Background: Malfunctions in the ubiquitin-proteasome system cause accumulation of non-functional, potentially toxic protein aggregates.
Results: The protein aggregates activate Nrf2 and are then excluded by autophagy in vivo.
Conclusion: Both Nrf2 and autophagy serve as in vivo cellular adaptations to impaired proteasome.
Significance: Cells contain networks of cellular defense mechanisms against defective proteostasis.
Keywords: Autophagy, Autophagy-related Protein 7 (ATG7), Nuclear Factor 2 (Erythroid-derived 2-Like Factor) (NFE2L2) (Nrf2), Proteasome, Protein Aggregation
Abstract
The ubiquitin-proteasome system and autophagy are crucially important for proteostasis in cells. These pathways are interdependent, and dysfunction in either pathway causes accumulation of ubiquitin-positive aggregates, a hallmark of human pathological conditions. To elucidate in vivo compensatory action(s) against proteasomal dysfunction, we developed mice with reduced proteasome activity in their livers. The mutant mice exhibited severe liver damage, accompanied by formation of aggregates positive for ubiquitin and p62/Sqstm1, an adaptor protein for both selective autophagy and the anti-oxidative Keap1-Nrf2 pathway. These aggregates were selectively entrapped by autophagosomes, and pathological features of livers with impaired proteasome activity were exacerbated by simultaneous suppression of autophagy. In contrast, concomitant loss of p62/Sqstm1 had no apparent effect on the liver pathology though p62/Sqstm1 was indispensable for the aggregates formation. Furthermore, defective proteasome function led to transcriptional activation of the Nrf2, which served as a physiological adaptation. Our in vivo data suggest that cells contain networks of cellular defense mechanisms against defective proteostasis.
Introduction
The 26S proteasome, in collaboration with the sophisticated ubiquitination system used for selection of target proteins, is responsible for degrading unnecessary or damaged proteins. Malfunctions in this pathway cause accumulation of non-functional, potentially toxic protein aggregates (1–3). The macroautophagy (hereafter referred to as autophagy) system serves as a supplier of molecular building blocks under starved conditions and also contributes to cellular renovation during cell differentiation (4, 5). Defects in this process can cause amino acid insufficiency, which impairs protein synthesis during adaptation to starvation, as well as energy production essential for cell survival and development (4, 5). Even under nutrient-rich conditions, autophagy occurs constitutively at low levels to mediate global turnover of cytoplasmic materials (6, 7).
Dysfunctions of autophagy coupled to the ubiquitin system have been directly linked to human conditions such as Parkinson disease and inflammatory disorders. Autophagy contributes to selective removal of aggregated proteins (aggrephagy), unnecessary or damaged mitochondria (mitophagy), and invading bacteria (xenophagy); these processes are usually mediated by ubiquitin signaling (8–10). When the ubiquitin-proteasome system is impaired due to accumulation of certain aggregation-prone proteins related to neurodegenerative disease, autophagy is responsible for eliminating ubiquitin-positive protein aggregates (11–13). In response to loss of mitochondrial membrane potential, the E3 ligase Parkin translocates to damaged mitochondria in a PINK1-dependent manner; once it is localized to mitochondria, it ubiquitinates outer membrane proteins, thereby inducing mitophagy (14, 15). Parkinson disease-related mutations of Parkin and PINK1 prevent induction of mitophagy, resulting in persistence of damaged mitochondria, which may play a role in the pathogenesis of Parkinson disease (14, 15). Invading bacteria in the cytosol and/or ruptured endosomal membranes are ubiquitinated by E3s, including Parkin (16) and LRSAM1 (17), which mediates autophagic sequestration of microbes to restrict their growth. Ubiquitin- and LC3-binding adaptor proteins, including p62/Sqstm1 (hereafter referred to as p62) (18), neighbor of BRCA1 gene 1 (Nbr1)2 (19), NDP52 (20), and optineurin (21), are translocated to these ubiquitinated cargos; this process is assumed to mediate sequestration of ubiquitinated cargos into autophagosomes. Among them, p62 and Nbr1 have been identified as major components of many types of aggregates or inclusions observed in various human diseases, including neurodegenerative diseases, liver disorders, and hepatocellular carcinomas (19, 22). But significance of such adaptor proteins on the aggregates, particularly in vivo, remains unclear.
The Keap1-Nrf2 pathway, one of the major cellular defense mechanisms against oxidative and electrophilic stresses (23, 24), is activated during selective autophagy (25–28). Under normal conditions, the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) is constitutively degraded through the ubiquitin-proteasome pathway; its binding partner, Keap1 (kelch-like ECH-associated protein 1), is an adaptor of the ubiquitin ligase complex that targets Nrf2. Exposure to electrophiles, reactive oxygen species, and nitric oxide instigates modification of the cysteine residues of Keap1, leading to its inactivation. As a result, Nrf2 is stabilized, and it subsequently translocates to the nucleus to induce the transcription of numerous cytoprotective genes through heterodimerization with small Maf proteins (23, 24). p62 also regulates the Keap1-Nrf2 pathway via a noncanonical mechanism (25–28). Under conditions of selective autophagy, Ser403 of the ubiquitin-associated domain of p62 is initially phosphorylated by casein kinase 2 or TANK-binding kinase 1, which promotes the translocation of p62 to cargos positive for ubiquitin (29, 30). Subsequently, Ser351 of the Keap1-interacting region of p62 is phosphorylated, followed by sequestration of Keap1 on the cargos. As a result, Nrf2 is stabilized; as in the canonical pathway, it then translocates into the nucleus to induce its cytoprotective target genes (25, 26). The ubiquitinated autophagic cargos, together with phosphorylated p62 and the Keap1 complex, are degraded by autophagy, leading to elimination of cytotoxic components (27). However, the physiological role of the coupling between the Keap1-Nrf2 system and selective autophagy in vivo has been not yet determined. In this study, we developed genetically modified mice with decreased 26S proteasome activity, which accumulate aggregate structures positive for both ubiquitin and p62 in their cells, and found that proteasome-dysfunction activates selective autophagy and the Keap1-Nrf2 pathway, both of which serve as cellular defense mechanisms.
