Background: The mechanisms whereby atypical PKCs (aPKCs) are silenced and activated by phosphatidylinositol 3,4,5-(PO4)3 are uncertain.
Results: Mutagenesis of arginine residues in the pseudosubstrate of aPKCs caused constitutive activation that was inhibited by exogenous pseudosubstrate. Pseudosubstrate-dependent inhibition was reversed by phosphatidylinositol 3,4,5-(PO4)3.
Conclusion: Pseudosubstrate arginine residues are required for autoinhibition and are targeted by phosphatidylinositol 3,4,5-(PO4)3 during aPKC activation.
Significance: Pseudosubstrate arginine residues are key regulators of aPKC.
Keywords: Diabetes, Insulin Resistance, Phosphatidylinositol Kinase (PI Kinase), Protein Kinase C (PKC), Signal Transduction
Abstract
Atypical PKC (aPKC) isoforms are activated by the phosphatidylinositol 3-kinase product phosphatidylinositol 3,4,5-(PO4)3 (PIP3). How PIP3 activates aPKC is unknown. Although Akt activation involves PIP3 binding to basic residues in the Akt pleckstrin homology domain, aPKCs lack this domain. Here we examined the role of basic arginine residues common to aPKC pseudosubstrate sequences. Replacement of all five (or certain) arginine residues in the pseudosubstrate sequence of PKC-ι by site-directed mutagenesis led to constitutive activation and unresponsiveness to PIP3 in vitro or insulin in vivo. However, with the addition of the exogenous arginine-containing pseudosubstrate tridecapeptide to inhibit this constitutively active PKC-ι, PIP3-activating effects were restored. A similar restoration of responsiveness to PIP3 was seen when exogenous pseudosubstrate was used to inhibit mouse liver PKC-λ/ζ maximally activated by insulin or ceramide and a truncated, constitutively active PKC-ζ mutant lacking all regulatory domain elements and containing “activating” glutamate residues at loop and autophosphorylation sites (Δ1–247/T410E/T560E-PKC-ζ). NMR studies suggest that PIP3 binds directly to the pseudosubstrate. The ability of PIP3 to counteract the inhibitory effects of the exogenous pseudosubstrate suggests that basic residues in the pseudosubstrate sequence are required for maintaining aPKCs in an inactive state and are targeted by PIP3 for displacement from the substrate-binding site during kinase activation.
Introduction
The atypical PKC (aPKC)2 isoforms PKC-ζ, PKC-λ, and primate-specific PKC-ι serve as critically important kinases in multiple cellular processes. Indeed, total knockout of PKC-λ/ι is embryonic lethal in mice, and alterations in aPKC in humans are important determinants in diverse disorders, including various cancers, where aPKC activity is increased frequently (1), and obesity and type 2 diabetes mellitus, where aPKC activity is diminished in muscle (2–4) and adipocytes (4), where aPKC and Akt are required for insulin-stimulated glucose transport, but inordinately increased in the liver (5), where aPKC and Akt are required for insulin-stimulated lipid synthesis. Despite their importance, little is known about the molecular mechanisms that are operative in aPKC activation.
Like Akt, aPKCs are activated by PI3K during actions of insulin (6) and other growth factors, e.g. insulin-like growth factor 1 (7). The activation of both aPKC and Akt by PI3K is effected by generation of phosphatidylinositol-3,4,5-(PO4)3 (PIP3) from phosphatidylinositol-4,5-(PO4)2 (PIP2) in the plasma membrane and other cellular compartments. Akt activation is thought to be initiated by an interaction of the acidic D3-phosphoinositide group of PIP3, with basic arginine and lysine residues in the pleckstrin homology (PH) domains contained in both Akt (8) and phosphoinositide-dependent protein kinase 1 (PDK1) (9). This colocalization allows PDK1 to phosphorylate threonine 308 in the Akt activation loop, which, in turn, facilitates phosphorylation of serine 473 in the C-terminal hydrophobic motif of Akt by “PDK2,” now identified as the mammalian target of rapamycin 2 (mTORC2) (10). Whether PIP3 increases the activity of PDK1 or mTORC2 is debatable (e.g. see Ref. 3). In any case, unlike Akt, aPKCs do not have a PH domain, and how PIP3 either localizes or activates aPKC at the molecular level is presently obscure. On the other hand, like Akt, the D3-PO4 group appears to mediate the activation of aPKC by PIP3 because PI-3,4-(PO4)2, but not PI-4,5-(PO4)2, increases aPKC phosphorylation (11).
As with Akt, as well as with conventional PKCs (α, β, and γ) and novel PKCs (δ, ϵ, η, and θ), aPKC activation requires phosphorylation of threonine residues in activation loops, viz. threonine 412 in PKC-ι, threonine 411 in PKC-λ, and threonine 410 in PKC-ζ by PDK1, and auto(trans)phosphorylation sites, viz. threonine 564 in PKC-ι, threonine 563 in PKC-λ, and threonine 560 in PKC-ζ in the turn regions of their catalytic domains (11, 12). In addition to required phosphorylations of loop and auto(trans)phosphorylation sites, PIP3 has been postulated to provoke allosteric effects during aPKC activation (12).
Like conventional and novel PKCs, aPKCs exist basally in a folded inactive state in which residues in the pseudosubstrate region of the regulatory domain bind to residues in the substrate-binding region of the catalytic domain. Therefore, it may be hypothesized that, in aPKCs, basic arginine residues in the pseudosubstrate sequence bind to acidic residues in the substrate-binding region and that disruption of this binding by an acidic ligand, such as PIP3, leads to molecular unfolding and exposure of the substrate-binding site not only to extrinsic substrates but also to the intrinsic auto- or trans-phosphorylation site. In this regard, note that, with conventional and novel PKCs, this unfolding and subsequent activation is effected by Ca2+, which binds to a site in the C2 region of the regulatory domain that is uniquely present in conventional PKCs, and diacylglycerol (DAG), which binds to sites in the C1 region of the regulatory domain that are present in both conventional and novel PKCs. Interestingly, it is thought that one potential DAG-binding site is also present in the C1 region of aPKCs but is functionally blocked by a set of four basic arginine residues that surrounds the opening to this pocketed activation site (13). Accordingly, these arginine residues in the “DAG pocket ring” could reasonably be a point of attack for PIP3.
