Background: Plasticity-related gene 5 (PRG5) is prominently expressed in neurons, but its neuronal function is unknown.
Results: PRG5 overexpression prematurely induces spine-like structures in immature hippocampal neurons, and PRG5 knockdown causes functionally relevant loss of excitatory synapses in dendrites of more mature neurons.
Conclusion: PRG5 expression is involved in proper spine formation.
Significance: We describe a new function of PRG5 in spinogenesis.
Keywords: Cell Biology, Cell Differentiation, Dendritic Spine, Membrane Protein, Phosphatidylinositol Phosphatase
Abstract
The transmembrane protein plasticity-related genes 3 and 5 (PRG3 and PRG5) increase filopodial formation in various cell lines, independently of Cdc42. However, information on the effects of PRG5 during neuronal development is sparse. Here, we present several lines of evidence for the involvement of PRG5 in the genesis and stabilization of dendritic spines. First, PRG5 was strongly expressed during mouse brain development from embryonic day 14 (E14), peaked around the time of birth, and remained stable at least until early adult stages (i.e. P30). Second, on a subcellular level, PRG5 expression shifted from an equal distribution along all neurites toward accumulation only along dendrites during hippocampal development in vitro. Third, overexpression of PRG5 in immature hippocampal neurons induced formation of spine-like structures ahead of time. Proper amino acid sequences in the extracellular domains (D1 to D3) of PRG5 were a prerequisite for trafficking and induction of spine-like structures, as shown by mutation analysis. Fourth, at stages when spines are present, knockdown of PRG5 reduced the number but not the length of protrusions. This was accompanied by a decrease in the number of excitatory synapses and, consequently, by a reduction of miniature excitatory postsynaptic current frequencies, although miniature excitatory postsynaptic current amplitudes remained similar. In turn, overexpressing PRG5 in mature neurons not only increased Homer-positive spine numbers but also augmented spine head diameters. Mechanistically, PRG5 interacts with phosphorylated phosphatidylinositols, phospholipids involved in dendritic spine formation by different lipid-protein assays. Taken together, our data propose that PRG5 promotes spine formation.
Introduction
Dendritic spines are postsynaptic components of most excitatory synapses. They are small actin-rich protrusions along dendrites and appear soon after dendritic processes are extended during neuronal differentiation. They are generally composed of a thin neck ending in a bulbous head (1–3). Particularly at the postnatal stage, when a multitude of synaptic connections is rapidly made, dendritic spines are highly dynamic (1, 3, 4), leading to changes in shape and motility essential for development and function of neuronal networks (5). Consequently, alterations in dendritic spine morphology and/or function are associated with pathological conditions, e.g. mental illnesses or age-related neurodegenerative diseases (6, 7).
Primary cultured hippocampal neurons are a well characterized and suited model for studying molecular mechanisms that regulate spine formation (8, 9). Spines in cultured hippocampal neurons form between 7 and 14 DIV.2 From DIV14 onward, most dendritic protrusions are spines (10). Based on their morphology, three different types of dendritic spines can be distinguished as follows: thin filopodium-like protrusions with a slender neck and a small head (“thin spines”), short spines without a well defined neck (“stubby spines”), and spines with a short neck and a large bulbous head (“mushroom spines”) (11, 12). Formation of dendritic filopodia/spines requires dramatic reorganization of planar regions of the plasma membrane to form a tightly curved membrane cylinder that sheaths the actin core (13). Almost nothing is known about the lipid composition of spine-like structures or about other types of filopodium/spine-localized integral membrane proteins that might play roles in membrane reorganization or in localization or tethering of the actin-based machinery responsible for filopodium/spine growth to the inner surface of the plasma membrane (14).
Recently, overexpression of PRG3 or PRG5 was found to increase the number of filopodia in different cell lines, as COS-7, HeLa, and N1E-115, using a mechanism that is independent on the canonical Cdc42-WASP-ARP2/3 pathway of filopodium protrusion (15, 16). PRGs are a vertebrate-specific class of, so far, five integral membrane proteins (also named lipid phosphatase-related proteins) and, in the case of PRG5, also referred as phosphatidic acid phosphatase type 2D (PAP2D)) (17–20). They are differentially expressed in the developing brain and regulated after brain injury (21, 22). Recently, we showed that PRG3 promotes neurite shaft protrusions and thereby regulates filopodial formation in immature primary neurons (23).
This study was designed to transfer the findings of PRG5 actions to a neuronal, physiologically relevant developmental context and to shed light on the functional importance of endogenous PRG5. Here, we showed that overexpression of mouse PRG5 prematurely induced spine-like structures on cultured young hippocampal neurons, whereas point mutations within the extracellular loops of PRG5 abolished this effect. Overexpression of PRG5 in more mature primary neurons increased not only the numbers of Homer-positive spines but also the diameters of the spine head significantly. By contrast, down-regulation of the endogenous expression level of PRG5 in more mature cultured hippocampal neurons reduced the density of excitatory synapses and, consequently, the frequency of mEPSCs.
EXPERIMENTAL PROCEDURES
Animals
Pregnant, postnatal, and adult C57BL/6 mice obtained from our central animal facility were kept under standard laboratory conditions (12-h light/dark cycle; 55 ± 15% humidity; 24 ± 2 °C room temperature, and water ad libitum, enriched and grouped), in accordance with German and European guidelines (2010/63/EU) for the use of laboratory animals. Approval of experiments was obtained from the local ethics body of Berlin (LAGeSO, T0108/11).
Construct
Database searches were performed using BLAST on the website of the National Centre for Biotechnology Information. Multiple sequence alignments (GenBankTM accession number ACJ60628, human PRG5; AAS80161, mouse PRG5; NP_001101190, rat PRG5) were performed using DNAsis Max Version 2.0 software (Hitachi, Olivet Cedex, France). The transmembrane structure was predicted with ProDom and SwissProt databases. Complementary DNA constructs of PRG5, containing a C-terminal FLAG or eGFP, were used to transiently transfect HEK-293 cells and primary neurons. For Western blot analyses, the following expression plasmids were used: mPRG1-eGFP, rPRG2-eGFP, mPRG3-eGFP, and mPRG4-eGFP. Complementary DNA from mouse brain was amplified by PCR using the primers: 5′-GAA TTC ATG CCC CTG CTG CCC GTG GCG CTC ATC AGC-3′ and 5′-GCG GCC GCT CAG TCG TCA TCG TCT TTG TAG TCT GTG ACT TCC GCA AAG GCA GTG ACG TGG TTC TGC AG-3′ containing the FLAG sequence. Oligonucleotides were synthesized by Metabion (Martinsried, Germany). PCR products were purified by spin columns (Qiagen, Hilden, Germany) and cloned in pCDNA3.1(+)zeo expression vector (Invitrogen). The PRG5-Δ C-terminal FLAG construct was generated by PCR from the full-length PRG5-FLAG construct without 37 amino acids at the C terminus. Site-directed mutants of mouse PRG5 were generated using the QuikChange II site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). The following forward primers were used to generate PRG5 S193W, PRG5 E195G, PRG5 E195H, and PRG5 R241E mutants, respectively: 5′-CGG AAA ACA TTC CCA TGG AAG GAA GCT GCC CTG-3′, 5′-ACA TTC CCA TCC AAG GGC GCT GCCC TGA GTG TC-3′, 5′-ACA TTC CCA TCC AAG CAC GCT GCC CTG AGT GTC-3′, 5′-CTT ACT GGA CTC AAC GAG GTA GCG GAA TAT CGA-3′, and the reverse primers 5′-CAG GGC AGC TTC CTT CCA TGG GAA TGT TTT CCG-3′, 5′-GAC ACT CAG GGC AGC GCC CTT GGA TGG GAA TGT-3′, 5′-GAC ACT CAG GGC AGC GTG CTT GGA TGG GAA TGT-3′, and 5′-TCG ATA TTC CGC TAC CTC GTT GAG TCC AGT AAG-3′ (mutated amino acids underlined). Expression plasmids for Western blot analyses were mPRG1-eGFP, rPRG2-eGFP, mPRG3-eGFP, mPRG4-eGFP, and rPRG5-eGFP.
In silico analysis of rat and mouse PRG5 revealed a putative consensus N-glycosylation site at amino acid 158 by using NetNGlyc 1.0. Point mutation of amino acid 158 of PRG5 from asparagine to glutamine within the expression plasmid PRG5-FLAG was performed with the QuikChange II site-directed mutagenesis kit (Stratagene) using the following primers: forward primer 5′-TG GCT TTG TGT AAG CCA CAG TAC ACA GCG CTC GGA TG-3′ and the reverse primer 5′-CA TCC GAG CGC TGT GTA CTG TGG CTT ACA CAA AGC CA-3′ (mutated amino acid underlined) (Metabion) as recommended by the manufacturer.
To suppress expression of endogenous PRG5, short hairpin RNAs (shRNA) specifically against mouse PRG5 were stably expressed. pSuper GFP vector (OligoEngine, Seattle) harboring shRNAs inserts was used (15). The shRNA target sequence against firefly luciferase served as control (23). For the PRG5 shRNA rescue plasmid, four silent mutations were introduced into the region targeting the shPRG5. Site-directed mutagenesis was carried out with the QuikChange II site-directed mutagenesis kit (Stratagene). Forward primer 5′-TA AAC CCA CTG GTA CGC CGA ACT GTA CGC TTC CTT GGA ATT TAT GCA TTT GGA C-3′ and the reverse primer 5′-G TCC AAA TGC ATA AAT TCC AAG GAA GCG TAC AGT TCG GCG TAC CAG TGG GTT TA-3′ (mutated amino acids underlined) (Metabion) were chosen. Sequences of all cDNAs were confirmed by sequencing. Verification of the shRNA-PRG5 construct was done in HEK-293 cells co-transfected with shRNA-PRG5 and PRG5-FLAG or the PRG5 rescue plasmid. The knockdown was analyzed by Western blotting using anti-FLAG antibody.
RNA Extraction and cDNA Synthesis
Organs were dissected from C57BL6/N mice and homogenized in TRIzol reagent (Invitrogen). Primary neurons, microglial cells, or astrocytes were scraped in 1× PBS and centrifuged for 5 min at 300 × g at 4 °C. Cell pellets were dissolved in 1 ml of TRIzol reagent. Total RNA was isolated according to the TRIzol protocol (Invitrogen). Following precipitation and drying, RNA was resuspended in an aliquot of RNase and DNase-free water quantified by A260 nm spectrophotometry (Biomate 3 spectrometer, Fisher) and stored at −80 °C. cDNA was synthesized with 5 μg of total RNA using the high capacity cDNA archive kit (Invitrogen) according to the manufacturer's protocol. As control, reaction was performed without MultiScribe reverse transcriptase. cDNA was diluted 1:5 with RNase- and DNase-free water and stored at −20 °C. The quality of amplified cDNA was controlled using β-actin PCR.