EXPERIMENTAL PROCEDURES
Mice
Rpt2flox/flox mice (31) were cross-bred with albumin-Cre transgenic mice (32) to generate Rpt2flox/flox;Alb-Cre mice. Atg7flox/flox, p62flox/flox, and the Nrf2-knock-out mice used in this study were described previously (33–35). Mice were housed in specific pathogen-free facilities, and the Ethics Review Committee for Animal Experimentation of the Tokyo Metropolitan Institute of Medical Science approved the experimental protocols.
Immunoblot Analysis
Immunoblots were carried out as described previously (26). Antibodies against p62 (Progen Biotechnik, GP62-C), ubiquitin (Santa Cruz Biotechnology, Inc., P4D1), Keap1 (Proteintech Group, Inc.), Nqo1 (Abcam, Inc.), Nrf2 (Santa Cruz Biotechnology, Inc., H-300), LC3B (Cell Signaling Technology, catalog no. 2775), Nbr1 (ProteinExpress Co., Ltd.), GFP (Invitrogen), actin (Chemicon Intl., Inc., MAB1501R), and lamin B (Santa Cruz Biotechnology, Inc., M-20) were purchased from the indicated suppliers. Anti-phosphorylated p62 polyclonal antibody was raised in rabbits using the peptide Cys+KEVDP(pS)TGELQSL as an antigen (26). The rabbit polyclonal antibodies against Atg7 and Rpt2 were described previously (36, 37).
Assay of Proteasome Activity
Peptidase activity was measured using a fluorescent peptide substrate, succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-MCA), as described previously (38).
Histological Examination
Fixation and embedding procedures for immunohistochemistry were described previously (39). Briefly, mouse livers were quickly excised, cut into small pieces, and then fixed by immersion in 4% paraformaldehyde/4% sucrose in 0.1 m phosphate buffer, pH 7.4 (PB). After rinsing, samples were embedded in paraffin (for H&E staining), or in OCT compound (for immunofluorescence). For immunofluorescence microscopy, sections were blocked and then incubated for 2–3 days at 4 °C with the following primary antibodies: guinea pig polyclonal antibody against p62 (Progen), rabbit polyclonal antibody against ubiquitin (DAKO), or rabbit polyclonal antibody against Keap1 (Proteintech Group). Immunofluorescence images were taken with an FV1000 laser scanning confocal microscope equipped with a UPlanSApo 40× numerical aperture 1.3 oil objective lens (Olympus). After image acquisition, contrast and brightness were adjusted using Photoshop CS4.
Electron Microscopy and Immunoelectron Microscopy
For conventional electron microscopy, livers were excised and fixed by immersion in 0.1 m PB containing 2% paraformaldehyde and 2% glutaraldehyde. Fixed samples were post-fixed with 1% OsO4, embedded in Epon812, and sectioned. Immunoelectron microscopy was carried out on ultrathin cryosections, as described previously (39). In brief, livers were fixed by cardiac perfusion with 0.1 m PB containing 4% paraformaldehyde and 4% sucrose and then frozen in PB with 2.3 m sucrose and 20% polyvinylpyrrolidone. Ultrathin sections were mounted on Formvar carbon-coated nickel grids, blocked with 1% bovine serum albumin in PBS, incubated with anti-ubiquitin (DAKO) and anti-p62 (Progen) antibodies, and then incubated with colloidal gold-conjugated secondary antibodies.
Quantitative Real-time PCR
Using the Transcriptor First-Strand cDNA Synthesis Kit (Roche Applied Science), cDNA was synthesized from 1 μg of total RNA. Quantitative PCR was performed using LightCycler® 480 Probes Master mix (Roche Applied Science) on a LightCycler® 480 (Roche Applied Science). Signals were normalized against that of β-glucuronidase (Gus). The sequences of the primers used were as follows: Nqo1 (left), AGCGTTCGGTATTACGATCC; Nqo1 (right), AGTACAATCAGGGCTCTTCTCG.
Statistical analysis
Values, including those displayed in the graphs, are means ± S.E. Statistical analysis was performed using the unpaired t test (Welch test). p values less than 0.05 denoted statistical significance.
RESULTS
Generation of Mice with Decreased Proteasome Activity
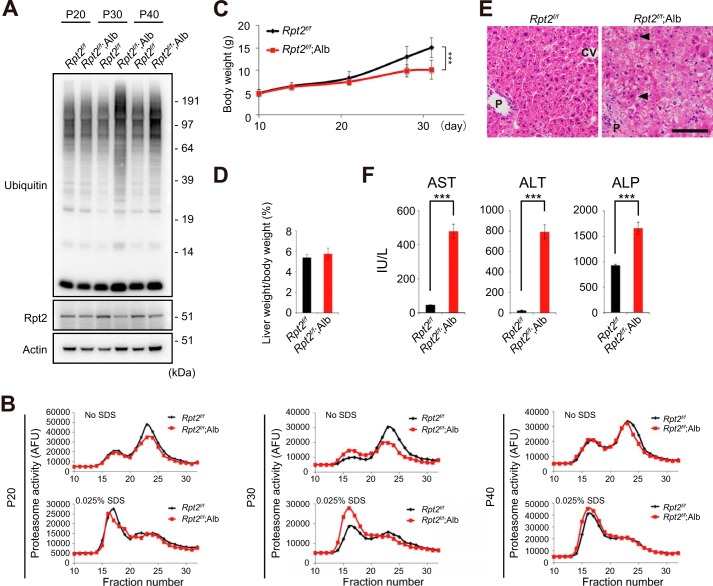
To investigate aggrephagy in vivo, we crossbred mice bearing a conditional knock-out of Rpt2, one of six ATPases of the 19S regulatory particle of the 26S proteasome (Rpt2flox/flox) (31), with albumin-Cre (Alb-Cre) transgenic mice (32). The Rpt2flox/flox;Alb-Cre (Rpt2f/f;Alb) mice were viable at birth and indistinguishable in appearance from their littermates. In Rpt2f/f;Alb mice, levels of Rpt2 protein in the liver started to decrease at postnatal day (P)30 and recovered at P40 (Fig. 1A); ubiquitinated proteins accumulated significantly in the liver at P30 (Fig. 1A). The chymotryptic activities of the 26S and 20S proteasomes (measured using Suc-LLVY-MCA as a substrate) in extracts from Rpt2f/f;Alb livers at P20 were comparable with those in age-matched control livers (Fig. 1B). The activity of the 26S proteasome decreased dramatically at P30 and recovered at P40, whereas the activity of the 20S proteasome increased at P30 only (Fig. 1B). Consistent with these kinetics, growth retardation was observed as early as at P30 (Fig. 1C). Although the ratio of liver weight to body weight was similar among genotypes even at P30 (Fig. 1D), decreased proteasome activity in Rpt2f/f;Alb livers was accompanied by signs of hepatic degeneration such as the presence of hypertrophic cells, dead cells, small regenerating cells, and inflammatory cells, as revealed by hematoxylin and eosin (H&E) staining (Fig. 1E) and by hepatocytic damage, as revealed by leakage of liver enzymes (Fig. 1F). The recovery of Rpt2 at P40 in the livers of Rpt2f/f;Alb mice might be attributed to rapid hepatocytic death due to impairment of proteasome activity, followed by compensatory regeneration of hepatocytes from oval cells, which express low levels of albumin (40). Collectively, these data indicate that at P30, Rpt2f/f;Alb mice exhibit liver injury accompanied by reduced proteasome activity in the liver.