In the case of aPKCs, it is currently uncertain how insulin or other PI3K activators use PIP3 to either localize aPKCs to the plasma membrane in juxtaposition with PDK1 or induce molecular unfolding and subsequent activation by auto(trans) phosphorylation. However, given the fact that the aPKC pseudosubstrate sequence, like the aPKC consensus substrate recognition sequence, contains basic arginine residues that flank a short sequence that, in substrates, contains phosphorylatable threonine or serine residues, we examined the importance of the five basic arginine residues that are common to pseudosubstrate sequences of all aPKCs for their ability to maintain aPKC in an inactive state and serve as a target used by PIP3 to provoke a dissociation of the pseudosubstrate from the substrate-binding site and, thereby, promote kinase activation.
EXPERIMENTAL PROCEDURES
Cell Culture and Incubation Conditions
3T3/L1 adipocytes were differentiated, cultured, and transfected with plasmids or infected with adenoviruses and incubated for 48–72 h to allow time for expression, as described previously (14). At the time of the experiment, the medium was removed, and cells were preincubated for 3 h in Krebs-Ringer phosphate buffer containing 5 mm d-glucose and then incubated for 30 min in glucose-free Krebs-Ringer phosphate buffer ± 100 nm insulin (Sigma). For studies of glucose transport, the cells were subsequently incubated for another 5 min with 0.05 mm d-glucose and a trace amount of [3H]2-deoxyglucose, as described previously (14). After incubation, the cells were washed three times with cold medium, released from incubation plates with 0.1% sodium dodecyl sulfate solution, and sonicated in buffer as described below. Portions of cell lysates were placed into Laemmli buffer for Western blot analysis, used for measurement of protein levels and [3H]2-deoxyglucose uptake, used for immunoprecipitation of epitope-tagged or total aPKCs, or used for harvesting of His6-tagged aPKCs by adsorption with Ni-NTA-coated beads (EMD Millipore, Billerica, MA).
Mouse Studies
In some experiments, we used aPKCs immunoprecipitated from livers of C57Bl/6-SV/129 mice that were treated intraperitoneally with maximally effective insulin (1 unit/kg body weight) or vehicle 15 min before killing, as described previously (15, 16).
Cell Lysate Preparations
As described previously (14–16), adipocytes were sonicated, and mouse liver was homogenized in ice-cold buffer containing 0.25 mm sucrose, 20 mm Tris-HCl (pH 7.5), 2 mm EGTA, 2 mm EDTA, 1 mm PMSF, 20 μg/ml leupeptin, 10 μg/ml aprotinin, 2 mm Na4P2O7, 2 mm Na3VO4, 2 mm NaF, and 1 μm microcystin; supplemented with 1% Triton X-100, 0.6% Nonidet, and 150 mm NaCl; and cleared by low-speed centrifugation.
aPKC Activation
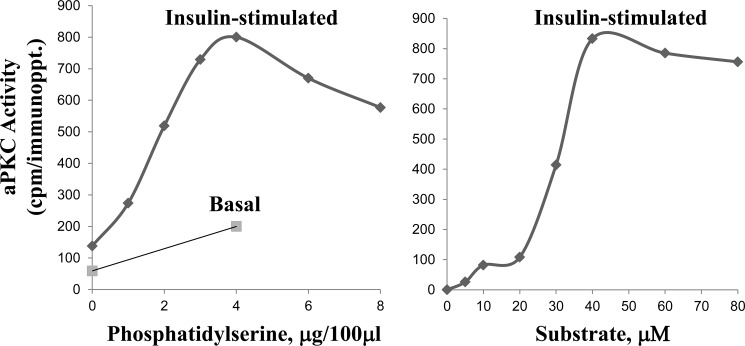
aPKC activity was measured essentially as described previously (6, 17) using His6-tagged aPKCs isolated from 3T3/L1 adipocyte lysates by adsorption onto Ni-NTA beads and eluted with assay buffer containing 100 mm EDTA, using HA-tagged aPKC isolated by immunoprecipitation with rabbit polyclonal anti-HA-purified IγG (Santa Cruz Biotechnology, Santa Cruz, CA, or using total aPKC isolated from lysates of 3T3/L1 adipocytes or mouse liver by immunoprecipitation with polyclonal antiserum (Santa Cruz Biotechnology) that recognizes the C termini of PKC-λ/ι and PKC ζ. Immunoprecipitated aPKC was collected on Sepharose-AG beads. Isolated aPKC was incubated for 8 min at 30 °C in 100 μl of buffer containing 50 mm Tris-HCl (pH7.5), 100 μm Na3VO4, 100 μm Na4P2O7, 1 mm NaF, 100 μm PMSF, 4 μg of phosphatidylserine (Sigma), 50 μm [γ-32P]ATP (NEN/Life Science Products, Waltham, MA), 5 mm MgCl2 and, as substrate, 40 μm serine analog of the PKC-ϵ pseudosubstrate (Enzo Life Sciences, Farmingdale, NY). After incubation, 32P-labeled substrate was trapped on P-81 filter paper and counted in a liquid scintillation counter. Note that phosphatidylserine serves as a cofactor by providing a phospholipid surface that binds and optimizes PKC activity during activation by diacylglycerol and other lipids (18–21); that, as shown in Fig. 1, both the phospholipid cofactor phosphatidylserine and the aPKC substrate are routinely present at optimal concentrations in the assay; and that insulin, via endogenous PIP3, provokes increases in activity that are additive to the dose-dependent effects of phosphatidylserine. In some cases, aPKC activation was also assessed by immunoblot analysis of phospho-Thr-563/560-PKC-ζ/λ.
FIGURE 1.
Dose-dependent requirements for phosphatidylserine (left panel) and substrate (right panel) during aPKC assay. Where indicated, mice were injected intraperitoneally with a maximally effective dose of insulin (1 unit/kg body weight) or saline vehicle 15 min before killing. aPKC was immunoprecipitated from liver lysates and assayed with increasing amounts of phosphatidylserine and a preferred aPKC substrate, i.e. the serine analog of the PKC-ϵ pseudosubstrate, along with other standard components of the aPKC assay (see “Experimental Procedures”). Data are mean of 2–4 replicates.
Western Blot Analyses
As described previously (2–5), harvested plasmids or cell lysates were immunoblotted for PKC-ζ/λ using a rabbit polyclonal antiserum (Santa Cruz Biotechnology) that recognizes the C termini of all aPKCs; λ, ι, and ζ; rabbit polyclonal anti-phospho-Thr-563/560-PKC-λ/ζ (Life Technology, Grand Island, NY); and rabbit polyclonal anti-phospho-Thr-411/410-PKC-λ/ζ (Cell Signaling Technology, Danvers, MA).