Quantitative Real Time PCR
Each PCR contained: 8 μl of H2O, 10 μl of TaqMan® Universal PCR Master Mix (Invitrogen), 1 μl of cDNA, and 1 μl of TaqMan gene expression assays for PRG5 (assay ID Mm01310525_m1), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; assay ID 4352932E), and β-actin (assay ID 4352933E). For hypoxanthine phosphoribosyltransferase, separate primer and probe were used (Primer Mix (forward 5′-ATC ATT ATG CCG AGG ATT TGG AA-3′; reverse 5′-TTG AGC ACA CAG AGG GCC A-3′) and probe (5′-TGG ACA GGA CTG AAA GAC TTG CTC GAG ATG-3′)). The reactions were run on the ABI PRISMTM 7700 sequence detection system (Invitrogen). Standard curves were produced with serial dilutions of cDNA from mouse brain at postnatal day 5 (P5) with amplification efficiency between 90 and 100%. Graphic and statistical analyses were performed using 7500 Fast System Software (Invitrogen). Each value is the average of at least three separate experiments. Identity and purity of primary cells were confirmed by quantitative real time PCR with neuron-specific class III β-tubulin (tuj1) as neuronal marker, glial fibrillary acidic protein as astrocyte marker, and ionized calcium-binding adaptor molecule 1 (Iba1) as microglia marker.
Hippocampal Primary Neurons, Preparation, and Transfection
Hippocampal primary neurons were isolated from C57BL6/N mouse embryos at embryonic day 18 (E18). Hippocampi from several embryos were collected and washed twice in ice-cold Hanks' buffered salt solution (Invitrogen). The tissue was incubated in 4 ml of Hanks' buffered salt solution and 400 μl of trypsin (Invitrogen) for 15 min at 37 °C, resuspended in plating medium (minimum essential medium) supplemented with 10% horse serum (Invitrogen), 0.6% glucose, 100 units/ml penicillin and 100 μg/ml streptomycin. After dissociation, neurons for immunocytochemical experiments were plated at a density of 100,000 cells/cm2 onto poly-l-lysine (Sigma)-coated coverslips into 12-well plates, and neurons for Western blot experiments were plated into 12-cm diameter dishes. Three hours after plating, cells were washed twice with phosphate-buffered saline (1× PBS) and incubated in Neurobasal A medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 0.5 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin at 37 °C, and 5% CO2. Primary cells were routinely maintained at 37 °C with 5% CO2 in Neurobasal A medium (Invitrogen) supplemented as described above. For transient transfection, Effectene (Qiagen) was used at DIV1 or DIV12–13, respectively. For electrophysiological recordings, primary neurons from the 1st day were cultured in Neurobasal A medium (Invitrogen) supplemented as described above, but penicillin and streptomycin were omitted.
Primary Mouse Astrocyte and Microglia Cultures
Astrocytes and microglia were prepared as described previously (24). Briefly, mouse cortex (P1–3) was isolated and carefully homogenized. Digestion was performed by 10 min of trypsin incubation. Dissociated glial cells were plated on poly-l-lysine-coated dishes and incubated at 37 °C with 5% CO2. After 2 days, the medium was refreshed, and the cells had detached from the astrocytes. The cells were shaken for 2 h at 37 °C, and the remaining free-floating microglia cells were removed.
HEK-293 Cells, Preparation, and Transfection
HEK-293 cells were routinely maintained at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin, in dishes at a cell density of 10,000–20,000 cells/cm2 and transfected the next day with calcium phosphate precipitation; 4 μg of DNA, 67.5 μl of sterile water, 7.5 μl of 2 m calcium chloride, and 67.5 μl of 2× HEPES-buffered saline were mixed and added to the dishes. One to 2 days after transfection, cells were used for further analyses.
Preparation of Total Protein Lysates and Recombinant Proteins
HEK-293 cells were washed with ice-cold PBS, lysed in a detergent-containing lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 25 mm MgCl2, 10% glycerol) containing protease inhibitors (Sigma), and sonicated three times for 3 s. Subsequently, the lysates were centrifuged for 10 min at 14,000 × g at 4 °C. Protein concentration of the supernatant was determined by a BCA protein assay kit (Thermo Scientific, Rockford, IL). To obtain recombinant protein, PRG1-eGFP, PRG2-eGFP, PRG3-eGFP, PRG4-eGFP, and PRG5-eGFP were purified from transfected HEK-293 cells using the μMACS epitope-tagged protein isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany), as recommended by the manufacturer.
Immunocytochemistry
Primary neurons were fixed 24 h after transfection in 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 5 min, washed three times with 1× PBS for 10 min, and incubated overnight at 4 °C with block solution, containing 10% FCS in 1× PBS. Neurons were incubated with first antibodies for 2–4 h at room temperature and then washed three times with 1× PBS for 10 min. Antibodies were used at the following dilutions: anti-Homer (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) 1:500; anti-PAP2D (Abcam, Cambridge, MA) 1:250; anti-MAP2 (Sigma) 1:1000; anti-FLAG (Sigma) 1:500; anti-α-tubulin (Synaptic System, Göttingen, Germany) 1:1500; anti-VGlut1 (Synaptic Systems) 1:500; anti-GluR2 (Synaptic Systems) 1:450; anti-JL-8 (Sigma) 1:1000; anti-Tau1 (Millipore, Schwalbach, Germany) 1:200. The second antibodies, Alexa Fluor 633, Alexa Fluor 568, Alexa Fluor 488, Alexa Rat 633, or Cy3 550, were then applied for 1–2 h at room temperature. Phalloidin-TRITC (Sigma) was used to visualize F-actin. Glasses were mounted in FluorSaveTM reagent.
Morphological Analysis
Primary neurons were transfected at DIV1 or after DIV13 with shRNA-PRG5, shRNA-luciferase, PRG5-FLAG, and eGFP or double-transfected with shRNA-PRG5 + PRG5 shRNA rescue plasmid, PRG5-FLAG or PRG3-FLAG + eGFP empty vector using Effectene (Qiagen), and analyzed 1–3 days later. For morphological analysis, confocal images of primary neurons were acquired with an upright laser microscope (Leica DM 2500) equipped with a ×63 objective (oil immersion, 1.4 NA) and a ×40 objective (oil immersion, 1.25 NA) using the 488 nm line of an argon-ion laser and the 543- and 633-nm line of a helium-neon laser. Background correction and adjustment of brightness and contrast were performed using Leica confocal software (Leica Microsystems, Wetzlar, Germany).
For co-localization studies, z-stacks of both fluorophores were sequentially acquired and average projected. For immunofluorescence quantification, the cellSens Dimension Desktop (Version 1.4.1 XV 3.4, Build 8624) software from Olympus was used. The fluorescence intensity (Fig. 4) of the endogenous PRG5 was measured in a region of interest (4–5 μm2) as maximum pixel fluorescence intensity drawn over three distinct MAP2-positive or Tau1-positive areas. All measurements were performed using three different immunostainings. Data analysis was performed using GraphPad Prism 5 (GraphPad Software Inc, La Jolla, CA) from n = 12 for MAP2 und n = 69 for Tau1-positive structures. Data are reported as means ± S.E.
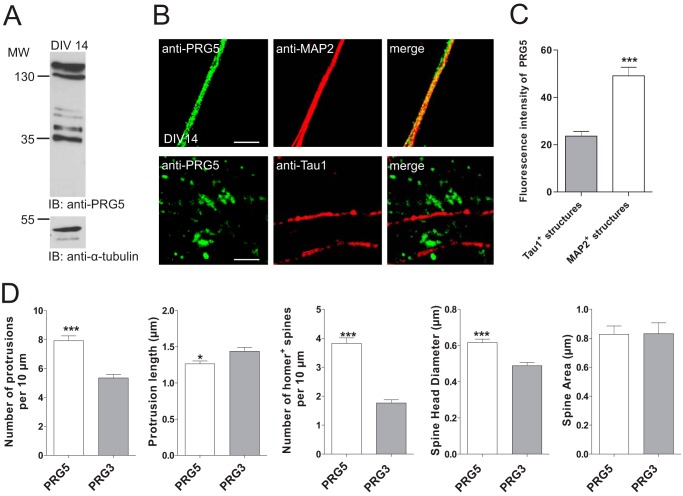
FIGURE 4.
PRG5 contributes to regulation of spine density and morphology in mature hippocampal neurons. A, Western blot analysis of membrane protein lysates from primary neurons at DIV14 shows endogenous membrane expression of PRG5. As loading control α-tubulin was used. IB, immunoblot. B, confocal images of dendritic and axonal structures from hippocampal neurons (DIV14) stained with anti-PRG5 (green) and anti-MAP2 (red, dendrites) or anti-Tau1 (red, axons) revealed a clear co-localization of PRG5 and MAP2 compared with Tau1. Scale bars, 10 μm. C, analyses of the PRG5 abundance on MAP2- and Tau1-positive structures measured by fluorescence signal intensity. Fluorescence intensity of PRG5 in MAP2-positive structures (n = 12) is significantly higher compared with the one in Tau1-positive structures (n = 69) (p ≤0.0001). D, morphology analyses in mature primary neurons transfected either with PRG5-FLAG or PRG3-FLAG 1 to 2 days after transfection. Determination of the numbers of protrusions, number of Homer-positive spines, and the spine head diameter revealed a significant increase compared with PRG3 control. *, 0.01 ≤ p ≥ 0.05, and ***, 0.001 ≤ p ≥ 0.01.
The fluorescence of PRG5-FLAG intensity was calculated as mean pixel fluorescence intensity drawn over three distinct areas to quantify the knockdown efficiency in neurons (DIV4 or DIV14 at Figs. 2 and 5, respectively). All measurements were performed using three independent immunostainings. Data analysis was performed using GraphPad Prism 5 (GraphPad Software Inc., Prism5 Version 5.02) from DIV4 neurons, n = 93 for PRG5 shRNA and n = 111 for control shRNA, and from DIV14 neurons, n = 201 for PRG5 shRNA and n = 123 for control shRNA transfected neurons. Data are reported as mean ± S.E.