FIGURE 1.
Time course analysis of Rpt2f/f;Alb mice. A, liver homogenates were prepared from mice of the indicated genotypes at P20, P30, and P40 and subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. B, proteasome activity in Rpt2f/f;Alb livers at P20, P30, and P40. C, growth curve of Rpt2f/f (control) and Rpt2f/f;Alb mice. Data are means ± S.E. of Rpt2f/f (n = 33) and Rpt2f/f;Alb (n = 24) mice. ***, p < 0.001. D, ratio of liver weight to body weight of Rpt2f/f (control) and Rpt2f/f;Alb mice at P30. Data are means ± S.E. of Rpt2f/f (n = 5) and Rpt2f/f;Alb (n = 8) mice. E, H&E staining of livers of indicated genotypes at P30. Mitotic cells or abnormal mitosis (arrowheads) were often observed in Rpt2-deficient hepatocytes. CV, central vein; P, portal triad. Bar, 100 μm. F, serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were measured. Data are means ± S.E. of Rpt2f/f (n = 3) and Rpt2f/f;Alb (n = 5) mice. ***, p < 0.001. IU/L, international units/liter. AFU, arbitrary fluorescent unit.
Characterization of Ubiquitinated Aggregates in Rpt2f/f;Alb Livers
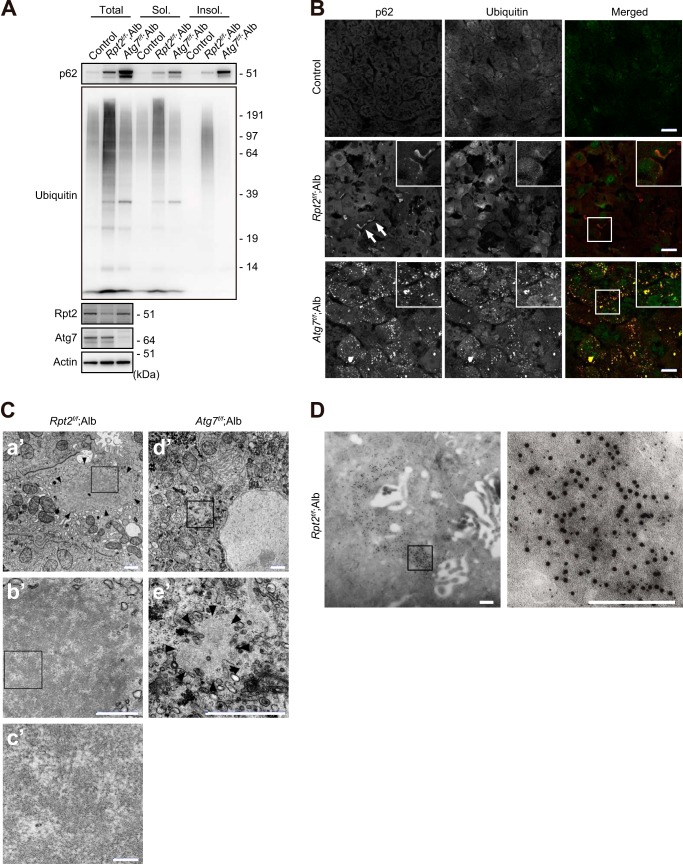
Immunoblot analysis revealed elevated levels of both soluble and insoluble ubiquitinated proteins in the livers of Rpt2f/f;Alb mice (Fig. 2A). The level of insoluble ubiquitinated proteins was significantly higher in Rpt2f/f;Alb than in Atg7flox/flox;Alb-Cre (Atg7f/f;Alb) livers, in which autophagy is impaired (Fig. 2A) (33). We also observed significant accumulation of p62 protein in both detergent-soluble and -insoluble fractions from Rpt2f/f;Alb livers, albeit less than in Atg7f/f;Alb liver fractions (Fig. 2A). Consistent with these biochemical data, immunofluorescence analysis revealed co-localization of ubiquitin and p62 on large pleomorphic aggregated structures and small punctate structures in Rpt2f/f;Alb hepatocytes (Fig. 2B). As in a previous study (33), aggregates observed in Atg7f/f;Alb hepatocytes were also positive for ubiquitin and p62 (Fig. 2B). Electron microscopy revealed that Rpt2f/f;Alb hepatocytes contained large pleomorphic structures mainly consisting of electron-lucent areas and scattered patchy electron-dense areas; this pattern appeared to be formed by the clustering of fibrillar elements (Fig. 2C). These features were distinct from the large circular or elliptical structures often observed in Atg7f/f;Alb hepatocytes (Fig. 2C). By double-immunoelectron microscopy, we confirmed the co-localization of p62 and ubiquitin in the cytoplasmic aggregated structures in Rpt2f/f;Alb hepatocytes (Fig. 2D). Colloidal gold particles representing ubiquitin were distributed in highly electron-dense areas, whereas particles representing p62 were distributed in areas of both high and low electron density.