Site-directed Mutagenesis and Preparation of Plasmids and Adenoviruses
HA-tagged, constitutively active, truncated mutant HA-Δ1–247/T410E/T560E-PKC-ζ, which lacks all regulatory domain elements and contains acidic glutamate residues at loop and autophosphorylation sites, was generated and cloned into the pCDNA3 plasmid as described previously (12).
For the generation of arginine-to-alanine mutations in the pseudosubstrate sequence and in the ring of arginine residues surrounding a pocket containing a potential DAG-binding site in the C1 region of PKC-ι (13), a clone of human PKC-ι was generated from a cDNA library prepared from human brain tissue using standard PCR techniques. The following primers were used to generate a PCR product suitable for direct recombination into the Gateway pDONR entry vector via BP Clonase II (Invitrogen): 5′, GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGCATCATCATCATCATCATGCCCCGACCCAGAGGGACAGCAGCACC; 3′, GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACTTCTCGAACTGGGGGTGGGACCTTCCTCCGACACATTCTTCTGCAGACATCAAAAGAGG. To minimize extraneous amino acids in the expressed protein, Shine-Dalgarno and Kozak translation initiation sequences and a start codon were included, along with coding for a His6 tag in the 5′ primer. In addition, the 3′ primer contained coding for a StrepII tag as well as a stop codon. PCR conditions included using 100 ng of cDNA for 35 cycles with denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 68 °C for 2 min.
Mutations of the human PKC-ι clone in the pDONR vector were generated using standard PCR techniques as follows: R147A/R150A/R151A/R160A (i.e. four arginine residues surrounding a putative DAG activation site) mutant, CCACACTTTCCAAGCCAAGGCTTTCAACGCGGCTGCTCACTGTGCCATCTGCACAGACGCAATATGGGGACTTGGACG (5′) and CGTCCAAGTCCCCATATTGCGTCTGTGCAGATGGCACAGTGAGCAGCCGCGTTGAAAGCCTTGGCTTGGAAAGTGTGG (3′); R126A (i.e. a single arginine residue in the pseudosubstrate sequence) mutant, GATAAATCCATCTACGCTAGAGGTGCACGCCGC (5′) and GCGGCGTGCACCTCTAGCGTAGATGGATTTATCTTCTCC (3′); R131A (i.e. a single arginine residue in the pseudosubstrate sequence) mutant, CGTAGAGGTGCACGCGCCTGGAGAAAGCTTTATTGTGC (5′) and ATAAAGCTTTCTCCAGGCGCGTGCACCTCTACG (3′); and R126A/R127A/R130A/R131A/R133A (all five arginine residues in the pseudosubstrate sequence) mutant, TAAATCCATCTACGCTGCAGGTGCAGCCGCCTGGGCAAAGCTTTATTGTGCC (5′) and GGCACAATAAAGCTTTCCCCAGGCGGCTGCACCTGCAGCGTAGATGGATTTATCTTCTCC (3′). PCR conditions included using 1 ng of vector for 40 cycles with denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 68 °C for 10 min. All primers for cloning and mutating the PKC-ι gene were supplied by Eurofins MWG Operon (Huntsville, AL).
For direct, plasmid-based expression in mammalian cell culture, wild-type and mutant versions of PKC-ι were transferred to the Gateway vector pcDNA3.2 using LR Clonase II (Invitrogen). To generate recombinant adenoviruses for expression of the PKC-ι clones, wild-type and mutant versions were transferred to the pAD vector using LR Clonase II (Invitrogen). Standard transfection and viral amplification techniques were used for large scale expression of each clone.
NMR Studies of PIP3 Binding to the aPKC Pseudosubstrate
NMR spectra were collected at 25 °C on a Varian VNMRS 600 MHz spectrometer equipped with a triple resonance pulse field z axis gradient cold probe. Two-dimensional 1H-H1 TOCSY spectra had widths of 12 ppm in both dimensions with 2048 (t2) × 120 (t1) complex data points. NMR spectra were processed and analyzed with VnmrJ 3.2A (Varian). Samples (pseudosubstrate peptides (Genscript, Piscataway, NJ) and PIP3 (Matreya)) were prepared in phosphate-buffered saline (pH 7.4) in 90% H2O, 10% D2O. NMR studies were performed by the Florida Center of Excellence for Drug Discovery at the University of South Florida.
Statistical Methods
Data are expressed as mean ± S.E. Statistical differences between two and three or more groups were determined by Student's t test and analysis of variance (Sigma Stat statistical software), respectively.
RESULTS
Activation of aPKC by PIP3 versus PIP2
In keeping with previous findings of increased overall 32PO4 labeling of aPKC during incubation of immunoprecipitated aPKC with PIP3, but not PIP2 (11), we found that PIP3, but not PIP2, increased both enzyme activity and autophosphorylation of aPKC immunoprecipitated from mouse liver (Fig. 2). These findings show that the D3-PO4 group of PIP3 is responsible for aPKC activation.
FIGURE 2.
Effects of PIP3 and PIP2 on activity and phosphorylation of aPKC. aPKC was immunoprecipitated from lysates of livers harvested from basal (i.e. not insulin-stimulated) mice, and assayed in the presence of increasing concentrations of PIP3 (white columns) or PIP2 (black columns). Data are mean ± S.E. of triplicate determinations. Also shown are representative immunoblots of alterations in autophosphorylation (phospho-thr-563/560-PKC-λ/ζ).
