implicating MRM2 in modification of U1369. Treatment with either 3 nm or 6 nm siRNA directed against MRM2 resulted in a general suppression of cell growth. The total cell number after 3 days of siRNA targeting MRM2 was decreased to 72% of the control using siRNA targeting a scrambled sequence.
DISCUSSION
Mammalian mitochondria possess three well defined 2′-O-methyltransferase family members. We used the DNAzyme and Northern blotting approach to confirm our previous indication that RNMTL1 is required for the methylation at G1370 of 16 S rRNA (38). We also provide novel evidence that MRM1 and MRM2 are required for the methylation at G1145 and U1369, respectively, which is consistent with the conservation of these methyltransferases and target residues in bacteria and yeast mitochondria. While this manuscript was under review an article appeared on-line supporting our previous observation that RNMTL1 is responsible for methylation of G1370 and suggesting that MRM2 was involved in modification of U1369 (53). This study, like that of Lee et al. (38), used a primer extension approach to detect 2′-O-methylation, which is difficult to interpret quantitatively due to variable polymerase bypass of the modified residue. The primer extension method is also complicated by local RNA sequence features. In our previous work (38), we found that the GGGG sequence encompassing G1145 has a tendency to block primer extension by reverse transcriptase. Second, detection of methylation sites using the primer extension method can be obscured by modification of a closely spaced residue, as in the case of two adjacent methylations occurring at U1369 and G1370. The DNAzyme approach circumvents these limitations of the primer extension assay.
The DNAzyme-mediated RNA cleavage approach described here is not limited to cases where modified RNA sites and RNA-modifying suspects have already been identified. One may apply DNAzymes to scan transcripts for 2′-OH modifications by shifting the annealing arms to redirect the interrogation site. Because the annealing arms will likely limit the analysis of modifications near the 5′ or 3′ end of transcripts, one may consider ligating single-stranded RNA to the ends or circularizing the RNA. DNAzymes have been developed to cleave at nearly all dinucleotide RNA junctions; thus they are not limited by the target RNA sequence. DNAzymes can also detect pseudouridylation sites, although less effectively than 2′-O-methylation sites (34).
The occurrence of adjacent 2′-O-methylation sites in mammalian 16 S rRNA seems unusual, given that the Saccharomyces cerevisiae LSU mitochondrial rRNA lacks one of these modifications. However, the corresponding G residue in the A-loop of yeast cytoplasmic 23 S rRNA is modified by 2′-O-methylation (54). A comprehensive evolutionary analysis of the conservation of the three mitochondrial RNA methyltransferases has recently appeared (53). Because 16 S rRNA from cells with normal levels of mitochondrial methyltransferases has some susceptibility to DNAzyme cleavage at G1145 and U1369, it is likely that at steady state, a significant fraction of 16 S rRNA molecules is not completely methylated and/or there is a structured order of methylation events, as with nucleo-cytoplasmic ribosomes (55, 56). It is possible that modification of either U1369 or G1370 may be sufficient to support mitoribosome assembly and translation, so that there may be some redundancy in the retention of both enzymes. We have not ruled out the possibility that some of the methylations may be dynamic (reversible) as with m6A in cytoplasmic mRNA (57–59).
Our in vitro methylation assays with purified protein and in vitro-synthesized RNA have not been successful (38). Although technical and biological explanations for this exist, we have not ruled out the remote possibility that RNMTL1, MRM1, and MRM2 act by an indirect mechanism. However, it is likely that the mitochondrial methyltransferases resemble bacterial methyltransferases by acting as stand-alone proteins that recognize and modify specific sites on RNA, in contrast to eukaryotic nucleo-cytoplasmic ribosomes, which require guide RNAs within small nucleolar ribonucleoprotein complexes to specify the modification sites.
Although the mechanism of mitoribosome assembly is not well understood, we are beginning to accumulate evidence that mitoribosome biogenesis begins at the nucleoid on nascent transcripts by incorporating newly synthesized MRPs. In addition to finding methyltransferases localized near nucleoids, we have recently shown that newly synthesized MRPs are localized in the nucleoid complex to encounter newly synthesized rRNA using pulse-chase stable isotope labeling by/with amino acids in cell culture labeling (29). It is also known that GTPases are important for mitoribosome assembly, just as they are for bacteria (60). Human NOA1 (hNOA1), also known as MTG3 or C4orf14, is associated with the nucleoid and is necessary for SSU assembly (61–63). We have also identified ERAL1 in nucleoids (29, 64). ERAL1 is a GTPase that functions as an RNA chaperone, binding to the TFB1M-mediated dimethylation region of 12 S rRNA (65, 66), potentially folding the RNA into the proper conformation needed for SSU assembly. The relationship between the nucleoid and ribosome assembly is becoming clearer, and more experiments in the future will solidify this relationship. It is also noteworthy that ∼30 bacterial rRNA modifications and modifying proteins have not been retained in mitochondria (67). The lack of modifications may be explained by an expansion of mitochondrial ribosomal proteins and truncation of rRNA structures. The retained modifications and modifying enzymes that have not been eliminated through evolution most likely play important roles in mitoribosome assembly and function.