FIGURE 2.
Characterization of ubiquitinated aggregates in Rpt2f/f;Alb livers. A, liver homogenates were prepared from mice of the indicated genotypes at P30. Total, soluble (Sol.), and insoluble (Insol.) fractions were subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. B, liver cryosections from mice of the indicated genotypes at P30 were double-immunostained with p62 and ubiquitin antibodies. A portion of each image is magnified and shown in the inset. Arrows indicate large pleomorphic aggregated structures. Merged images are shown at the right column (red, p62; green, ubiquitin). Bars, 20 μm. C, electron micrographs of hepatocytes of the indicated genotypes. The boxed regions in a′, b′, and d′ are enlarged and shown in b′, c′, and e′, respectively. Arrowheads indicate aggregated structures. Bars, a′, 1 μm; b′ and c′, 0.5 μm; d′, 0.1 μm. D, immunoelectron micrographs showing double labeling of ubiquitin (12-nm colloidal gold particles) and p62 (6-nm colloidal gold particles) in hepatocytes of Rpt2f/f;Alb mice at P30. The boxed region is enlarged and shown at the right. Bars, 0.2 μm.
Induction of Aggrephagy in Rpt2f/f;Alb Livers
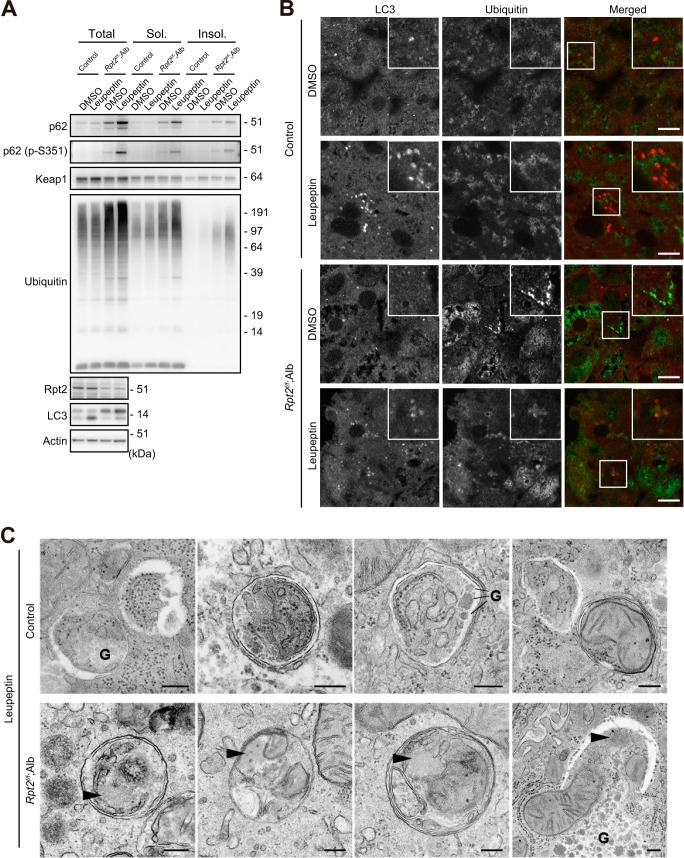
We next examined autophagic flux in livers of Rpt2f/f;Alb mice. Intraperitoneal (i.p.) injection of leupeptin, a lysosomal cysteine proteinase inhibitor, resulted in elevation of LC3-II (an indicator of autophagic flux) (41) in livers of both control and Rpt2f/f;Alb mice, although the effect was smaller in the mutants (Fig. 3A). The levels of ubiquitinated proteins and p62 in mutant livers further increased upon intraperitoneal injection of leupeptin, but this effect was not observed in control livers (Fig. 3A). Phosphorylation of p62 at Ser351, which signifies induction of selective autophagy (26), increased only in the intraperitoneally injected mutant livers (Fig. 3A), implying increased selective autophagy of aggregate structures (aggrephagy). Indeed, immunofluorescence analysis revealed that LC3/ubiquitin double-positive structures were frequently observed in leupeptin-treated Rpt2f/f;Alb mice (8.23%), whereas they could barely be detected in leupeptin-treated control hepatocytes (2.68%) (Fig. 3B). Furthermore, we confirmed by electron microscopy that autophagosomes in the mutant hepatocytes occasionally (arrowheads; 7/46, 15.2%) contained aggregate-like amorphous structures, whereas those in control hepatocytes did not (0/64; 0%) (Fig. 3C).
FIGURE 3.
Autophagic degradation of ubiquitin-positive aggregates accumulated due to impaired proteasome activity. A, mice of the indicated genotypes were subjected to intraperitoneal injection of leupeptin at P30. One hour after the injection, liver homogenates were prepared and subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. A graph indicates quantitative densitometry of immunoblotting data (n = 3) and the ratios of insoluble (Insol.) ubiquitinated proteins relative to that of dimethyl sulfoxide (DMSO)-treated control mice. B, liver cryosections from mice of the indicated genotypes treated as described in A were double-immunostained with LC3 (red in the merged image) and ubiquitin (green in the merged image) antibodies. The boxed region is magnified and shown in the inset. Bars, 10 μm. C, electron micrographs of hepatocytes from mice of the indicated genotypes, treated as described in A. Arrowheads, aggregate-like amorphous structures. G, glycogen granules. Bar, 0.2 μm. Sol., soluble.