Effects of Mutagenesis of Pseudosubstrate Arginine Residues versus DAG Pocket Arginine Residues on the Activity of His6-tagged Forms of PKC-ι: Plasmid Studies
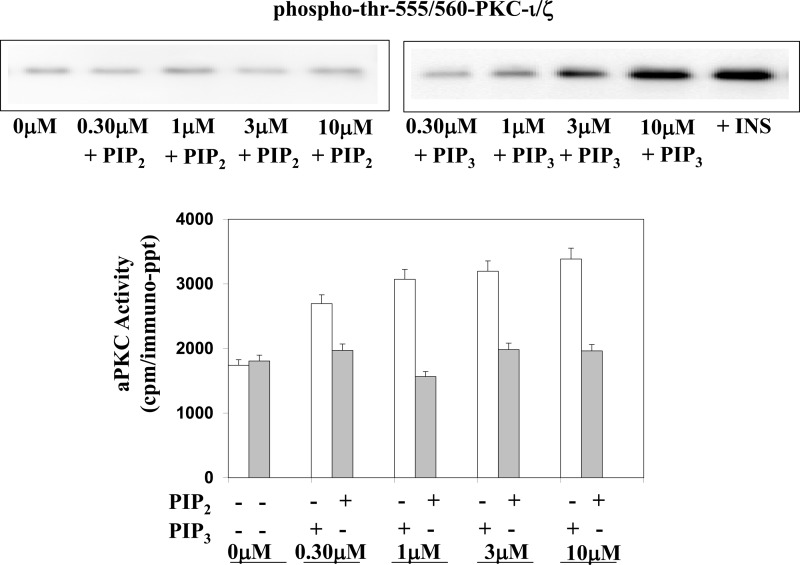
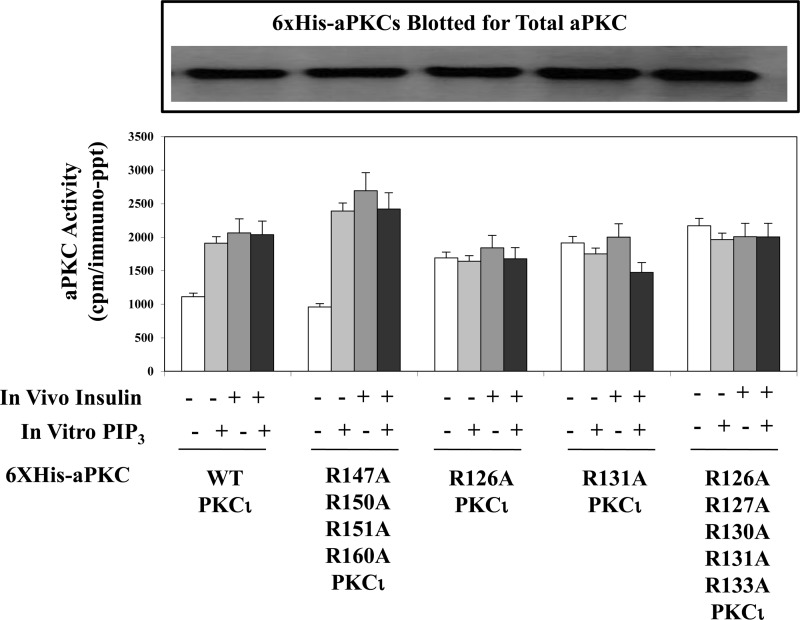
We questioned whether the cluster of five basic arginine residues common to pseudosubstrate sequences of all aPKCs ζ/λ, viz. the nearly identical sequences of SIYRRGARRWRKLYCANG for PKC-ι/λ and SIYRRGARRWRKLYRANG for PKC-ζ, may be important for maintaining aPKCs in an inactive state and for activation by PIP3. For this purpose, we thought it important to compare these five arginine residues of the PKC-ι/λ pseudosubstrate sequence to another cluster of nearby arginine residues in the regulatory domain of aPKC that are known to have “activating potential.” Accordingly, we chose four arginine residues that reportedly (13) form a positively charged ring that denies DAG access to a pocket containing a putative DAG-binding site located in the C1 region of the regulatory domain 14–27 residues beyond the pseudosubstrate sequence. Note that the four arginine residues that comprise this protective ring are present in the C1 regions of both PKC-ι/λ and PKC-ζ, therefore preventing activation of aPKC by DAG. In contrast, they are absent in conventional and novel PKCs, therefore, presumably, allowing an activation by DAG (13). Also note that the removal of these four arginine residues from aPKC has been reported to allow DAG activation of this mutagenized aPKC (13). Therefore, we mutated arginine residues Arg-126, Arg-127, Arg-130, Arg-131, and Arg-133 in the pseudosubstrate region and arginine residues Arg-147, Arg-150, Arg-151, and Arg-160 in the DAG pocket ring area of His6-tagged PKC-ι to neutral alanine residues. After transfection and expression of plasmids containing these mutant constructs and wild-type His6-tagged PKC-ι in 3T3/L1 adipocytes, the wild-type and mutated forms of His6-tagged PKC-ι were harvested on Ni-NTA-coated beads, eluted, and assayed for aPKC activity. As seen in the representative immunoblot shown in Fig. 3 and as confirmed by the finding of similar maximal kinase activities, it is clear that comparable amounts of enzyme were recovered in the harvested aPKCs and assayed subsequently.
FIGURE 3.
Effects of mutation of arginine residues in the pseudosubstrate sequence of PKC-ι (R126A, R127A, R130A, R131A, and R133A) versus mutation of arginine residues contained within a ring surrounding a putative diacylglycerol activation pocket of PKC-ι (R147A, R150A, R151A, and R160A) on resting/basal aPKC activity and its activation by insulin in vivo and PIP3in vitro. Plasmids encoding WT and mutated forms of His6-tagged PKC-ι were transfected into 3T3/L1 adipocytes, and, after allowing 48–72 h for expression, cells were treated for 30 min with 100 nm insulin or left untreated. After incubation, WT and mutated forms of His6-tagged PKC-ι were adsorbed from cell lysates onto Ni-NTA beads from which the His6-tagged aPKC was eluted and assayed for aPKC activity with or without maximally effective 10 μm PIP3 as indicated. Immunoblot analysis shows nearly equal expression of the wild-type and mutated forms of His6-tagged PKC-ι as recovered in eluates of Ni-NTA beads. Data are mean ± S.E. of duplicate incubations in a representative experiment. Comparable results were obtained in two separate experiments.
Following mutation of the five arginine residues of the aPKC pseudosubstrate sequence, the resultant mutant, His6-R126A/R127A/R130A/R131A/R133A-PKC-ι, was constitutively active, as judged by apparently maximal increases in “basal” activity and the failure of either insulin treatment in intact 3T3/L1 adipocytes or PIP3 in vitro (i.e. added directly to the enzyme assays) to further activate this mutant PKC-ι (Fig. 3). Moreover, mutagenesis of even a single arginine residue at either Arg-126 or Arg-131 (i.e. sites that could flank a central amino acid sequence that can harbor threonine or serine residues that can be phosphorylated) similarly yielded constitutively active PKC-ι mutants (Fig. 3). In marked contrast, mutagenesis of all four arginine residues in the ring surrounding the pocket containing a putative DAG activation site, i.e. His6-R147A/R150A/R151A/R160A-PKC-ι, had no effect on either the basal aPKC activity of this mutant or on the ability of either insulin in intact cells or PIP3 in vitro to activate this mutant (Fig. 3).