FIGURE 2.
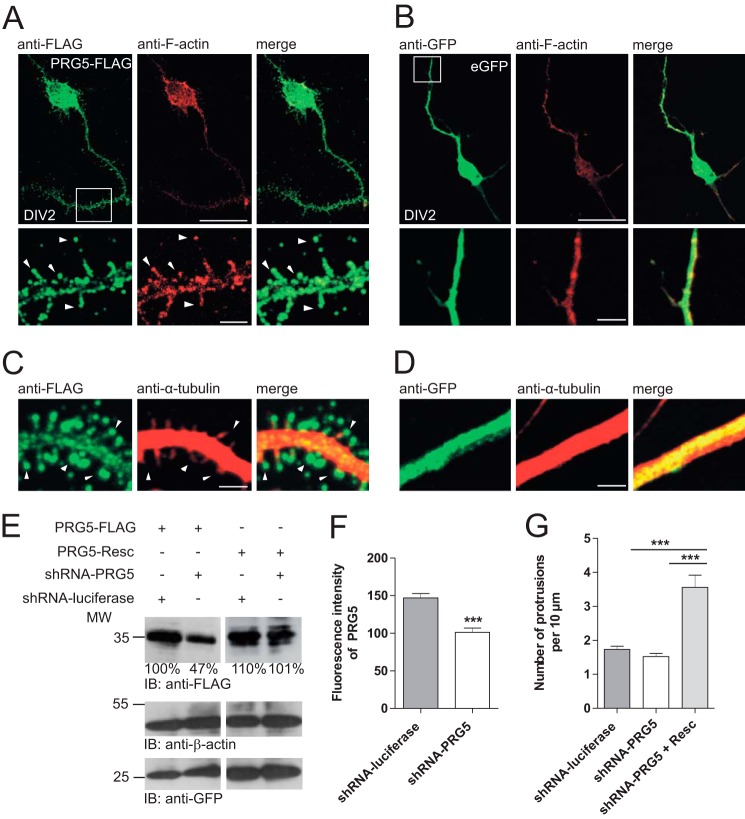
PRG5 induces spine-like structures in immature hippocampal neurons. A, hippocampal primary neurons transfected with PRG5-FLAG (left, green) and co-immunostained with anti-F-actin (middle, red). Magnification (white frame, lower images) shows that PRG5 overexpression induces spine-like structures along neurites. Arrowheads indicate PRG5 expression in the bulbous head. B, eGFP overexpressing hippocampal neurons have the typical morphology of immature neurons. There is no spine formation along the neurites. C and D, magnification of neurites from neurons transfected with PRG5-FLAG or eGFP and co-stained with anti-FLAG (green) and anti-α-tubulin (red). White arrowheads indicate heads of different spine-like structures induced by PRG5 overexpression in comparison with eGFP-transfected cells (C). Scale bars for the upper images in A and B represent 10 μm and for the magnified sections (lower images) in A and B, and 4 μm for C and D, respectively. E, verification of PRG5 shRNA and shRNA-resistant PRG5 construct by Western blot. PRG5-FLAG was reduced when compared with controls or with PRG5 shRNA-resistant protein levels. Ratios below the Western blot bands refer to densitometry measurements of the gray intensity of these bands in relation to the control band. Anti-β-actin was used as loading control and for normalization. Anti-GFP shows transfection efficiency. IB, immunoblot. F, quantification of shRNA-PRG5 efficiency. The shRNA-PRG5 (n = 94) expression reduced endogenous PRG5 expression more than one-third when compared with control (n = 112). G, PRG5 knockdown did not affect the numbers of protrusions/filopodia in immature neurons. Primary neurons were transfected with shRNA-PRG5 or controls at DIV1 and morphologically analyzed at DIV2. Numbers of protrusions/filopodia per 10 μm in immature neurons were counted. When co-transfected with PRG5, rescue plasmid (Resc.) protrusion/filopodia drastically increased. The mutations are silent so that the rescue plasmid is comparable with PRG5 overexpression. ***, p ≤ 0.0001.
FIGURE 5.
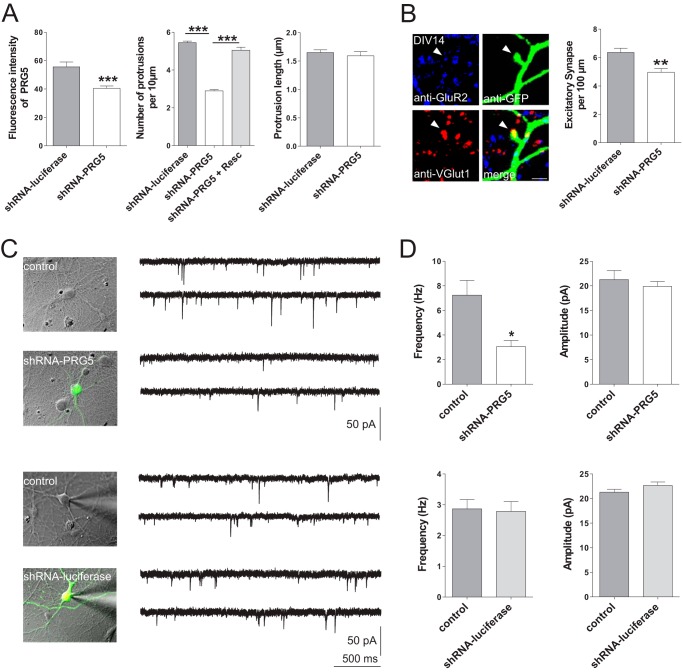
Alteration of PRG5 expression reduces functional synapses in mature hippocampal neurons. A, efficiency of shRNA-PRG5 in mature neurons. The shRNA-PRG5 (n = 201) expression reduced the endogenous PRG5 expression more than one-third when compared with control (n = 123). Such PRG5 knockdown in mature primary neurons reduced the numbers of protrusions (shRNA-PRG5 n = 274 versus shRNA-luciferase n = 205) but not their length (shRNA-PRG5 n = 90 versus shRNA-luciferase n = 102). B, quantitative analysis of excitatory synapses. Left, structures expressing anti-GFP, anti-VGlut1, as marker for presynaptic structures, and anti-GluR2, as marker for postsynaptic structures in close proximity, are regarded as synapses. Scale bar represents 2 μm. Right, population data indicate that down-regulating PRG5 reduces the number of excitatory synapses, compared with control (shRNA-luciferase). C, representative example of nontransfected (control) and transfected (shRNA-PRG5, upper row) or nontransfected (control) and transfected (shRNA-luciferase, lower row) primary cultured hippocampal neurons (DIV14–16). Current traces were recorded somatically under whole-cell patch clamp conditions at −60 mV holding potential. Negative deflections represent electrically and pharmacologically isolated mEPSCs. Note that shRNA-PRG5 and shRNA-luciferase were transfected in different culture dishes and that control represents recordings in respective culture dishes, suggesting that differences in control frequencies are due to culture specific variations. D, population data of frequencies and amplitudes of mEPSCs without (control, gray columns, n = 24) or with PRG5 silencing (shRNA-PRG5, white columns, n = 18, upper row) and from untransfected (control, gray columns, n = 19) primary neurons and shRNA-luciferase transfected primary neurons (shRNA-luciferase, white columns, n = 25, lower row). *, 0.01 ≤ p ≥ 0.05; **, 0.001 ≤ p ≥ 0.01; and ***, p ≤ 0.0001.
The number of protrusions or of excitatory synapses were counted using Leica confocal software (Leica Microsystems). For analyses of protrusion density, all protrusions per 10 μm of neurite (regions randomly chosen) were counted. Protrusions positive to Homer were classified as spines. Protrusion length, spine head diameter, and spine area were measured as described previously (25). Excitatory synapses in hippocampal neurons were stained with antibodies specific for synaptic proteins (GluR2, postsynaptic; VGlut1, presynaptic). Punctate staining was detected along GFP-positive neurons, and only puncta with obvious GluR2/VGlut1 overlap were counted as synapses. A potential bias during area selection was excluded by constructing a 100-μm circle around the cell body and counting all synapses in this area. All measurements were performed in four to six different transfected cultures.
Data analysis was performed using GraphPad Prism 5. Data are reported and presented as mean ± S.E. Significance was assessed using the two-tailed Student's t test for unpaired data at the given significance level (p) where ***, 0.0001 < p; **, 0.001 < p < 0.01; *, 0.01 < p < 0.05.
Protein Isolation and Western Blot Analysis
Mice were sacrificed and brains were removed and homogenized with an ULTRA-TURRAX homogenizer in STM buffer: 250 mm sucrose, 10 mm Tris, 1 mm MgCl2, with 1% Triton (v/v), and a protease inhibitor mixture (Sigma). Protein concentrations were determined by BCA protein assay (Thermo Fisher Scientific Inc.) using BSA as standard. Samples were separated by 10–12% SDS-PAGE, and proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare). The membrane was incubated with primary antibody anti-PAP2D (Abcam) 1:500, 1:1000 dilution of anti-JL-8 (Clontech), or anti-FLAG antibody (Sigma) 1:1000, β-actin (Sigma) 1:10,000, α-tubulin (Sigma) 1:1000 followed by horseradish peroxidase-conjugated secondary antibody. The immunoreactive bands were detected with SuperSignal West Pico chemiluminescent substrate (ECL) (GE Healthcare). Subcellular protein fractions (cytosol and membrane) were prepared using the kit PromoKine cell fractionation kit protocol (PromoKine, Heidelberg, Germany).
Deglycosylation Assay
For detection of potential N-glycosylation, PRG5 total protein lysates from transfected HEK-293 cells were treated with peptide:N-glycosidase F (PNGase F, Roche Applied Science) as recommended by the manufacturer. Controls were treated using the same process but without PNGase F.