Exacerbation of Liver Pathology in Rpt2f/f;Alb Mice by Suppression of Autophagy
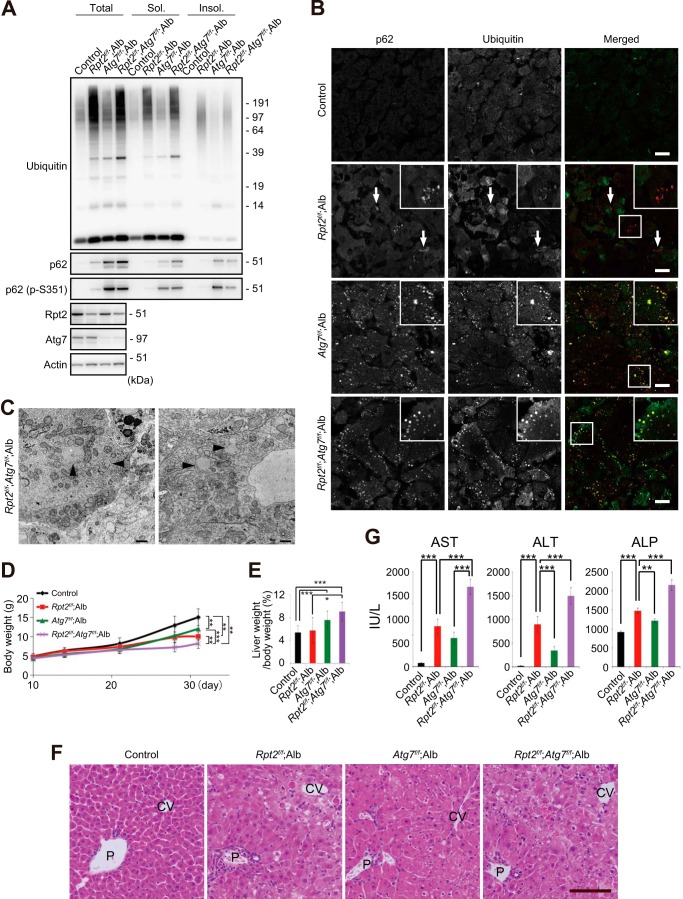
Next, to clarify the physiological significance of autophagy induced in Rpt2f/f;Alb liver, we generated hepatocyte-specific Rpt2 and Atg7 double-knock-out mice (Rpt2f/f;Atg7f/f;Alb). Concomitant loss of Atg7 in Rpt2f/f;Alb mice resulted in dramatic accumulation of soluble and insoluble p62 (Fig. 4A), supporting the evidence p62 is a selective substrate for autophagy (18, 33) and gene expression of p62 is induced by impairment of autophagy (25). Meanwhile, ubiquitinated proteins were present in detergent-soluble and -insoluble fractions of Rpt2f/f;Atg7f/f;Alb livers to a similar extent in those of Rpt2f/f;Alb livers (Fig. 4A). This observation might be attributed to more severe hepatic damage in the double-mutant livers (see below). In the absence of Atg7, the large pleomorphic aggregates positive for ubiquitin and p62 observed in Rpt2f/f;Alb hepatocytes became small, round, and scattered throughout the cytoplasm (Fig. 4, B and C), similar to structures observed in autophagy-deficient livers (Figs. 2B and 4B). In addition, liver damage due to impaired proteasome activity was more severe than in Atg7f/f;Alb mice, and the damage was exacerbated by simultaneous loss of Atg7. The growth delay in Rpt2f/f;Atg7f/f;Alb mice was more severe than in Rpt2f/f;Alb or Atg7f/f;Alb mice (Fig. 4D). Furthermore, we observed hepatomegaly accompanied by cellular hypertrophy and degeneration, both typical phenotypes of autophagy-deficient livers (33), in hepatocytes of Rpt2f/f;Atg7f/f;Alb mice (Fig. 4, E and F). The Rpt2f/f;Atg7f/f;Alb mice exhibited higher serum levels of aspartate aminotransferase, alanine aminotransferase, and alkaline phosphatase than single Atg7- or Rpt2-knock-out mice (Fig. 4G).
FIGURE 4.
Exacerbation of pathology in Rpt2f/f;Alb liver by concomitant loss of Atg7. A, liver homogenates were prepared from mice of the indicated genotypes at P30. Total, soluble (Sol.), and insoluble (Insol.) fractions were subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. B, liver cryosections of Rpt2f/f;Atg7f/f;Alb mice were double-immunostained with p62 and ubiquitin antibodies. A portion of each image was magnified and shown in the inset. Arrows indicate large pleomorphic aggregated structures. Merged images are shown in the right column (red, p62; green, ubiquitin). Bars, 20 μm. C, electron micrographs of Rpt2f/f;Atg7f/f;Alb hepatocytes. Arrowheads indicate aggregated structures. Bar, 1 μm. D, growth curves of mice of the indicated genotypes. Data are means ± S.E. of control (n = 35), Atg7f/f;Alb (n = 22), Rpt2f/f;Alb (n = 10), and Rpt2f/f;Atg7f/f;Alb (n = 17) mice. **, p < 0.01; ***, p < 0.001. E, ratio of liver weight to body weight of mice of the indicated genotype at P30. Data are means ± S.E. of control (n = 19), Atg7f/f;Alb (n = 13), Rpt2f/f;Alb (n = 10), and Rpt2f/f;Atg7f/f;Alb (n = 8) mice. *, p < 0.05; ***, p < 0.001. F, H&E staining of livers of the indicated genotypes at P30. P, portal triad; CV, central vein. Bar, 100 μm. G, serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were measured. Data are means ± S.E. of control (n = 17), Atg7f/f;Alb (n = 13), Rpt2f/f;Alb (n = 8), and Rpt2f/f;Atg7f/f;Alb (n = 16) mice. **, p < 0.01; ***, p < 0.001. IU/L, international units/liter.