Effects of Mutagenesis of Arginine Residues in the Pseudosubstrate versus DAG Pocket Sequences on Kinase and Functional Activities of His6-tagged Forms of PKC-ι: Adenovirus Studies
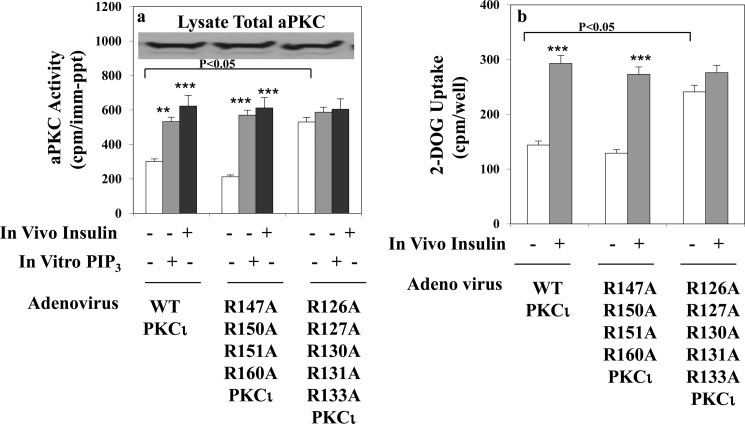
To examine the functional consequences in intact adipocytes of alterations in arginine residues in the pseudosubstrate sequence, compared with alterations of arginine residues in the ring area surrounding the putative DAG activation site, the plasmid constructs described above were placed into adenoviruses and inserted into 3T3/L1 adipocytes. In adipocytes infected with either an adenovirus encoding wild-type His6-tagged PKC-ι or an adenovirus encoding His6-tagged-PKC-ι mutated in the DAG pocket ring, i.e. the R147A/R150A/R151A/R160A mutant, insulin in intact adipocytes and PIP3 in vitro provoked 2- to 3-fold increases in total immunoprecipitable aPKC activity (Fig. 4a). Moreover, insulin provoked comparable 2- to 3-fold increases in glucose transport in intact adipocytes (Fig. 4b). In contrast, in adipocytes infected with an adenovirus encoding His6-tagged PKC-ι mutated in the pseudosubstrate sequence, i.e. the R126A/R127A/R130A/R131A/R133A mutant, basal aPKC activity (Fig. 4a) and glucose transport activity (Fig. 4b) were increased constitutively to the same level as that provoked by insulin in adipocytes infected with wild-type His6-tagged PKC-ι, and the addition of insulin in intact adipocytes or PIP3 in vitro was without further effect on either total immunoprecipitable aPKC activity (Fig. 4a) or glucose transport (Fig. 4b).
FIGURE 4.
Effects of mutation of arginine residues in the pseudosubstrate sequence of PKC-ι (R126A, R127A, R130A, R131A, and R133) versus mutation of arginine residues contained within a ring surrounding a putative diacylglycerol activation pocket of PKC-ι (R147A, R150A, R151A, and R160A) on resting/basal total cellular aPKC enzyme activity and its activation by insulin in vivo or PIP3in vitro (a) and insulin-stimulated glucose transport in intact 3T3/L1 adipocytes (b). a and b, cells were infected with multiplicity of 10 adenovirus-expressing WT or the indicated mutant forms of PKC-ι, and, after allowing 72 h for expression, the cells were treated for 30 min with 100 nm insulin or left untreated. b, cells were incubated for another 5 min, during which [3H]2-deoxyglucose uptake was measured (see “Experimental Procedures”). a, following incubation, total cellular aPKC was immunoprecipitated from cell lysates with anti-PKC-λ/ζ antiserum, and aPKC activity was measured with or without the addition of maximally effective10 μm PIP3 as indicated. Data are mean ± S.E. of four determinations. As shown in the immunoblot analysis, total cellular aPKC levels were comparable in adipocytes expressing WT and mutant forms of PKC-ι. Note that 3T3/L1 adipocytes only contain mouse PKC-ι, which is 98% homologous to human PKC-ι. **, p < 0.01; ***, p < 0.001.
PIP3 Reverses Exogenous Pseudosubstrate-induced Inhibition of Constitutively Active His6-R126A/R127A/R130A/R131A/R133A-PKC-ι
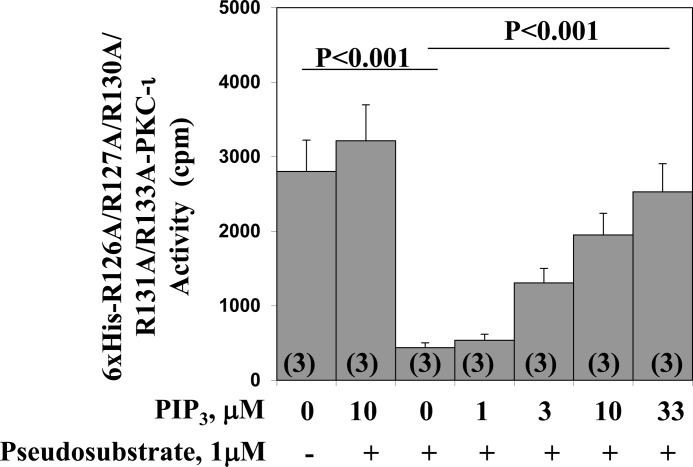
As shown in Fig. 3, PIP3 had no effect on the kinase activity of the His6-R126A/R127A/R130A/R131A/R133A-PKC-ι mutant used in Fig. 5, which was constitutively active because of the replacement of all five arginine residues in the endogenous pseudosubstrate sequence. However, with the addition of exogenous pseudosubstrate to incubation of the His6-R126A/R127A/R130A/R131A/R133A-PKC-ι mutant, the kinase activity of this mutant was reduced markedly (Fig. 5). Therefore, it was particularly interesting to find that, in this exogenous pseudosubstrate-inhibited state, the addition of PIP3 led to a dose-dependent activation to levels approaching that of the original constitutive mutant (Fig. 5).
FIGURE 5.
Dose-related stimulatory effects of PIP3 on pseudosubstrate-inhibited activity of constitutively active PKC-ι in which five arginine residues in the pseudosubstrate sequence were replaced by alanine residues. The constitutive mutant form of PKC-ι, His6-126A/R127A/R130A/R131A/R133-PKC-ι, was harvested from plasmid-transfected 3T3/L1 adipocytes as described in Fig. 3 and incubated with or without a maximally effective concentration of pseudosubstrate inhibitor (1 μm), the indicated concentrations of PIP3, and other components of the aPKC assay. Data are mean ± S.E. of the number of determinations shown in parentheses.