This research was supported by a Senior Scholar Award from the Ellison Medical Foundation (to D. F. B.).
- MRP
- mitochondrial ribosomal protein
- SSU
- small subunit
- LSU
- large subunit
- DEPC
- diethylpyrocarbonate.
REFERENCES
- 1. DiMauro S., Schon E. A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- 2. Vafai S. B., Mootha V. K. (2012) Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 [DOI] [PubMed] [Google Scholar]
- 3. Coulbault L., Deslandes B., Herlicoviez D., Read M. H., Leporrier N., Schaeffer S., Mouadil A., Lombès A., Chapon F., Jauzac P., Allouche S. (2007) A novel mutation 3090 G→A of the mitochondrial 16S ribosomal RNA associated with myopathy. Biochem. Biophys. Res. Commun. 362, 601–605 [DOI] [PubMed] [Google Scholar]
- 4. Haque M. E., Elmore K. B., Tripathy A., Koc H., Koc E. C., Spremulli L. L. (2010) Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J. Biol. Chem. 285, 28353–28362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z., Song Y., Li D., He X., Li S., Wu B., Wang W., Gu S., Zhu X., Wang X., Zhou Q., Dai Y., Yan Q. (2014) The novel mitochondrial 16S rRNA 2336T→C mutation is associated with hypertrophic cardiomyopathy. J. Med. Genet. 51, 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prezant T. R., Agapian J. V., Bohlman M. C., Bu X., Oztas S., Qiu W. Q., Arnos K. S., Cortopassi G. A., Jaber L., Rotter J. I., et al. (1993) Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4, 289–294 [DOI] [PubMed] [Google Scholar]
- 7. Galmiche L., Serre V., Beinat M., Assouline Z., Lebre A. S., Chretien D., Nietschke P., Benes V., Boddaert N., Sidi D., Brunelle F., Rio M., Munnich A., Rötig A. (2011) Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 32, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 8. Kenmochi N., Suzuki T., Uechi T., Magoori M., Kuniba M., Higa S., Watanabe K., Tanaka T. (2001) The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics 77, 65–70 [DOI] [PubMed] [Google Scholar]
- 9. O'Brien T. W., O'Brien B. J., Norman R. A. (2005) Nuclear MRP genes and mitochondrial disease. Gene 354, 147–151 [DOI] [PubMed] [Google Scholar]
- 10. Almajan E. R., Richter R., Paeger L., Martinelli P., Barth E., Decker T., Larsson N. G., Kloppenburg P., Langer T., Rugarli E. I. (2012) AFG3L2 supports mitochondrial protein synthesis and Purkinje cell survival. J. Clin. Invest. 122, 4048–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 12. Greber B. J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J. P., Leibundgut M., Aebersold R., Ban N. (2014) Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 [DOI] [PubMed] [Google Scholar]
- 13. Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., Watanabe K. (2001) Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome: systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 276, 21724–21736 [DOI] [PubMed] [Google Scholar]
- 14. Goldschmidt-Reisin S., Kitakawa M., Herfurth E., Wittmann-Liebold B., Grohmann L., Graack H. R. (1998) Mammalian mitochondrial ribosomal proteins. N-terminal amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 273, 34828–34836 [DOI] [PubMed] [Google Scholar]
- 15. Graack H. R., Bryant M. L., O'Brien T. W. (1999) Identification of mammalian mitochondrial ribosomal proteins (MRPs) by N-terminal sequencing of purified bovine MRPs and comparison to data bank sequences: the large subribosomal particle. Biochemistry 38, 16569–16577 [DOI] [PubMed] [Google Scholar]
- 16. Koc E. C., Burkhart W., Blackburn K., Koc H., Moseley A., Spremulli L. L. (2001) Identification of four proteins from the small subunit of the mammalian mitochondrial ribosome using a proteomics approach. Protein Sci 10, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koc E. C., Burkhart W., Blackburn K., Moyer M. B., Schlatzer D. M., Moseley A., Spremulli L. L. (2001) The large subunit of the mammalian mitochondrial ribosome: analysis of the complement of ribosomal proteins present. J. Biol. Chem. 276, 43958–43969 [DOI] [PubMed] [Google Scholar]
- 18. Koc E. C., Cimen H., Kumcuoglu B., Abu N., Akpinar G., Haque M. E., Spremulli L. L., Koc H. (2013) Identification and characterization of CHCHD1, AURKAIP1, and CRIF1 as new members of the mammalian mitochondrial ribosome. Front. Physiol. 4, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Brien T. W., Fiesler S. E., Denslow N. D., Thiede B., Wittmann-Liebold B., Mougey E. B., Sylvester J. E., Graack H. R. (1999) Mammalian mitochondrial ribosomal proteins (2): Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 274, 36043–36051 [DOI] [PubMed] [Google Scholar]