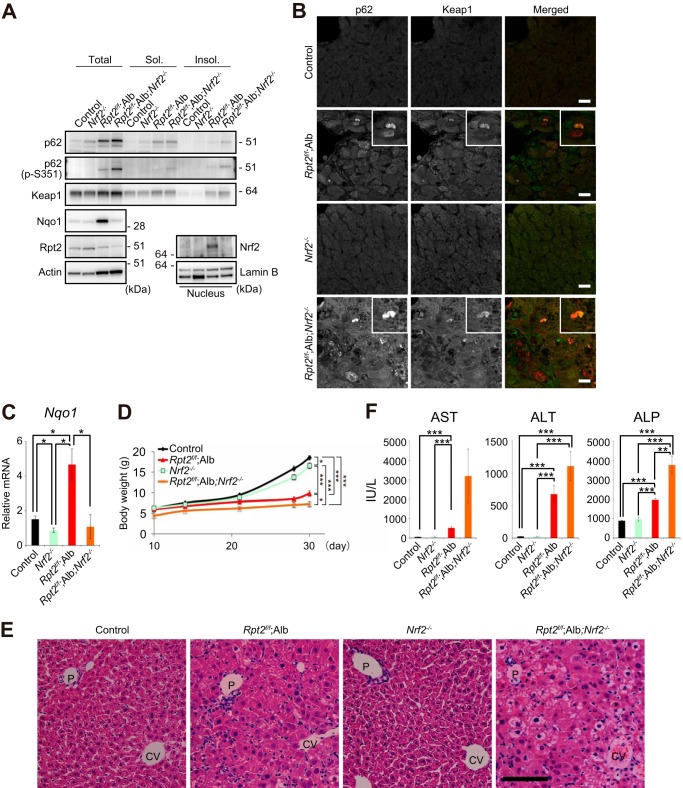
Feedback Activation of the Keap1-Nrf2 Pathway when the Proteasome Is Impaired
We postulated that under proteasome-defective conditions, the Keap1-Nrf2 pathway should be activated in a p62 phosphorylation-dependent manner. In fact, we observed accumulation of not only p62, but also its Ser351-phosphorylated form, in Rpt2f/f;Alb livers (Figs. 2A and 5A). A significant proportion of Ser351-phosphorylated p62 was recovered in the detergent-insoluble fraction (Figs. 2A and 5A), and Keap1 was also recovered in the detergent-insoluble fraction (Fig. 5A). At the same time, Nrf2 was stabilized in the mutant livers (Fig. 5A). Immunofluorescence analysis revealed extensive co-localization of p62 and Keap1 in the same aggregate structures (Fig. 5B). Consequently, gene expression of the Nrf2 target gene Nqo1 (NAD(P)H dehydrogenase quinone 1) in the livers of Rpt2f/f;Alb mice was markedly induced (Fig. 5C), and we also observed increased levels of Nqo1 protein (Fig. 5A). As expected, loss of Nrf2 in Rpt2f/f;Alb mice suppressed induction of Nrf2 targets (Fig. 5, A and C). p62 was present at higher levels in Rpt2flox/flox;Nrf2−/−;Alb-Cre (Rpt2f/f;Nrf2−/−;Alb) than in control livers (Fig. 5A). Therefore, as in Rpt2f/f;Alb livers, aggregate structures positive for both p62 and Keap1 were detected in hepatocytes of Rpt2f/f;Nrf2−/−;Alb mice (Fig. 5B). The double mutant mice exhibited slower growth than Rpt2f/f;Alb mice, whereas single knock-out of Nrf2 hardly affected growth, at least at P30 (Fig. 5D). H&E staining revealed no significant difference between control and Nrf2 single knock-out livers; by contrast, simultaneous loss of Nrf2 and Rpt2 in the liver caused degenerative alterations more severe than those observed in Rpt2 single knock-out livers (Fig. 5E), and leakage of hepatic enzymes into sera was more severe in double knock-out (Rpt2f/f;Nrf2−/−;Alb) mice than in Rpt2f/f;Alb mice (Fig. 5F).
FIGURE 5.
Feedback activation of the Keap1-Nrf2 pathway serves as a physiological adaptation to impaired proteasome function. A, total lysates, detergent-soluble (Sol.) and -insoluble (Insol.) fractions, and nuclear fractions from livers of the indicated genotypes were subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. B, liver cryosections from mice of the indicated genotypes were double-immunostained with antibodies against p62 and Keap1 antibodies. Arrows indicate large pleomorphic aggregated structures. Merged images are shown in the right column of each panel (red, p62; green, Keap1). Bars, 20 μm. C, total RNAs were prepared from livers of the indicated genotypes. Values were normalized to the amount of mRNA in the livers of control mice. Data are means ± S.E. of control (n = 14), Nrf2−/− (n = 10), Rpt2f/f;Alb (n = 5), and Rpt2f/f;Alb;Nrf2−/− (n = 7) mice. *, p < 0.05. D, growth curves of mice of the indicated genotypes. Data are means ± S.E. of control (n = 15), Nrf2−/− (n = 13), Rpt2f/f;Alb (n = 6), and Rpt2f/f;Alb;Nrf2−/− (n = 6) mice. *, p < 0.05; ***, p < 0.001. E, H&E staining of livers of the indicated genotypes at P30. P, portal triad; CV, central vein. Bar, 100 μm. F, serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were measured. Data are means ± S.E. of control (n = 13), Nrf2−/− (n = 11), Rpt2f/f;Alb (n = 6), and Rpt2f/f;Alb;Nrf2−/− (n = 7) mice. **, p < 0.01; ***, p < 0.001. IU/L, international units/liter.