PIP3 Reverses Exogenous Pseudosubstrate-induced Inhibition of Full-length Native Forms of Mouse Liver aPKC
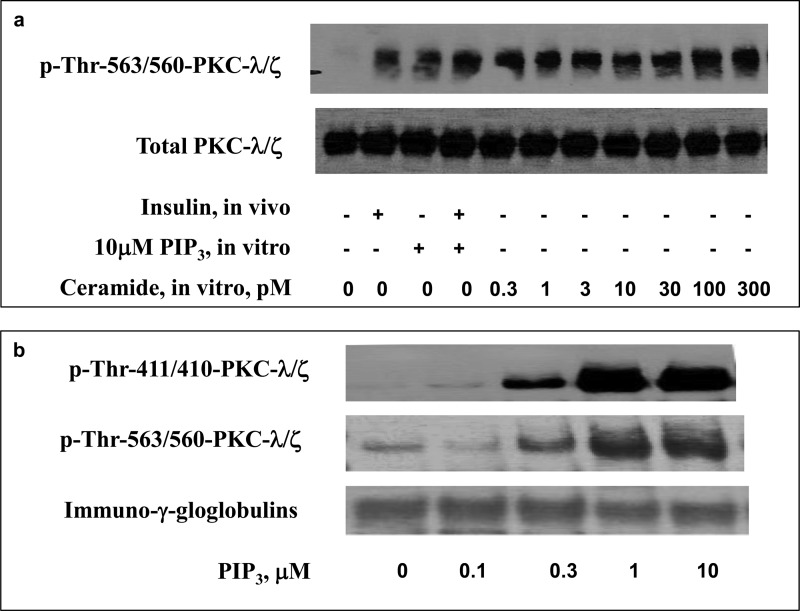
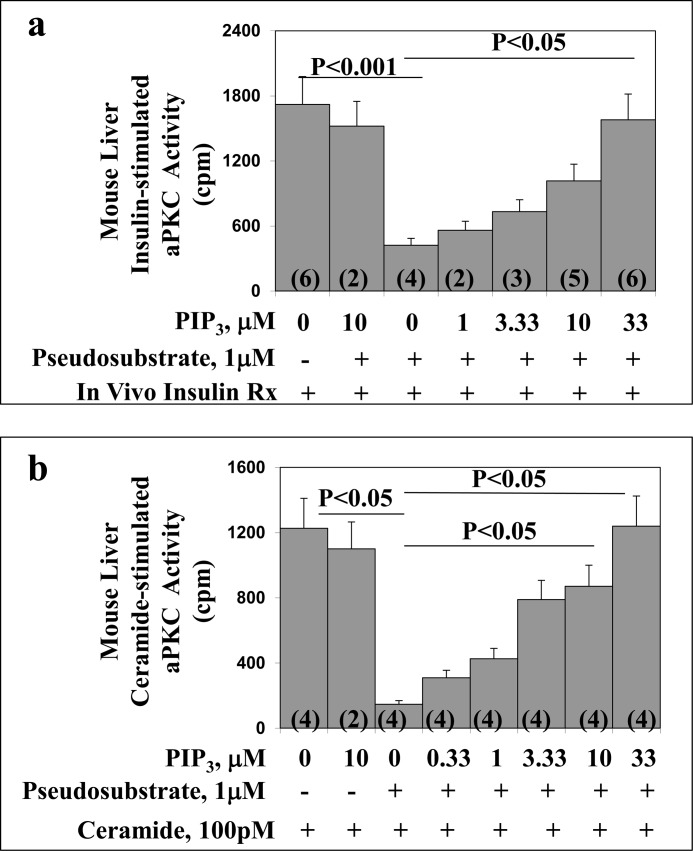
In addition to using PIP3 to reverse the inhibitory effects of exogenous pseudosubstrate on the His6-R126A/R127A/R130A/R131A/R133A-PKC-ι mutant, we employed a comparable approach with activated forms of native aPKC immunoprecipitated from lysates of mouse liver (which contains comparable amounts of PKC-λ and PKC-ζ) activated in vivo by insulin or in vitro by direct addition of 100 pm ceramide. As seen in Fig. 6, insulin treatment in vivo provoked a strong increase in phosphorylation of the aPKC autophosphorylation site, Thr-563/560-PKC-λ/ζ, that was comparable with that elicited by direct addition of PIP3 to the assay. Addition of PIP3 did not elicit a significant increase in auto(trans)phosphorylation above that induced by insulin alone. Similarly, low to high picomolar concentrations of ceramide provoked increases in phosphorylation of Thr-563/560-PKC-λ/ζ comparable with that elicited by insulin and/or PIP3 (Fig. 6). Further evidence that aPKC was maximally or near maximally activated by these insulin and ceramide treatments was confirmed by the finding that the addition of exogenous PIP3 in vitro (in the absence of added pseudosubstrate) did not provoke further increases in aPKC activity in aPKC stimulated with either insulin in vivo or ceramide in vitro (Fig. 7). In addition to increasing phosphorylation at the auto(trans)phosphorylation site, PIP3 increased phosphorylation of the activation loop site thr-411/410-PKC-λ/ζ (Fig. 6). Of the greatest importance was the finding that, when these activated mouse liver aPKCs were incubated with exogenous pseudosubstrate, their kinase activities were suppressed markedly and that, regardless of whether insulin or ceramide was used to activate this aPKC, the inhibitory effect of pseudosubstrate on the stimulated aPKC was largely reversed by PIP3 in a dose-dependent manner (Fig. 7).
FIGURE 6.
Activation of mouse liver aPKC by treatment with insulin in vivo and/or PIP3 and ceramide in vitro (a) and by PIP3in vitro (b). a, where indicated, mice were treated with vehicle or insulin (1 unit/kg body weight) intraperitoneally 15 min before killing. Liver lysates were obtained, and aPKC was collected by immunoprecipitation and incubated with the indicated concentrations of ceramide and PIP3 and other components of the aPKC assay. b, aPKCs were immunoprecipitated from liver lysates of basal/untreated mice and incubated with the indicated concentrations of PIP3 and other components of the aPKC assay. After incubation, reaction mixtures were subjected to Western blot analysis for the indicated phosphoproteins and proteins. Representative blots are shown.
FIGURE 7.
Dose-related stimulatory effects of PIP3 on pseudosubstrate-inhibited activity of mouse liver aPKC activated in vivo by insulin (panel a) or in vitro by ceramide (panel b). In panel a, liver was obtained from mice treated in vivo with insulin (1units/kg body weight) given intraperitoneally 15 min before killing. Rx, treatment. In panel b, liver was obtained from basal/untreated mice and activated during subsequent in vitro assay by addition of maximally effective 100 pm ceramide-16:0. In both cases, total aPKC was immunoprecipitated and assayed in the presence of maximally effective 1 μm pseudosubstrate and indicated concentrations of PIP3. Values are mean ± S.E. of the number of determinations shown in parentheses.