- 20. O'Brien T. W., Liu J., Sylvester J. E., Mougey E. B., Fischel-Ghodsian N., Thiede B., Wittmann-Liebold B., Graack H. R. (2000) Mammalian mitochondrial ribosomal proteins (4): Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 275, 18153–18159 [DOI] [PubMed] [Google Scholar]
- 21. Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., Watanabe K. (2001) Proteomic analysis of the mammalian mitochondrial ribosome: identification of protein components in the 28 S small subunit. J. Biol. Chem. 276, 33181–33195 [DOI] [PubMed] [Google Scholar]
- 22. Cavdar Koc E., Ranasinghe A., Burkhart W., Blackburn K., Koc H., Moseley A., Spremulli L. L. (2001) A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 492, 166–170 [DOI] [PubMed] [Google Scholar]
- 23. Miyazaki T., Shen M., Fujikura D., Tosa N., Kim H. R., Kon S., Uede T., Reed J. C. (2004) Functional role of death-associated protein 3 (DAP3) in anoikis. J. Biol. Chem. 279, 44667–44672 [DOI] [PubMed] [Google Scholar]
- 24. Kaushal P. S., Sharma M. R., Booth T. M., Haque E. M., Tung C. S., Sanbonmatsu K. Y., Spremulli L. L., Agrawal R. K. (2014) Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc. Natl. Acad. Sci. U.S.A. 111, 7284–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agrawal R. K., Sharma M. R. (2012) Structural aspects of mitochondrial translational apparatus. Curr. Opin. Struct. Biol. 22, 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amunts A., Brown A., Bai X.-C., Llácer J. L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S. H. W., Ramakrishnan V. (2014) Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mears J. A., Sharma M. R., Gutell R. R., McCook A. S., Richardson P. E., Caulfield T. R., Agrawal R. K., Harvey S. C. (2006) A structural model for the large subunit of the mammalian mitochondrial ribosome. J. Mol. Biol. 358, 193–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma M. R., Koc E. C., Datta P. P., Booth T. M., Spremulli L. L., Agrawal R. K. (2003) Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108 [DOI] [PubMed] [Google Scholar]
- 29. Bogenhagen D. F., Martin D. W., Koller A. (2014) Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 19, 618–629 [DOI] [PubMed] [Google Scholar]
- 30. Seidel-Rogol B. L., McCulloch V., Shadel G. S. (2003) Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33, 23–24 [DOI] [PubMed] [Google Scholar]
- 31. Metodiev M. D., Spåhr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N. G., Ruzzenente B. (2014) NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 10, e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metodiev M. D., Lesko N., Park C. B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., Larsson N. G. (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9, 386–397 [DOI] [PubMed] [Google Scholar]
- 33. Maden B. E., Hughes J. M. (1997) Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma 105, 391–400 [DOI] [PubMed] [Google Scholar]
- 34. Buchhaupt M., Peifer C., Entian K. D. (2007) Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 361, 102–108 [DOI] [PubMed] [Google Scholar]
- 35. Baer R. J., Dubin D. T. (1981) Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 9, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ofengand J., Bakin A. (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266, 246–268 [DOI] [PubMed] [Google Scholar]
- 37. Rorbach J., Minczuk M. (2012) The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J. 444, 357–373 [DOI] [PubMed] [Google Scholar]
- 38. Lee K. W., Okot-Kotber C., LaComb J. F., Bogenhagen D. F. (2013) Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J. Biol. Chem. 288, 31386–31399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lövgren J. M., Wikström P. M. (2001) The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 183, 6957–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sirum-Connolly K., Peltier J. M., Crain P. F., McCloskey J. A., Mason T. L. (1995) Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie 77, 30–39 [DOI] [PubMed] [Google Scholar]
- 41. Caldas T., Binet E., Bouloc P., Costa A., Desgres J., Richarme G. (2000) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16414–16419 [DOI] [PubMed] [Google Scholar]
- 42. Pintard L., Bujnicki J. M., Lapeyre B., Bonnerot C. (2002) MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 21, 1139–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu Y. T., Shu M. D., Steitz J. A. (1997) A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3, 324–331 [PMC free article] [PubMed] [Google Scholar]