Electrophysiological Recordings
Primary cultured hippocampal pyramidal neurons were recorded 48–96 h after transfection, between DIV14 and 16. mEPSCs were acquired at 32 °C in somatic whole-cell patch clamp technique using an EPC-10 amplifier (HEKA, Lambrecht/Pfalz, Germany). All recordings were performed at a holding potential of −60 mV in extracellular solution containing (in mm) the following: 124 NaCl, 4 KCl, 3 CaCl2, 2 MgCl2, 25 HEPES, 10 glucose, and pH was adjusted with NaOH to 7.3. Addition of 1 μm tetradoxin (R&D Systems, Inc., Minneapolis, MN) and 10 μm bicuculline (R&D Systems, Inc.) pharmacologically isolated mEPSCs. Recording electrodes were pulled with a micropipette puller P-97 (Sutter Instruments, Novato, CA) and had a resistance of 2.5–4 megohms when filled with a solution containing (in mm) the following: 120 potassium gluconate, 10 KCl, 10 sodium phosphocreatine, 1 MgCl2, 1 CaCl2, 11 EGTA, 10 HEPES, 2 Mg2+-ATP and 0.3 GTP (osmolarity 290 mosmol, pH adjusted to 7.2 with KOH). Signals were sampled at a rate of 6.25 kHz using Patchmaster2.3 software (HEKA Electronics). For analyses of mEPSCs, MiniAnalysis Version 6.0.9 (Synaptosoft, Decatur, GA) was used. Statistics were performed using Origin7 (OriginLab, Northampton, MA); two-tailed Student's t tests were used in the case of normal distributed data (Shapiro-Wilk test) and nonparametric Mann-Whitney U tests for not normal distributed data. All data are presented as mean ± S.E.
Lipid Binding Assays
Nitrocellulose membranes spotted with a variety of lipids were used from Echelon Biosciences (PIP StripsTM and PIP ArraysTM), according to the manufacturer's protocol. The PIP strip and PIP array were incubated overnight at 4 °C with total protein lysates of HEK 293 cells either overexpressing PRG5-FLAG or PRG5-Δ C-terminal FLAG. Bound protein was detected using the anti-FLAG antibody and visualized by ECL.
PIP beads (Echelon) were washed twice with 5 volumes of wash/binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 1% BSA) for 1 h. Control beads without cross-linked lipid were prepared in an identical manner. Protein lysate overexpressing PRG5-FLAG was incubated with the beads rotating overnight at 4 °C. The beads were washed five times with 8 volumes of wash/binding buffer by pelleting them by centrifugation at 1000 rpm at 4 °C for 1 min, and the bound proteins were eluted by boiling the beads in 100 μl of Laemmli sample buffer. The proteins were then separated on a 12% SDS-polyacrylamide gel. PRG5-FLAG protein was detected using anti-FLAG antibody by Western blot.
RESULTS
PRG5 Is Brain-enriched and Prominently Expressed in Neurons
Previous work revealed that PRG5 is highly expressed in adult mouse and human brain tissue (15, 20). Here, we comparatively analyzed mouse PRG5 mRNA expression in various organs at P5 by qRT-PCR (Fig. 1A). Levels of PRG5 mRNA were highest in brain and at least 10 times lower in heart, lung, and testis. We did not detect any PRG5 mRNA expression in thymus, liver, spleen, and kidney. Because PRG5 was previously shown to be strongly expressed in the hippocampal region (15), we evaluated PRG5 mRNA expression during hippocampal development (Fig. 1B). PRG5 mRNA present as early as embryonic day 14 (E14) peaked around the time of birth and remained on relatively high levels thereafter, at least until the early adult stage (P30) (Fig. 1B). To further specify its cellular abundance, we analyzed PRG5 mRNA expression in primary cultured neurons, astrocytes, and microglia. Primary cultured neurons showed highest PRG5 mRNA levels, whereas the signal was below detection threshold in astrocytes and microglia cells (Fig. 1C).
FIGURE 1.
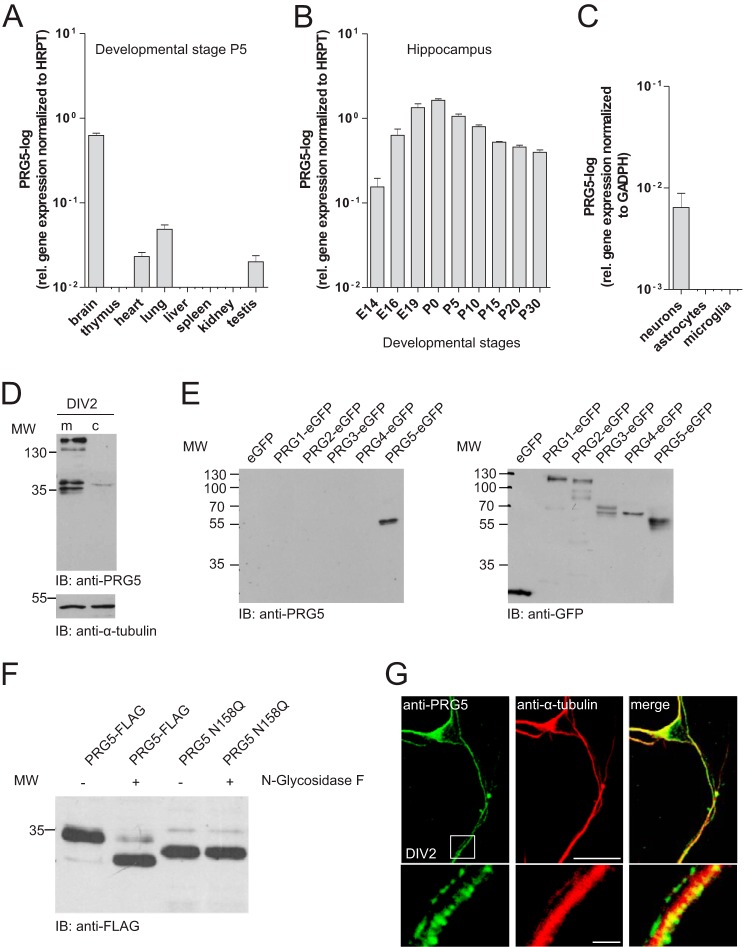
PRG5 is mainly expressed in the brain. A, qRT-PCR of different mouse tissues (harvested at postnatal day 5 (P5)) revealed the most prominent PRG5 mRNA expression in the brain. Data were normalized to hypoxanthine phosphoribosyltransferase (HPRT). B, age-dependent changes in PRG5 expression levels revealed by qRT-PCR of mouse hippocampal tissue between E14 and P30. The highest PRG5 mRNA expression was detected around birth. Data were normalized to hypoxanthine phosphoribosyltransferase. C, PRG5 mRNA expression in neurons, astrocytes, and microglia normalized to GAPDH. qRT-PCR analysis reveals a robust neuronal expression of PRG5. D, Western blot analysis of membrane (m) and cytosolic (c) protein fractions demonstrate endogenous membrane expression of PRG5 in immature primary hippocampal neurons (DIV2). Anti-α-tubulin served as loading control. IB, immunoblot. E, specificity analysis of the anti-PRG5 antibody revealed no cross-reactivity to other PRG family members. Fusion proteins of several PRG family members with eGFP (PRG1, PRG2, PRG3, PRG4, and PRG5) were used in the Western blot analyses. The anti-PRG5 antibody detected protein exclusively in the PRG5 lane. F, Western blot of HEK-293 cells after transfection with two different PRG5 constructs (PRG5-FLAG and PRG5 N158Q). Native lysates (1st and 3rd lanes) or lysates treated with N-glycosidase F (2nd and 4th lanes) were probed with an anti-FLAG antibody. When WT PRG5 protein lysate treated with N-glycosidase F, a band is detected at a lower molecular weight compared with untreated lysate (1st and 2nd lanes). This points out that PRG5 is N-glycosylated. Mutation of asparagine 158 to glutamine (3rd and 4th lanes) shifts the PRG5 band to a lower molecular weight with and without treatment. Note that the mutant PRG5 protein band is not at the same height than the WT PRG5-treated band. G, confocal images of immature neurons endogenously expressing PRG5 (green) and anti-α-tubulin (red). Magnification of structures within the white frame shows a strong dotted PRG5 signal along the neurites. Scale bars for the upper images represent 10 μm and for the magnified sections (lower images) 4 μm, respectively.
In line with these data, Western blot and immunocytochemical analysis revealed neuronal presence of PRG5 protein. In detail, Western blot analysis confirmed endogenous expression in immature primary neurons (Fig. 1D). We found two double bands, at about 35 and 130 kDa, respectively, in native membrane protein lysates, whereas the cytosolic fraction almost showed a signal (Fig. 1D). The PRG5 antibody we used did not cross-react with other PRG family members (Fig. 1E). Here, the recombinant PRG5-eGFP showed only the double band at the lower molecular level. The existence of double bands in the immunoblot suggested glycosylation of the PRG5 protein. Indeed, hydrolyzation of N-linked glycan chains from PRG5-FLAG fusion protein with peptide:N-glycosidase F (PNGase F) resulted in a prominent band with lower molecular weight (Fig. 1F). Furthermore, we identified a putative consensus N-glycosylation site at amino acids 158 within the second extracellular loop via in silico analysis of rat and mouse PRG5. To abolish N-glycosylation we point-mutated PRG5 (N158Q) by substituting asparagine for glutamine (26). This N158Q mutation in its native form showed a prominent band at a lower molecular weight than the wild type native PRG5 protein band. PNGase F treatment caused no further alteration compared with the N158Q mutation in its native form. This result confirms an N-glycosylation at amino acids 158 in PRG5 protein, but it also indicates that the PRG5 protein may undergo more than one post-translational modification. We further investigated the subcellular distribution of PRG5 in immature primary neurons. After 2 days in culture, hippocampal neurons exhibited several undifferentiated α-tubulin-containing processes of similar length (Fig. 1G). During this early phase of neuronal differentiation, PRG5 appeared in the cell body and along all neurites as a clear and strong dotted signal (Fig. 1G). Taken together, these data indicate the expression of PRG5 in the membranes of early postnatal hippocampal neurons and suggest an involvement in developmental regulation.
PRG5 Prematurely Induces Spine-like Structures in Immature Hippocampal Neurons
Given the prominent neuronal membrane expression of endogenous PRG5 and its previously described role in filopodial formation (15), we hypothesized a contribution of PRG5 during neuronal differentiation of hippocampal neurons. To test for an early influence, we overexpressed PRG5 or reduced the PRG5 level by shRNA in primary neurons starting from DIV1. Overexpression of PRG5 led to a marked increase in that protein all over the neurons (Fig. 2A), i.e. on the soma and in both proximal and distal regions of neurites at DIV2. As in the native neurons (Fig. 1G), PRG5 protein appeared to be enriched in the plasma membrane. More interesting, PRG5 overexpression led to the appearance of spine-like structures all along the neurite plasma membrane. Most of them resemble mushroom, stubby, or thin spine shapes (Fig. 2, A and C, higher magnification, white arrowheads). Such spine-like structures are usually not present at this stage as corroborated by its lack in nontransfected neurons (Fig. 1G). The transfection procedure itself was unlikely to cause the appearance of spine-like structures because overexpressing only eGFP did not alter the morphology of neurites (Fig. 2, B and D, higher magnification). Most of the PRG5-induced premature protrusions ended in a large bulbous head, containing polymerized F-actin (Fig. 2A, higher magnification), although α-tubulin structures remained restricted to main neurite shafts (Fig. 2C). This indicates that the structures might become spines rather than branching points. Moreover, PRG5 showed a prominent presence on the head of each single protrusion (Fig. 2, A and C).