PIP3 Reverses Pseudosubstrate-induced Inhibition of the Catalytic Domain of the Truncated Δ1–247-T410A/T560A-PKC-ζ Mutant
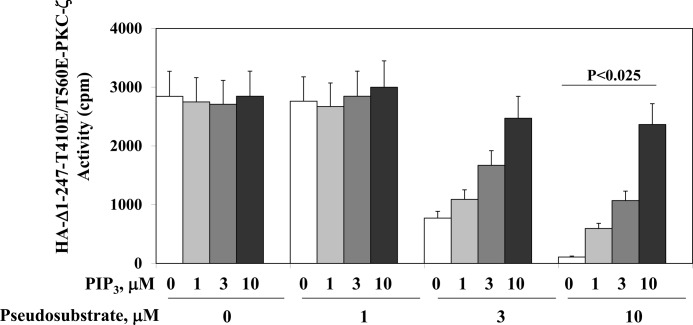
In addition to using the full-length aPKCs forms described above, we also used a form of aPKC that would not only be fully active in the absence of an activating ligand that operates through the regulatory domain but would also be resistant to activation by loop or autophosphorylation. Accordingly, we used a truncated (Δ1–247) form of aPKC that not only lacks the entire regulatory domain, in which extrinsic activators such as PIP3 are thought to operate, but that, additionally, contains T410E and T560E; i.e. glutamate “PO4-mimicking/activating” mutations in the catalytic domain. Use of this construct allowed us to be more certain that the inhibition of aPKC activity afforded by the addition of exogenous pseudosubstrate was most likely attributable to its binding to a site in the catalytic domain, presumably the substrate-binding site, and that, therefore, the PIP3-dependent reversal of the inhibitory action of exogenous pseudosubstrate was most likely attributable to the displacement of the pseudosubstrate from the substrate-binding site.
As expected, the Δ1–247/T410E/T560E-PKC-ζ mutant was constitutively active and could not be activated by PIP3. On the other hand, this mutant was inhibited dose-dependently by exogenous pseudosubstrate (Fig. 8). Most importantly, PIP3 reversed the inhibitory effects of exogenous pseudosubstrate in a dose-dependent manner (Fig. 8).
FIGURE 8.
Dose-related stimulatory effects of PIP3 on pseudosubstrate-inhibited activity of a truncated, glutamate-pseudophosphorylated, constitutively active PKC-ζ. 3T3/L1 adipocytes were grown for 72 h with a plasmid encoding HA-tagged Δ1–247-T410E/T560E-PKC-ζ, and then the PKC-ζ mutant was harvested with anti-HA antiserum and incubated with increasing concentrations of pseudosubstrate to diminish activity of the constitutive Δ1–247-T410E/T560E-PKC-ζ mutant and increasing concentrations of PIP3 to counteract the inhibitory effects of the pseudosubstrate. Data are mean ± S.E. of two to three determinations.
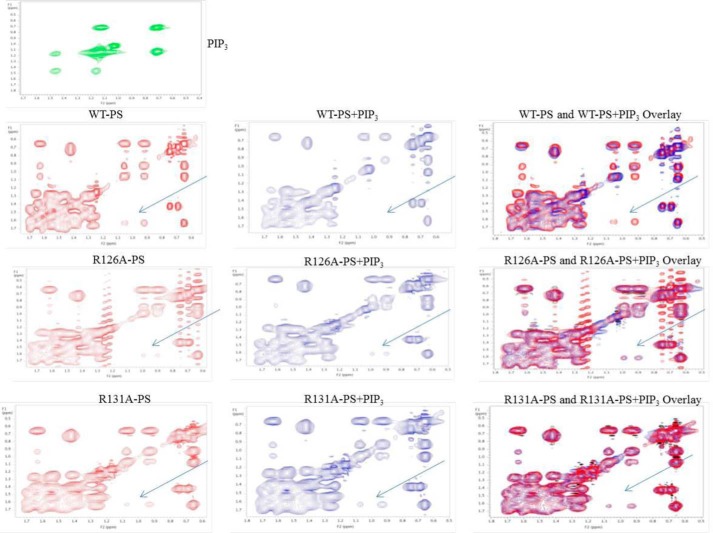
PIP3 Binding to Isolated Pseudosubstrate and Lack Thereof to Arginine-Substituted (R126A and R131A) Pseudosubstrates: NMR Studies
We used NMR to show direct binding of PIP3 to the isolated pseudosubstrate peptide. As seen in Fig. 9, the native pseudosubstrate sequence of PKC-ι/λ, SIYRRGARRWRKLYCANG, yielded an NMR spectrum that was altered (arrows) by PIP3, indicating binding of PIP3 to this pseudosubstrate sequence. In contrast, using pseudosubstrate sequences in which either arginine 126 or arginine 131 was replaced by alanine, i.e. SIYARGARRWRKLYCANG and SIYRRGARAWRKLYCANG, the ability of PIP3 to alter the NMR spectrum was lost (Fig. 9). These findings suggested that both arginine 126 and arginine 131 were required for binding of PIP3 to the pseudosubstrate. This dovetails with the fact that replacement of either arginine 126 or arginine 131 with alanine in the pseudosubstrate led to constitutive activation of aPKC and loss of the ability of PIP3 to cause further activation, as shown above.
FIGURE 9.
Shown are the effects of PIP3 on the NMR spectra of WT pseudosubstrate (PS) peptide (SIYRRGARRWRKLYCANG) and arginine-replaced pseudosubstrate peptide sequences (SIYARGARRWRKLYCANG and SIYRRGARAWRKLYCANG); i.e. alanine substituted for arginine at Arg-126 and Arg-131 (R126A and R131A). Peptides were analyzed without (left column) or without PIP3 (center column). The right column shows merged spectra. Also shown is the NMR spectrum of PIP3.
DISCUSSION
These findings show that arginine residues in the pseudosubstrate sequence play a key role in maintaining aPKC in an inactive state under basal conditions. Indeed, the fact that removal of even a single arginine residue at position Arg-126 or Arg-131 led to full constitutive activation suggests that the maintenance of autoinhibition by a suitably tight interaction between the pseudosubstrate in the regulatory domain and the substrate-binding site in the catalytic domain requires the participation of at least two arginine contact sites that flank a central amino acid sequence that, in a substrate, could harbor a phosphorylation site. In this regard, note that placement of an acidic PO4-mimicking aspartate residue between the arginine residues Arg-126 and Arg-131, i.e. mimicking a phosphorylated aPKC substrate that should theoretically abrogate binding of the mutated pseudosubstrate to the substrate-binding site, does in fact lead to constitutive activation of PKC-ζ (20).