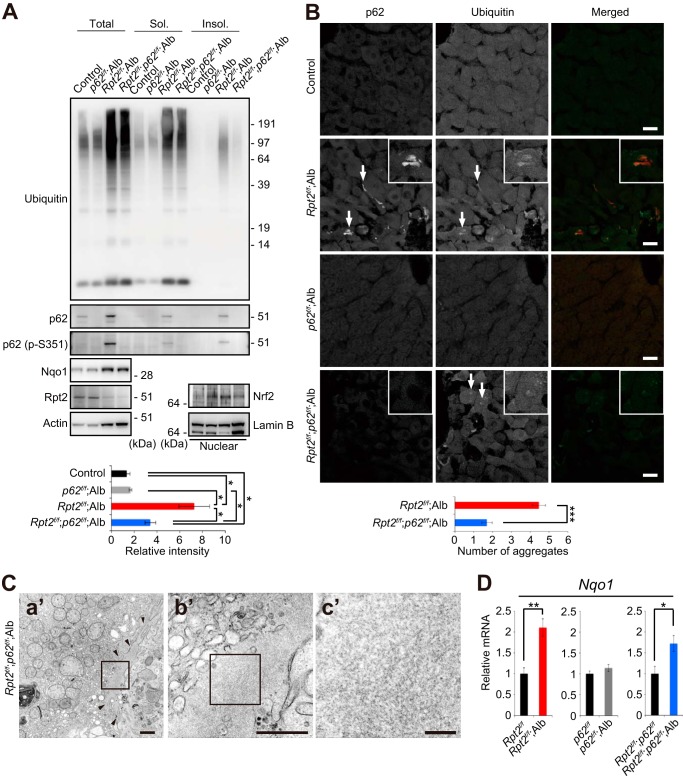
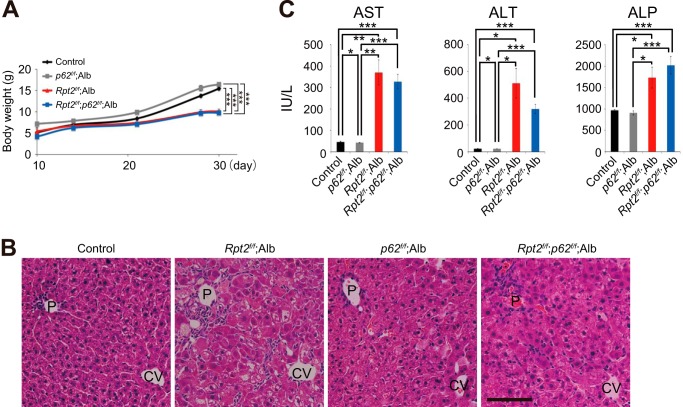
Role of p62 in Formation of Aggregate-containing Structures in Rpt2f/f;Alb Hepatocytes
To investigate the effect of loss of p62 on aggregate formation, Nrf2 activation and pathology in mice with decreased proteasome activity, we generated hepatocyte-specific Rpt2 and p62 double-knock-out mice (Rpt2f/f;p62f/f;Alb). The levels of total ubiquitinated proteins in livers of Rpt2f/f;p62f/f;Alb mice were similar to those in Rpt2f/f;Alb livers (Fig. 6A). The accumulation of insoluble ubiquitinated proteins in Rpt2f/f;Alb mice was dramatically suppressed by loss of p62 (Fig. 6A). Although the number of aggregates was reduced by deletion of p62, we still occasionally detected large pleomorphic aggregates positive for ubiquitin, even in Rpt2f/f;p62f/f;Alb hepatocytes, by immunofluorescence staining (Fig. 6B). In electron micrographs, the aggregated structures appeared rather homogeneous, containing less electron-dense areas than those in Rpt2f/f;Alb hepatocytes (Fig. 6C). We speculated that Nrf2-activation observed in single Rpt2-deficient livers was abrogated by concomitant loss of p62. The nuclear translocation as well as induction of Nqo1 tended to be inhibited by simultaneous loss of p62, but we did not recognize any significant differences (Fig. 6, A and D). Rpt2f/f;p62f/f;Alb pups were born at Mendelian frequency, and their growth was delayed similarly to that of Rpt2f/f;Alb mice (Fig. 7A). Histological analysis revealed degenerated features in Rpt2f/f;p62f/f;Alb livers similar to those observed in Rpt2-deficient livers, although hepatocytic hypertrophy tended to be ameliorated (Fig. 7B). Leakage of hepatic enzymes in Rpt2f/f;p62f/f;Alb was detected at a level similar to that in single Rpt2 knock-out mice (Fig. 7C).
FIGURE 6.
Role of p62 in formation of aggregate-containing structures and Nrf2 activation in Rpt2f/f;Alb hepatocytes. A, total lysates, detergent-soluble (Sol.) and -insoluble (Insol.) fractions, and nuclear fractions from livers of the indicated genotypes were subjected to immunoblotting with the indicated antibodies. Data were obtained from three independent experiments. B, liver cryosections from mice of the indicated genotypes were double-immunostained with p62 and ubiquitin antibodies. A portion of each image is magnified and shown in the inset. Arrows indicate large pleomorphic aggregated structures. Merged images are shown in the right column (red, p62; green, ubiquitin). Bars, 20 μm. The graph shows the average number (± S.E.) of ubiquitin-positive large aggregates counted in an area of 210 × 210 μm in liver sections from three animals for each genotype (n = 30). C, electron micrograph of Rpt2f/f;p62f/f;Alb hepatocytes. The boxed regions in a′ and b′ are enlarged and shown in b′ and c′, respectively. Arrowheads indicate aggregated structures. Bars, a′, 1 μm; b′, 0.5 μm; c′, 0.1 μm. D, total RNAs were prepared from livers of the indicated genotypes. Values were normalized to the amount of mRNA in the livers of control mice. Data are means ± S.E. of Rpt2f/f (n = 4), Rpt2f/f;Alb (n = 8), p62f/f (n = 13), p62f/f;Alb (n = 10), Rpt2f/f;p62f/f (n = 11), and Rpt2f/f;p62f/f;Alb (n = 12) mice. *, p < 0.05; **, p < 0.01.
FIGURE 7.
Pathology in Rpt2f/f;Alb liver by concomitant loss of p62. A, growth curves of mice of the indicated genotypes. Data are means ± S.E. of control (n = 44), p62f/f;Alb (n = 9), Rpt2f/f;Alb (n = 6), and Rpt2f/f;p62f/f;Alb (n = 15) mice. ***, p < 0.001. B, H&E staining of livers of the indicated genotypes at P30. P, portal triad; CV, central vein. Bar, 100 μm. C, serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were measured. Data are means ± S.E. of control (n = 31), p62f/f;Alb (n = 5), Rpt2f/f;Alb (n = 5), and Rpt2f/f;p62f/f;Alb (n = 12) mice. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
In this study, we showed that reduced proteasome activity caused formation of aggregate structures positive for ubiquitin and p62 (Fig. 2) and then activated not only aggrephagy but also the Keap1-Nrf2 pathway (Figs. 3 and 5). Simultaneous suppression of autophagy in proteasome-suppressed livers induced accumulation of p62; in addition, the large pleomorphic p62- and ubiquitin-positive aggregates found in proteasome-suppressed livers became small and round (Fig. 4, B and C). Meanwhile, additional loss of p62 in hepatocytes with impaired proteasome activity greatly reduced the level of ubiquitin-positive aggregates with altered morphological compositions (Fig. 6, A–C). Because the fibril-like structures were recognized even in Rpt2/p62 double-deficient hepatocytes (Fig. 6C), we concluded that cellular levels of p62 determine the morphological characteristics of ubiquitin aggregates but not the primarily formation of fibril-like structures.