To investigate the effect of endogenous PRG5, we first confirmed the PRG5 shRNA efficiency by its ability to reduce overexpressed PRG5 and specificity by the lack of effect on PRG5 shRNA-resistant construct in HEK-293 cells (Fig. 2E). In primary hippocampal neurons, PRG5 shRNA reduced endogenous PRG5 level by one-third as estimated by immunocytochemistry (shRNA-luciferase 146.9 ± 5.7, n = 111; shRNA-PRG5 101.3 ± 5.6, n = 93, p < 0.0001, Fig. 2F). Despite this reduction, the morphology of the neurons was similar. In particular, the numbers of protrusions, which are predecessors of dendritic spines (shRNA-luciferase: 1.7 ± 0.1, n = 104; shRNA-PRG5: 1.5 ± 0.1 n = 100, p < 0.166, Fig. 2G), was comparable. Additional overexpression of the PRG5-shRNA-resistant construct drastically increased protrusion numbers (shRNA-PRG5 + rescue plasmid: 3.6 ± 0.4 n = 74, p < 0.0001, Fig. 2G).
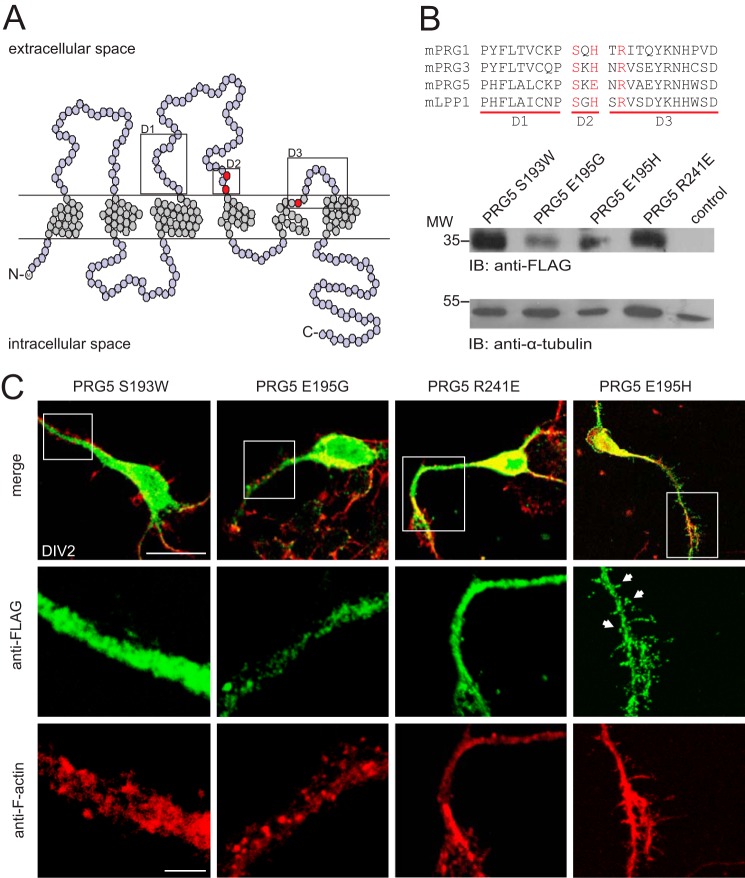
Residues within the Extracellular Loops Are Essential for PRG5-mediated Induction of Spine-like Structures in Immature Neurons
As a member of the PRG family, PRG5 is characterized by six membrane-spanning domains with three extracellular loops containing highly conserved enzymatically active domains (Fig. 3A). Because these extracellular loops were important for the PRG3 induction of filopodia (16), we here generated a series of mutant constructs, exchanging amino acids in the second and third extracellular loop (Fig. 3A, red dots). In detail, we replaced the conserved amino acids PRG5 Ser-193 and Arg-241 with Trp-193 and Glu-241, respectively. The nonconserved PRG5 Glu-195 we exchanged with Gly-195 or with His-195 to reproduce the PRG3 sequence in the D2 domain of PRG5. All PRG5 mutants were sufficiently expressed as shown by immunoblots of total protein lysate from the PRG5 mutant expressing HEK-293 cells (Fig. 3B).
FIGURE 3.
Residues within the extracellular loops are essential for PRG5-mediated spine-like structure. A, structure model based on the amino acid sequence of human PRG5. PRG5, like the LPPs, contains six putative transmembrane regions and intracellularly located C and N termini. The three mutated amino acids Ser-193, Glu-195, and Arg-241 are marked in red. Boxes are positioned at D1, D2, and D3 regions, respectively. B, alignment of the domain sequences from mouse PRGs with mouse LPP-1 (mPRG1 (Q7TMEO), mPRG3 (NP_848871), mPRG5 (AAS80161), and mLPP-1 (NP_032273)). In red are residues of conserved amino acids that have been shown to be important for the PRG3 induction of filopodia. Western blot analysis is shown of protein lysates from HEK-293 cells transfected with PRG5-FLAG mutants or empty vector control and detected with an anti-FLAG. α-Tubulin served as loading control. C, immature primary neurons (DIV2) transfected with mutants S193W, E195G, R241E, or E195H and co-stained with anti-FLAG and anti-F-actin. Neurons, overexpressing PRG5 S193W, R241E, or E195G, did not show any protrusions at the plasma membrane. On the contrary, overexpression of PRG5-FLAG E195H induced a considerable rearrangement of the plasma membrane, with a high number of spine-like structures. White arrowheads point to the spine-like structures. Scale bars represent 10 or 3 μm (higher magnification images), respectively.
Immature hippocampal neurons overexpressing PRG5 S193W, R241E, or E195G did not show any protrusions at the plasma membrane; in particular, swollen or spine-like structures were absent (Fig. 3C). Moreover, these mutant constructs displayed a dispersed fluorescence signal in membrane and cytosolic regions of neurites and soma, indicating that the majority of the mutated protein remains intracellular (Fig. 3C).
In contrast, overexpression of PRG5 E195H in immature primary neurons induced a considerable rearrangement of the plasma membrane, with a high number of spine-like structures along the soma and the neuronal processes (Fig. 3C), thus mimicking the situation after overexpression of nonmutated PRG5 (Fig. 2, A and C). Again, each protrusion was rich in F-actin, and a clear fluorescent signal from the induced mutant protein was distinguishable, especially at the bulbous head-like structure (Fig. 3C). This confirms the causal connection of PRG5 and spine-like structure formation in young primary neurons and suggests that membrane insertion of PRG5 is a prerequisite of spine-like structure formation.
In More Mature Hippocampal Neurons Endogenous PRG5 Accumulates in Dendrites, and PRG5 Overexpression Alters Dendritic Spine Formation
Because synapses are not regularly formed as early as DIV2, we temporally and efficiently down-regulated endogenous levels of PRG5 or overexpressed PRG5 in later stage hippocampal primary neurons. We chose DIV14, because at this stage PRG5 was still prominent in the membrane fraction (Fig. 4). In mature neurons, when an extensive network of synaptic connections is established, PRG5 was predominantly expressed in dendrites (Fig. 4B), as shown by co-localization with MAP2. In contrast, neurites that were positive for Tau1 showed only a weak PRG5 expression at this time point (Fig. 4B). We quantified the abundance of PRG5 in axons and dendrites by PRG5 fluorescence intensity measurements on Tau1- or MAP2-positive structures. The PRG5 expression on Tau1-positive structures was less than half the one on MAP2-positive structures (fluorescence intensity of PRG5 at Tau1-positive structures: 23.6 ± 2.0, n = 69; fluorescence intensity of PRG5 at MAP2-positive structures: 49.1 ± 3.6, n = 12, Fig. 4C). We next examined the effect of PRG5 overexpression on spine morphology. To minimize an overestimation of spine structures, we used PRG3, a transmembrane construct similar to PRG5, as control. PRG5 increases the number of protrusions per 10 μm (7.9 ± 0.3, n = 62) compared with neurons transfected with PRG3 (5.3 ± 0.2 n = 76, p < 0.0001; Fig. 4D). However, the protrusion length was reduced by PRG5 (1.3 μm ± 0.04, n = 167), in comparison with PRG3-transfected neurons (1.4 μm ± 0.05, n = 173, p < 0.02). This suggests that PRG5 is more involved in spine formation and not as much in filopodial formation as PRG3. Therefore, we analyzed Homer-positive spines in more detail. The number of Homer-positive structures (spine density) and the spine head diameter was significantly increased in PRG5-transfected neurons (spine density, 3.8 ± 0.2, n = 62; spine head diameter, 0.6 ± 0.02, n = 121) compared with PRG3-overexpressing neurons (spine density, 1.8 ± 0.11, n = 76, p < 0.0001; spine head diameter, 0.5 μm ± 0.02, n = 86, p < 0.0001). In contrast, the spine area did not differ between PRG5- and PRG3-transfected neurons (PRG5 0.8 μm ± 0.06, n = 91; PRG3 0.8 μm ± 0.074, n = 69 p < 0.96). On the contrary, knocking down endogenous PRG5 levels with PRG5 shRNA in mature primary neurons, which reduced endogenous PRG5 levels by one-third (shRNA-luciferase 55.6 ± 3.4, n = 123; shRNA-PRG5 40.5 ± 1.6, n = 201, p < 0.0001, Fig. 5A), decreased the number of protrusions per 10 μm (2.9 ± 0.06, n = 274) compared with neurons transfected with control shRNA (5.4 ± 0.1, n = 205, p < 0.0001). In addition, the PRG5-shRNA-resistant construct rescued the PRG5 knockdown effects (shRNA-PRG5 + rescue plasmid 5.04 ± 0.2, n = 93, p < 0.0001, Fig. 4E). We did not observe a difference in the protrusion length after reduction of endogenous PRG5 (shRNA-luciferase, 1.6 μm ± 0.05, n = 102; shRNA-PRG5, 1.6 μm ± 0.07, n = 90, p < 0.18, Fig. 5A).