- 44. Saikia M., Dai Q., Decatur W. A., Fournier M. J., Piccirilli J. A., Pan T. (2006) A systematic, ligation-based approach to study RNA modifications. RNA 12, 2025–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dong Z. W., Shao P., Diao L. T., Zhou H., Yu C. H., Qu L. H. (2012) RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Behm-Ansmant I., Helm M., Motorin Y. (2011) Use of specific chemical reagents for detection of modified nucleotides in RNA. J. Nucleic Acids 2011, 408053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maden B. E., Corbett M. E., Heeney P. A., Pugh K., Ajuh P. M. (1995) Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77, 22–29 [DOI] [PubMed] [Google Scholar]
- 48. Cruz R. P., Withers J. B., Li Y. (2004) Dinucleotide junction cleavage versatility of 8-17 deoxyribozyme. Chem. Biol. 11, 57–67 [DOI] [PubMed] [Google Scholar]
- 49. Vincze E., Bowra S. (2005) Northerns revisited: a protocol that eliminates formaldehyde from the gel while enhancing resolution and sensitivity. Anal. Biochem. 342, 356–357 [DOI] [PubMed] [Google Scholar]
- 50. Dubin D. T., Taylor R. H., Davenport L. W. (1978) Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 5, 4385–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blanchard S. C., Puglisi J. D. (2001) Solution structure of the A loop of 23S ribosomal RNA. Proc. Natl. Acad. Sci. 98, 3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. King M. P., Attardi G. (1993) Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 268, 10228–10237 [PubMed] [Google Scholar]
- 53. Rorbach J., Boesch P., Gammage P. A., Nicholls T. J. J., Pearce S. F., Patel D., Hauser A., Perocchi F., Minczuk M. (2014) MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell 10.1091/mbc.E14-01-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Decatur W. A., Fournier M. J. (2002) rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 [DOI] [PubMed] [Google Scholar]
- 55. Hengesbach M., Schwalbe H. (2014) Structural basis for regulation of ribosomal RNA 2′-o-methylation. Angew Chem. Int. Ed. Engl. 53, 1742–1744 [DOI] [PubMed] [Google Scholar]
- 56. Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T. (2013) The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature 502, 519–523 [DOI] [PubMed] [Google Scholar]
- 57. Chen B., Ye F., Yu L., Jia G., Huang X., Zhang X., Peng S., Chen K., Wang M., Gong S., Zhang R., Yin J., Li H., Yang Y., Liu H., Zhang J., Zhang H., Zhang A., Jiang H., Luo C., Yang C. G. (2012) Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971 [DOI] [PubMed] [Google Scholar]
- 58. Jia G., Fu Y., He C. (2013) Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., Vågbø C. B., Shi Y., Wang W. L., Song S. H., Lu Z., Bosmans R. P., Dai Q., Hao Y. J., Yang X., Zhao W. M., Tong W. M., Wang X. J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H. E., Klungland A., Yang Y. G., He C. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Britton R. A. (2009) Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63, 155–176 [DOI] [PubMed] [Google Scholar]
- 61. He J., Cooper H. M., Reyes A., Di Re M., Kazak L., Wood S. R., Mao C. C., Fearnley I. M., Walker J. E., Holt I. J. (2012) Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 40, 6097–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kolanczyk M., Pech M., Zemojtel T., Yamamoto H., Mikula I., Calvaruso M. A., van den Brand M., Richter R., Fischer B., Ritz A., Kossler N., Thurisch B., Spoerle R., Smeitink J., Kornak U., Chan D., Vingron M., Martasek P., Lightowlers R. N., Nijtmans L., Schuelke M., Nierhaus K. H., Mundlos S. (2011) NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell 22, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paul M. F., Alushin G. M., Barros M. H., Rak M., Tzagoloff A. (2012) The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. J. Biol. Chem. 287, 24346–24355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bogenhagen D. F., Rousseau D., Burke S. (2008) The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]
- 65. Dennerlein S., Rozanska A., Wydro M., Chrzanowska-Lightowlers Z. M., Lightowlers R. N. (2010) Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 430, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uchiumi T., Ohgaki K., Yagi M., Aoki Y., Sakai A., Matsumoto S., Kang D. (2010) ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 38, 5554–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Purta E., O'Connor M., Bujnicki J. M., Douthwaite S. (2009) YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 72, 1147–1158 [DOI] [PubMed] [Google Scholar]