What is the physiological significance of p62 on these aggregates? One possibility is that p62 serves a receptor function in aggrephagy; this idea is supported by the observation that ubiquitin aggregates positive for p62 were degraded in an autophagy-dependent manner (Fig. 3). However, additional loss of Atg7, but not p62, exacerbated the pathology in proteasome-defective liver (Figs. 4 and 7), suggesting that p62 is not involved in recognition of the aggregates. Although there remains a possibility that Nbr1, whose domain structure is quite similar to that of p62 (8), compensates the function of p62, simultaneous loss of Nbr1 in p62-deficient livers did not exhibit any accumulation of ubiquitinated proteins in contrast to defective autophagy.3 Another possibility is that p62 serves as a scaffold for Nrf2 activation. Ser351 of p62 is phosphorylated on cargos destined for autophagy, such as ubiquitin-positive aggregates, and this phosphorylation is followed by robust Nrf2 activation (26). Indeed, we observed p62 phosphorylation and subsequent Nrf2 activation in livers with decreased proteasome activity (Fig. 5, A and C). However, additional loss of Nrf2 (Fig. 5), but not of p62 (Fig. 7), exacerbated the pathological state caused by inhibition of proteasome activity. This discrepancy can be explained by the fact that Nrf2 degradation is dependent on the ubiquitin-proteasome system (23, 24). In Rpt2/p62 double knock-out livers, reduced proteasome activity could directly activate Nrf2 even in the absence of p62 (Fig. 6, A and D). In other words, the effect of phosphorylated p62 on Nrf2 activation might be hidden by the robust activation of Nrf2 that occurs in response to impairment of the ubiquitin-proteasome system. In conclusion, our data show for the first time that both elimination of aggregate structures by autophagy and activation of Nrf2 under proteasome-defective conditions serve as physiological adaptations to impaired proteasome function in vivo.
Acknowledgments
We thank Y. Yang (Tokyo Metropolitan Institute of Medical Science) for excellent technical assistance and K. Kanno and A. Yabashi (Fukushima Medical University School of Medicine) for assistance with histological studies.
This work was supported by Grant-in-aid for Scientific Research on Innovative Areas 25111006 (to M. K. and S. W.), Funding Program for Next Generation World Leading Researchers Grant LS132 (to M. K.), the Takeda Science Foundation (to T. K. and M. K.), and a Global Research Laboratory grant (to M-S. L. and M. K.).
Y.-S. Sou and M. Komatsu, unpublished data.
- Nbr1
- neighbor of BRCA1 gene 1
- Keap1
- kelch-like ECH-associated protein 1
- Nqo1
- NAD(P)H dehydrogenase quinone 1
- Nrf2
- nuclear factor erythroid 2-related factor 2
- P
- postnatal day.
REFERENCES
- 1. Goldberg A. L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 2. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 4. Mizushima N., Levine B. (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol. 12, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 6. Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 7. Rubinsztein D. C., Mariño G., Kroemer G. (2011) Autophagy and aging. Cell 146, 682–695 [DOI] [PubMed] [Google Scholar]
- 8. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 9. Youle R. J., Narendra D. P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deretic V., Levine B. (2009) Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 12. Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 14. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manzanillo P. S., Ayres J. S., Watson R. O., Collins A. C., Souza G., Rae C. S., Schneider D. S., Nakamura K., Shiloh M. U., Cox J. S. (2013) The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huett A., Heath R. J., Begun J., Sassi S. O., Baxt L. A., Vyas J. M., Goldberg M. B., Xavier R. J. (2012) The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe 12, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 20. Thurston T. L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 21. Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayes J. D., McMahon M. (2009) NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 24. Jaramillo M. C., Zhang D. D. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 26. Ichimura Y., Waguri S., Sou Y. S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M. S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. (2013) Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 51, 618–631 [DOI] [PubMed] [Google Scholar]
- 27. Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. (2012) Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. U.S.A. 109, 13561–13566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bae S. H., Sung S. H., Oh S. Y., Lim J. M., Lee S. K., Park Y. N., Lee H. E., Kang D., Rhee S. G. (2013) Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 17, 73–84 [DOI] [PubMed] [Google Scholar]
- 29. Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N. (2011) Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 [DOI] [PubMed] [Google Scholar]
- 30. Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M. A., Dupont N., Ornatowski W., Jiang S., Bradfute S. B., Bruun J. A., Hansen T. E., Johansen T., Deretic V. (2012) TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bedford L., Hay D., Devoy A., Paine S., Powe D. G., Seth R., Gray T., Topham I., Fone K., Rezvani N., Mee M., Soane T., Layfield R., Sheppard P. W., Ebendal T., Usoskin D., Lowe J., Mayer R. J. (2008) Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 28, 8189–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Postic C., Shiota M., Niswender K. D., Jetton T. L., Chen Y., Moates J. M., Shelton K. D., Lindner J., Cherrington A. D., Magnuson M. A. (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 [DOI] [PubMed] [Google Scholar]
- 33. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 34. Harada H., Warabi E., Matsuki T., Yanagawa T., Okada K., Uwayama J., Ikeda A., Nakaso K., Kirii K., Noguchi N., Bukawa H., Siow R. C., Mann G. E., Shoda J., Ishii T., Sakurai T. (2013) Deficiency of p62/sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J. Neurosci. 33, 14767–14777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322 [DOI] [PubMed] [Google Scholar]
- 36. Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. (1999) Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 38. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 39. Waguri S., Komatsu M. (2009) Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 453, 181–196 [DOI] [PubMed] [Google Scholar]
- 40. Wang E. Y., Yeh S. H., Tsai T. F., Huang H. P., Jeng Y. M., Lin W. H., Chen W. C., Yeh K. H., Chen P. J., Chen D. S. (2011) Depletion of beta-catenin from mature hepatocytes of mice promotes expansion of hepatic progenitor cells and tumor development. Proc. Natl. Acad. Sci. U.S.A. 108, 18384–18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]