Thus, PRG5 was present in all neurites in immature neurons but shifted toward expression in dendrites before DIV14. At this stage when spines are present, PRG5 levels regulate density and head diameter of spines.
Down-regulating PRG5 Expression Reduces Functional Synapses
Given that morphological formation and maturation of spines is directly correlated with synapse formation in cultured hippocampal neurons (27), we hypothesized less excitatory synapses accompanying the down-regulation of the PRG5 protein level. Indeed, counting structures expressing VGlut1 as pre-synaptic and GluR2 as post-synaptic markers in close proximity revealed about 20% reduced synapses in neurons, where PRG5 was down-regulated (shRNA-PRG5, 4.96 ± 0.26, n = 55; shRNA-luciferase, 6.35 ± 0.30, n = 51, p < 0.002; Fig. 5A). This reduction was reflected in our somatic whole-cell patch clamp recordings of cultured hippocampal neurons at DIV14–16. In an initial series of experiments, PRG5 knockdown resulted in a significant reduction of the mean frequency of mEPSCs (shRNA-PRG5, 3.05 ± 0.49 Hz, n = 18) when compared with untransfected neurons (7.24 ± 1.19 Hz, n = 24; U test, p < 0.02; Fig. 5, B and C). No changes in mEPSC amplitudes were observed (shRNA-PRG5 19.85 ± 1.03 pA, n = 18; control 21.26 ± 1.82 pA, n = 24, U test, p = 0.95; Fig. 5C).
To further confirm that the frequency reduction in neurons was induced by shRNA-PRG5, we tested the effect of shRNA-luciferase instead of untransfected neurons as control. Both the amplitude (shRNA-luciferase, 22.65 ± 0.70 pA, n = 25; control, 21.28 ± 0.58 pA, n = 19; U test, p = 0.15) and the frequency of mEPSCs (shRNA-luciferase, 2.78 ± 0.32 Hz, n = 25; control, 2.86 ± 0.3 Hz, n = 19; U test, p = 0.82) were not affected by shRNA-luciferase vector transfection (Fig. 5, B and C).
Taken together, these functional data corroborate our morphological findings by indicating that PRG5 reduction in mature dendrites attenuated the number of spines accompanied by a decrease in number of excitatory synapses and, consequently, reduced mEPSC frequencies, although mEPSC amplitudes remained similar.
PRG5 Binds PIPs Most Likely at the C Terminus, and the C Terminus Is Necessary for Spine Formation
It has been shown that PRG5 attenuates lysophosphatidic acid-induced neurite collapse and that this effect is mediated by the C terminus of PRG5 (15). Moreover, there is ample evidence that lipids are involved in spine formation (28–30). Hence, we hypothesized that the PRG5 C terminus is capable of interacting directly or indirectly with lipids. Therefore, we first used a protein-lipid overlay assay applied to a membrane. We observed binding of PRG5 to phosphatidic acid (PA), phosphatidylserine (PS), and to all phosphorylated phosphatidylinositols (PtdInsP) but no binding to various other lipids, including nonphosphorylated phosphatidylinositol, phosphatidylcholine, phosphatidylethanolamine, lysophosphatidic acid, and others (Fig. 6B). Incubating total lysate protein without overexpression did not produce any signal (data not shown).
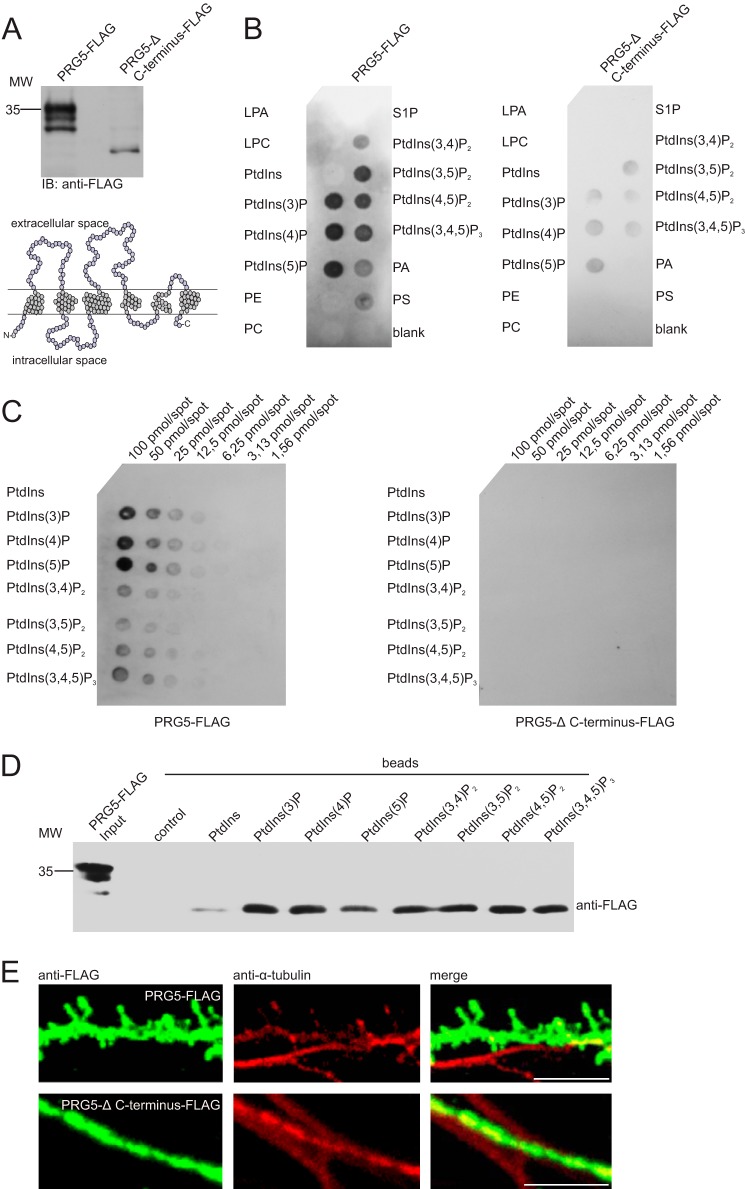
FIGURE 6.
PRG5 C terminus is essential for spine formation and binds phospholipids. A, Western blot analysis of protein lysates from HEK-293 cells transfected with PRG5-FLAG or PRG5-Δ C-terminal FLAG, both detected with an anti-FLAG. The structure model is based on amino acid sequence of human PRG5 and shows the truncation of PRG5. Removal of 37 amino acids at the C terminus led to an ∼10-kDa shift as shown by Western blotting. Note that the PRG5-Δ C-terminal FLAG construct showed almost only one prominent band. B, protein-lipid overlay assay using 5 μg/ml total protein lysate of HEK-293 cells overexpressed with PRG5-FLAG or PRG5-Δ C-terminal FLAG incubated with immobilized phosphatidylinositols and other lipids at 4 °C overnight. Membranes were immunoblotted with anti-FLAG antibody, and bound protein was visualized by ECL. PRG5 bound to phosphatidic acid (PA), phosphatidylserine (PS) and to all phosphorylated phosphatidylinositols (PtdInsP), but not to nonphosphorylated-phosphatidylinositol (Pins), phosphatidylcholine (PC), phosphatidylethanolamine (PE), lysophosphatidic acid (LPA), lysophosphocholine (LPC), or sphingosine 1-phosphate (S1P). In contrast, PRG5-Δ C terminus did not bind to any lipid of the strip except weakly to PtdInsP. C, binding specificity of these lipids was assessed by PIP ArraysTM. The array experiments probed with PRG5-FLAG again showed signals for all PtdInsP. Interestingly, PRG5 bound preferentially to monophosphorylated headgroups, and position of phosphate on the inositol ring had only a minor effect on binding. The array probed with the PRG5-Δ C-terminal FLAG showed no signal, pointing to the C terminus as region of interaction. D, PIP pulldown assay for confirming PRG5-PtdInsP binding. As in lipid strip results (B), PRG5 bound to all PtdInsP. A weak signal is also present for PtdIns. E, hippocampal primary neurons transfected with PRG5-FLAG (upper row, left, green) or PRG5-Δ C-terminal FLAG (lower row, left green) and co-immunostained with anti-tubulin (middle red). PRG5 overexpression induced spine-like structures, whereas PRG5-Δ C-terminal FLAG overexpressing hippocampal neurons showed no alteration. Scale bar, 5 μm.
The cardinal PRG5 binding partners, PtdInsPs, are localized at the inner sheet of cellular membranes and thus putatively in close proximity to intracellular components of PRG5, such as the C terminus. To study the involvement of the C terminus of PRG5 in lipid binding, we generated a truncated PRG5 version (PRG5-Δ C terminus), which lacks 37 amino acids C-terminally. Such truncation led to a shift of about 10 kDa compared with full-length PRG5 in the Western blot (Fig. 6A).
The lack of major parts of the C terminus in PRG5 substantially weakened the binding of PtdInsP and disabled binding to all other lipids on the lipid strip (Fig. 6B). This indicates that the PRG5 C terminus is strongly involved in protein-lipid interactions.
To assess their binding specificity, PRG5 or PRG5-Δ C terminus was probed against all eight phosphoinositides (PIP ArrayTM) spotted with concentration gradients from 100 to 1.56 pmol/spot. PRG5-FLAG showed some preference for monophosphorylated headgroups, because a di- and triphosphorylated headgroup gave consistently lower binding signals. However, the phosphate position on the inositol ring only slightly affected the binding signal, suggesting some plasticity of the binding site (Fig. 6, B and C). The array probed with the PRG5-Δ C-terminal FLAG lacked specific signals, although the lipid strips showed a signal of PtdInsP, even if it was substantially weakened. One can assume that the C terminus is the PRG5 protein region where the protein-lipid binding takes place.
To investigate whether these findings are due to direct interactions between PRG5 and phospholipids, we used a pulldown assay. In this assay, beads coated with different phosphoinositides and control beads were incubated with protein lysate overexpressing PRG5-FLAG. This in vitro assay also demonstrated that PRG5 bound to all PtdInsP (Fig. 6D). A weak band was also detectable with PtdIns. To analyze the function of the C terminus and therefore the protein-lipid interaction, we transfected PRG5-FLAG and PRG5-Δ C-terminal FLAG into primary neurons. In line with above results, the PRG5-Δ C-terminal FLAG did not alter the morphology of neuritis, although the whole PRG5 overexpression led to the appearance of the spine-like structures along the neurite, as we observed by immunocytochemistry (Fig. 6E).
These protein-lipid binding studies showed that PRG5 interacts with PtdInsP at the C terminus. The PRG5 C terminus and respective lipid bindings are essential for the induction of the spine-like structures on neurites of primary neurons.
DISCUSSION
Extending previous descriptions on plasma membrane predominance of overexpressed PRG5 fusion protein (15), we demonstrate here the following: first, that endogenous PRG5 protein was primarily present in membranes of immature and mature primary neurons. Second, we showed on the subcellular expression level that PRG5 was present in all neurites of immature neurons in vitro, but it accumulates at dendrites and was co-localized with post-synaptic markers, as GluR2 in more mature neurons. Third, in immature neurons PRG5 overexpression induced formation of spine-like structures ahead of time. In contrast, knockdown of PRG5 did not affect numbers of protrusions/filopodia, suggesting that PRG5 was more involved in spine development than in neurite growth. For that, proper amino acid sequences of the extracellular domains were essential. Fourth, overexpressing PRG5 in more mature neurons increased the total number of protrusions. Specifically the number of Homer-positive spines more than doubled compared with controls and PRG5 overexpression increases the spine head diameter. However, acute reduction of PRG5 levels in more mature neurons by shRNA caused reduction of protrusions, relevant loss of excitatory synapses, and consequently, reduction of mEPSC frequencies.
PRG5 Is Timely and Spatially Regulated in the Brain
Similar to other members of the PRG superfamily, such as PRG1 and PRG3 (18), PRG5 is enriched in brain neurons of several mammalian species, including mouse (15, 20). In the region of our actual study, the hippocampus PRG expression patterns are differently regulated throughout development. Although PRG3 is first and strongly expressed already from early embryonic (E14) stages and decreased after birth (23), the expression of its closest relative PRG5 (73% homolog at the nucleotide and 55% on the amino acid level (15)) peaked, although it was somewhat delayed at P0 and only slightly decreased postnatally (this study). Finally, PRG1 expression starts around birth to increase during early postnatal development and remains on maximum levels until adulthood (21). In addition, the developmental changes of subcellular expression patterns of PRGs differ remarkably, although PRG expression is apparently equally distributed along all neurites in early developmental stages and during differentiation PRG3 (23); on the one hand, it accumulates in axonal structures, PRG5 (this study) and PRG1 (31), and on the other hand, it accumulates at postsynaptic dendritic sites. These temporal and spatial expression characteristics suggest distinct functional roles of all three proteins at different stages of neuronal differentiation.
Post-translational Modification of PRG5
PRG5 appeared as a glycoprotein after transfection in HEK-293 cells. PRG5 is N-glycosylated, as predicted by in silico analysis. The N-glycosylation sequence in the second extracellular loop of PRG5 was also identified in other members of the lipid phosphatase/phosphotransferase family as a consensus glycosylation sequence (23, 26). In addition, our results show that the PRG5 protein may undergo more than one post-translational modification, which could be, for instance, O-glycosylation, phosphorylation, or palmitoylation. Post-translational modifications perform critical biological functions such as cell adhesion, molecular trafficking, clearance, and signal transduction (32). More analyses are required to identify further modifications and their effects on PRG5 protein.
PRG5 Plays a Role in Spine Formation and Synaptic Changes
Our data further increase the body of evidence for complementary action of PRG3 and PRG5 in shaping neurons (18). Both PRG3 (23) and PRG5 (this study) initiate protrusions in the neuronal membrane. So far, PRG5 was related to induction of filopodia in different cell lines and to formation of neurites and branches in cortical immature neurons (15). Filopodia at early neuronal developmental stage neurons are just similar to those emerging from non-neuronal cells and do not show any morphological plasticity (33). Mature neurons have dendritic spines capable of morphological plasticity depending on synaptic activity, whereas immature neurons, cultured up to DIV7, have thin and long (filopodium-like) protrusions (12, 13). In our study, we found that immature neurons overexpressing PRG5 were littered with membrane protrusions all over the cell, and in contrast to PRG3-induced protrusions, they not only had thin and long morphologies but were shaped as mushroom and stubby spines. Although reduction of PRG3 in immature neurons decreased the number of neurites between 2- and 5-μm lengths at the neurite shaft (23), PRG5 reduction showed no changes in the numbers of protrusions/filopodia (this study). That suggests a major involvement of PRG5 in spine formation in contrast to PRG3, which seems principally involved in regulation of neurite consolidation (23).
In addition, PRG3 expression in mature neurons shifted to the axonal side and stabilized the axon (23), whereas PRG5 (this study) and PRG1 (31) were localized mostly on the postsynaptic dendritic site. In mature neurons, spines are grouped in several morphological classes that are thought to correlate with different developmental stages of the associated synapses (11, 34). In this view, spines with large heads are stable, express large numbers of AMPA-type glutamate receptors, and contribute to strong synaptic connections (5, 7, 35).
With PRG5 we added a new and unexpected player to the certain synaptic molecules that recruit or influence necessary cytoskeletal and membrane components, which in turn enable the postsynaptic membrane to reorganize and protrude into a spine-like structure. Such synaptic molecules include members of the Rho family GTPases that control the actin cytoskeleton (36, 37). Actin filaments provide a foundation for synapse shape, motility, and stability. Several actin-regulatory pathways activated by transmembrane signals influence the properties of dendritic spines (7, 38). One signaling cascade involves an active EphB-intersection complex together with N-WASP. This, in turn, activates the Rho family GTPases Cdc42 and triggers actin polymerization via the Arp2/3 complex, which is proposed to result in expansion of dendritic spine heads (39). Even so, PRG5 induces filopodial formation independently of mDia1, a Rho effector molecule, and Cdc42 (15). Interestingly, overexpression of syndecan-2 in immature neurons partially mimics the PRG5 effects found in our study (40). Syndecan-2 promotes spine-like structures via the neurofibromin-PKA-Ena/VASP pathway (41). The molecular mechanisms involved in PRG5 actions are still elusive. Our results, however, showed that overexpression of PRG5 increased the density of spines and their head diameter, suggesting a direct involvement in the organization of spine morphology, and an influence on modulation of excitatory transmission by PRG1 (31). Furthermore, down-regulation of endogenous PRG5 protein levels resulted in a loss of excitatory synapses and, consequently, in an evident decrease in mEPSC frequency. This illustrates that the amount and localization of PRG5 participate in the development and function of the hippocampal circuitry by putatively affecting spine formation in an all-or-none manner.
How Might PRG5 Promote and Stabilize Dendritic Spines?
The generation of any kind of protrusions up to and including spines requires a tight spatiotemporal coordination of many different factors. One of them is the dynamic membrane remodeling achieved by the interplay between lipids and proteins (13, 42). Such interplay is most likely when both players are located in the neuronal membrane. PRG5 membrane insertion depends on extracellular domains, as we demonstrate here by PRG5 mutated in the extracellular domains D2 or D3 displaying a more intracellular localization. When not present in the membrane, PRG5 failed to induce spine-like structures at the plasma membrane. Interestingly, the PRG5 mutant reproducing the D2 domain of PRG3 is located in the plasma membrane and induces thin and long as well as spine-like structures. Although replaced amino acids are important for substrate recognition in enzymatically active lipid phosphate phosphatase family members, enzymatic activity is unlikely to be involved here, because comparison of the relevant regions of the PRG5 protein with those of LPP1 reveals that a number of crucial catalytic residues are missing, and up to now enzymatic activity was not reported for PRG5. However, PRG1 interacts with bioactive phospholipids such as lysophosphatidic acid at the synapse. It has been suggested that PRG1 regulates excitatory transmission depending on its interaction with and uptake of bioactive lipids mediated by their extracellular domains (31).
In addition to extracellular domains, the C terminus of PRG5 contributes to filopodial formation, at least in cell lines (15). Proteins that include the lipid-binding domain I-BAR cause curvatures in the cell membrane and are involved in the generation of filopodia (14). In particular, PtdIns(4,5)P2 sufficiently recruits one key I-BAR family member, IRSp53, which is involved in generating membrane curvature (43, 44). Although PRG5 lacks I-BAR domains and induces filopodial formation independent of Cdc42, it binds several lipids as PA, PS, and PtdInsP, and this binding was suspended when the C terminus was absent. Therefore, the C terminus appears essential for these lipid interactions. Furthermore, the C terminus, and thus presumably lipid binding, is involved in spine formation. Interestingly, PA favors membrane curvatures (28, 45). The phospholipid bilayer can be deformed causing positive or negative membrane curvature. Changes in membrane curvature accompany, for example, endocytosis, exocytosis, or spine formation. Its notable within this context that PA binding activates the serine/threonine kinase PAK1, independent of small GTPases (i.e. Rac1 or Cdc42), and that activated PAK1 maintains dendritic spines (46). Another study showed that hydrolysis of PtdIns(4,5)P2 by phospholipase C is required for spine actin depolymerization and thus for structural and functional changes in spines (47). It seems that PRG5 preferably interacts with phosphorylated headgroups of PtdInsP. In general, lipid headgroups are the attachment sites for peripheral membrane proteins and therefore aid the recruitment of proteins necessary to generate curvature. PtdInsP are particularly important as their headgroups are easily modified (13, 48). Further investigation is needed, in particular interaction studies close to the in vivo situation, to understand the different function of the PRG5-PtdInsP interactions.
In conclusion, our results suggest that endogenous levels of PRG5 play a role in proper spine formation, morphology of the spine, and stabilization of functional excitatory synapses. PRG5 action involves the C terminus, most likely by local lipid interaction. Because spine abnormalities are found in many pathological conditions, including mental illnesses and age-related neurodegenerative diseases (6, 7), PRG5 levels might be relevant for the susceptibility, pathophysiology, or therapy for such diseases.
Acknowledgments
Rike Dannenberg, Nora Ebermann, Jan Csupor, and Bettina Brokowski are acknowledged for excellent technical assistance and Tanja Velmans for performing the qRT-PCR analysis. We thank Jutta Schüler for help with the confocal analyses and Eugenia Rojas-Puente from AG Eickholt (Cluster of Excellence NeuroCure and Institute of Biochemistry) and Dr. Markus Höltje (Institute of Integrative Neuroanatomy) for providing mouse hippocampi or mouse hippocampal neurons.
This work was supported by Deutsche Forschungsgemeinschaft Grant BR 2345/1-1 (to A. U. B.) and the Sonnenfeld-Stiftung for sponsoring technical equipment for A. U. B. and U. S. and the NaFöG sponsored P. C.
- DIV
- days in vitro
- mEPSC
- miniature excitatory postsynaptic current
- qRT
- quantitative RT
- PIP
- phosphoinositide
- PtdInsP
- phosphorylated phosphatidylinositol
- PA
- phosphatidic acid
- PS
- phosphatidylserine
- eGFP
- enhanced GFP
- PRG
- plasticity-related gene
- PAP2D
- phosphatidic acid phosphatase type 2D
- P
- postnatal day
- E
- embryonic day
- TRITC
- tetramethylrhodamine isothiocyanate
- PNGase F
- peptide:N-glycosidase.
REFERENCES
- 1. Bhatt D. H., Zhang S., Gan W. B. (2009) Dendritic spine dynamics. Annu. Rev. Physiol. 71, 261–282 [DOI] [PubMed] [Google Scholar]
- 2. Hotulainen P., Hoogenraad C. C. (2010) Actin in dendritic spines: connecting dynamics to function. J. Cell Biol. 189, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunaevsky A., Tashiro A., Majewska A., Mason C., Yuste R. (1999) Developmental regulation of spine motility in the mammalian central nervous system. Proc. Natl. Acad. Sci. U.S.A. 96, 13438–13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dailey M. E., Smith S. J. (1996) The dynamics of dendritic structure in developing hippocampal slices. J. Neurosci. 16, 2983–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kasai H., Matsuzaki M., Noguchi J., Yasumatsu N., Nakahara H. (2003) Structure-stability-function relationships of dendritic spines. Trends Neurosci. 26, 360–368 [DOI] [PubMed] [Google Scholar]
- 6. Fiala J. C., Spacek J., Harris K. M. (2002) Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 39, 29–54 [DOI] [PubMed] [Google Scholar]
- 7. Calabrese B., Wilson M. S., Halpain S. (2006) Development and regulation of dendritic spine synapses. Physiology 21, 38–47 [DOI] [PubMed] [Google Scholar]
- 8. Schwamborn J. C., Li Y., Püschel A. W. (2006) GTPases and the control of neuronal polarity. Methods Enzymol. 406, 715–727 [DOI] [PubMed] [Google Scholar]
- 9. Dent E. W., Barnes A. M., Tang F., Kalil K. (2004) Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J. Neurosci. 24, 3002–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henkemeyer M., Itkis O. S., Ngo M., Hickmott P. W., Ethell I. M. (2003) Multiple EphB receptor tyrosine kinases shape dendritic spines in the hippocampus. J. Cell Biol. 163, 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bourne J. N., Harris K. M. (2008) Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 31, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sekino Y., Kojima N., Shirao T. (2007) Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem. Int. 51, 92–104 [DOI] [PubMed] [Google Scholar]
- 13. McMahon H. T., Gallop J. L. (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 14. Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P. K., Kinnunen P. K., Butcher S. J., Lappalainen P. (2009) Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95–107 [DOI] [PubMed] [Google Scholar]
- 15. Broggini T., Nitsch R., Savaskan N. E. (2010) Plasticity-related gene 5 (PRG5) induces filopodia and neurite growth and impedes lysophosphatidic acid- and nogo-A-mediated axonal retraction. Mol. Biol. Cell 21, 521–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sigal Y. J., Quintero O. A., Cheney R. E., Morris A. J. (2007) Cdc42 and ARP2/3-independent regulation of filopodia by an integral membrane lipid-phosphatase-related protein. J. Cell Sci. 120, 340–352 [DOI] [PubMed] [Google Scholar]
- 17. Bräuer A. U., Nitsch R. (2008) Plasticity-related genes (PRGs/LRPs): a brain-specific class of lysophospholipid-modifying proteins. Biochim. Biophys. Acta 1781, 595–600 [DOI] [PubMed] [Google Scholar]
- 18. Strauss U., Bräuer A. U. (2013) Current views on regulation and function of plasticity-related genes (PRGs/LPPRs) in the brain. Biochim. Biophys. Acta 1831, 133–138 [DOI] [PubMed] [Google Scholar]
- 19. Brindley D. N. (2004) Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J. Cell Biochem. 92, 900–912 [DOI] [PubMed] [Google Scholar]
- 20. Sun L., Gu S., Sun Y., Zheng D., Wu Q., Li X., Dai J., Dai J., Ji C., Xie Y., Mao Y. (2005) Cloning and characterization of a novel human phosphatidic acid phosphatase type 2, PAP2d, with two different transcripts PAP2d_v1 and PAP2d_v2. Mol. Cell. Biochem. 272, 91–96 [DOI] [PubMed] [Google Scholar]
- 21. Bräuer A. U., Savaskan N. E., Kühn H., Prehn S., Ninnemann O., Nitsch R. (2003) A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat. Neurosci. 6, 572–578 [DOI] [PubMed] [Google Scholar]
- 22. Savaskan N. E., Bräuer A. U., Nitsch R. (2004) Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur. J. Neurosci. 19, 212–220 [DOI] [PubMed] [Google Scholar]
- 23. Velmans T., Battefeld A., Geist B., Farrés A. S., Strauss U., Bräuer A. U. (2013) Plasticity-related gene 3 promotes neurite shaft protrusion. BMC Neurosci. 14, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Savaskan N. E., Rocha L., Kotter M. R., Baer A., Lubec G., van Meeteren L. A., Kishi Y., Aoki J., Moolenaar W. H., Nitsch R., Bräuer A. U. (2007) Autotaxin (NPP-2) in the brain: cell type-specific expression and regulation during development and after neurotrauma. Cell. Mol. Life Sci. 64, 230–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith D. L., Pozueta J., Gong B., Arancio O., Shelanski M. (2009) Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc. Natl. Acad. Sci. U.S.A. 106, 16877–16882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Q. X., Pilquil C. S., Dewald J., Berthiaume L. G., Brindley D. N. (2000) Identification of structurally important domains of lipid phosphate phosphatase-1: implications for its sites of action. Biochem. J. 345, 181–184 [PMC free article] [PubMed] [Google Scholar]
- 27. Abe R., Shimizu T., Yamagishi S., Shibaki A., Amano S., Inagaki Y., Watanabe H., Sugawara H., Nakamura H., Takeuchi M., Imaizumi T., Shimizu H. (2004) Overexpression of pigment epithelium-derived factor decreases angiogenesis and inhibits the growth of human malignant melanoma cells in vivo. Am. J. Pathol. 164, 1225–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mattila P. K., Pykäläinen A., Saarikangas J., Paavilainen V. O., Vihinen H., Jokitalo E., Lappalainen P. (2007) Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuesto G., Enriquez-Barreto L., Caramés C., Cantarero M., Gasull X., Sandi C., Ferrús A., Acebes Á., Morales M. (2011) Phosphoinositide-3-kinase activation controls synaptogenesis and spinogenesis in hippocampal neurons. J. Neurosci. 31, 2721–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim K., Yang J., Kim E. (2010) Diacylglycerol kinases in the regulation of dendritic spines. J. Neurochem. 112, 577–587 [DOI] [PubMed] [Google Scholar]
- 31. Trimbuch T., Beed P., Vogt J., Schuchmann S., Maier N., Kintscher M., Breustedt J., Schuelke M., Streu N., Kieselmann O., Brunk I., Laube G., Strauss U., Battefeld A., Wende H., Birchmeier C., Wiese S., Sendtner M., Kawabe H., Kishimoto-Suga M., Brose N., Baumgart J., Geist B., Aoki J., Savaskan N. E., Bräuer A. U., Chun J., Ninnemann O., Schmitz D., Nitsch R. (2009) Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell 138, 1222–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 33. Shirao T., González-Billault C. (2013) Actin filaments and microtubules in dendritic spines. J. Neurochem. 126, 155–164 [DOI] [PubMed] [Google Scholar]
- 34. Alvarez V. A., Sabatini B. L. (2007) Anatomical and physiological plasticity of dendritic spines. Annu. Rev. Neurosci. 30, 79–97 [DOI] [PubMed] [Google Scholar]
- 35. Haeckel A., Ahuja R., Gundelfinger E. D., Qualmann B., Kessels M. M. (2008) The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J. Neurosci. 28, 10031–10044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 37. Hall A., Nobes C. D. (2000) Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshihara Y., De Roo M., Muller D. (2009) Dendritic spine formation and stabilization. Curr. Opin. Neurobiol. 19, 146–153 [DOI] [PubMed] [Google Scholar]
- 39. Irie F., Yamaguchi Y. (2004) EPHB receptor signaling in dendritic spine development. Front. Biosci. 9, 1365–1373 [DOI] [PubMed] [Google Scholar]
- 40. Ethell I. M., Yamaguchi Y. (1999) Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J. Cell Biol. 144, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin Y. L., Lei Y. T., Hong C. J., Hsueh Y. P. (2007) Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J. Cell Biol. 177, 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arstikaitis P., Gauthier-Campbell C., Carolina Gutierrez Herrera R., Huang K., Levinson J. N., Murphy T. H., Kilimann M. W., Sala C., Colicos M. A., El-Husseini A. (2008) Paralemmin-1, a modulator of filopodia induction is required for spine maturation. Mol. Biol. Cell 19, 2026–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scita G., Confalonieri S., Lappalainen P., Suetsugu S. (2008) IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 18, 52–60 [DOI] [PubMed] [Google Scholar]
- 44. Lim K. B., Bu W., Goh W. I., Koh E., Ong S. H., Pawson T., Sudhaharan T., Ahmed S. (2008) The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 [DOI] [PubMed] [Google Scholar]
- 45. Kooijman E. E., Carter K. M., van Laar E. G., Chupin V., Burger K. N., de Kruijff B. (2005) What makes the bioactive lipids phosphatidic acid and lysophosphatidic acid so special? Biochemistry 44, 17007–17015 [DOI] [PubMed] [Google Scholar]
- 46. Bokoch G. M., Reilly A. M., Daniels R. H., King C. C., Olivera A., Spiegel S., Knaus U. G. (1998) A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J. Biol. Chem. 273, 8137–8144 [DOI] [PubMed] [Google Scholar]
- 47. Horne E. A., Dell'Acqua M. L. (2007) Phospholipase C is required for changes in postsynaptic structure and function associated with NMDA receptor-dependent long-term depression. J. Neurosci. 27, 3523–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Behnia R., Munro S. (2005) Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 [DOI] [PubMed] [Google Scholar]