The specificity of arginine residues in the pseudosubstrate sequence as critical regulators of aPKC activity is suggested by the finding that the loss of all four arginine residues that form a functional ring that reportedly (13) denies DAG access to a potential activation site had no effect on basal aPKC activity or its activation in vivo by insulin, which increases endogenous PIP3 production, or its activation in vitro by direct addition of exogenous PIP3 to the assay. We did not evaluate other arginine sites to see whether they influence aPKC activity because it appeared that the arginine residues in the pseudosubstrate sequence were sufficient to satisfy the requirements for both autoinhibition and, as discussed below, activation of aPKC by PIP3.
In addition to providing evidence that arginine residues in the pseudosubstrate are required for autoinhibition of aPKC, these findings provide strong evidence that PIP3 activates aPKC by counteracting arginine-dependent autoinhibitory effects of the pseudosubstrate. The most likely explanation for this activating effect is that the acidic D3-PO4 group of PIP3 binds to basic arginine residues in the pseudosubstrate sequence and causes their dissociation from acidic residues in the substrate-binding site. The strongest evidence for this suggestion was perhaps our findings in studies of the Δ1–247/T410E/T560E-PKC-ζ mutant in which PIP3 had no activating effect unless the activity of this construct was inhibited by addition of pseudosubstrate. In this pseudosubstrate-inhibited state, PIP3 was a very effective activator of this Δ1–247/T410E/T560E-PKC-ζ mutant, despite the absence of any regulatory domain components other than the added pseudosubstrate. The most reasonable conclusion for this finding is that the autoinhibitory pseudosubstrate is targeted by PIP3.
Further support for the idea that PIP3 activates aPKC by reversing pseudosubstrate-dependent autoinhibition was our finding that stimulatory effects of PIP3 were seen only in conjunction with pseudosubstrate-induced inhibition of the full-length His6-R126A/R127A/R130A/R131A/R133A-PKC-ι mutant, which is otherwise constitutively active as a result of loss of all five arginine residues in its pseudosubstrate sequence. In addition, the fact that similar stimulatory effects of PIP3 were seen following exogenous pseudosubstrate-induced inhibition of insulin- and ceramide-activated, full-length forms of mouse liver aPKC suggests that the findings seen with the two constitutively active mutants, viz. His6-R126A/R127A/R130A/R131A/R133A-PKC-ι and Δ1–247/T410E/T560E-PKC-ζ, were not particular to these mutants and could be seen in native aPKC forms. This confirmation seemed important because, for uncertain reasons, higher concentrations of pseudosubstrate were needed to inhibit the truncated Δ1–247/T410E/T560E-PKC-ζ as compared with activated full-length forms of aPKC.
It was interesting to find that the addition of PIP3 to the in vitro assay of aPKC immunoprecipitated from control (unstimulated) mouse liver provoked increases in the phosphorylation of the activation loop site, Thr-411/410-PKC-λ/ζ, and the auto(trans)phosphorylation site, Thr-563/560-PKC-λ/ζ, which are required for, and indicative of, activation (11, 12). This demonstrates that the allosteric alteration of aPKC (presumably molecular unfolding because of pseudosubstrate dissociation from the substrate-binding site) induced by PIP3 can precede and trigger these phosphorylations. In this regard, it is noteworthy that PDK1 is known to be present in aPKC immunoprecipitates (21).
It is interesting that findings in crystallographic and solution structure studies suggest that basic arginine and lysine residues within the PH domains of both Akt and PDK1 bind to the acidic D3-PO4 groups of PIP3 and phosphatidylinositol-3,4-(PO4)2 (22, 23). This interaction between acidic phosphoinositides and basic residues in the PH domains of Akt and PDK1 is similar to what appears to occur during PIP3-induced aPKC activation, viz. an interaction between the acidic D3-PO4 group of PIP3 and one or more of the basic arginine residues in the pseudosubstrate sequence. In this regard, it is noteworthy that phosphatidylinositol-4,5-(PO4)2 is much less effective than PIP3 for activating Akt and aPKC (6, 11, 12) and that it seems likely that the D3-PO4 group of PIP3 largely accounts for its activating properties.
Relative to these findings, Lopez-Garcia et al. (24) have suggested that, along with the attachment of the pseudosubstrate to the substrate-binding site, there is an additional mechanism for inhibition of aPKC activity that involves binding of the C1 region to the C-terminal PIF-pocket. In addition, Graybill et al. (25) have suggested that a region of C1 just beyond the pseudosubstrate sequence is required along with the pseudosubstrate for autoinhibition of aPKC and, interestingly, that this C1 region is targeted by Par-6 and, thereby, activates aPKC by a mechanism involving displacing the endogenous pseudosubstrate. Accordingly, it appears that there is more than one mechanism for activation of aPKC. By analogy, it may be hypothesized that the activation of aPKC by ceramide, details of which are currently uncertain but appear to involve direct binding to aPKC (26), is mediated by an interaction with a region of C1 that induces displacement of the pseudosubstrate. In any case, here we show that ceramide-induced activation can be overcome by addition of an exogenous pseudosubstrate-containing peptide and that this inhibitory effect of exogenous pseudosubstrate can be reversed by PIP3.
To summarize, these findings show that removal of arginine residues from the pseudosubstrate sequence of PKC-ι by site-directed mutagenesis leads to constitutive activation and loss of ability of PIP3 to further activate the mutagenized aPKC. These findings also show that, with addition of exogenous pseudosubstrate to inhibit the constitutively active form of PKC-ι that lacks all five arginine residues in the pseudosubstrate sequence, viz. His6-R126A/R127A/R130A/R131A/R133A-PKC-ι, fully activated forms of full-length native mouse liver aPKC, and a constitutively active, truncated mutant form of PKC-ζ that lacks all regulatory domain elements and, additionally, cannot be phosphorylated at loop or autophosphorylation sites, viz. Δ1–247/T410E/T560E-PKC-ζ, the addition of PIP3 resulted in nearly full activation. These findings suggest that basic arginine residues in the pseudosubstrate sequence of the regulatory domain, by presumably binding to acidic residues in the substrate-binding site in the catalytic domain, maintain aPKC in a folded, inactive state and, moreover, are targeted by PIP3 for displacement from the substrate-binding site during enzyme activation.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1DK 065969-09 (to R. V. F.). This work was also supported by funds from the Department of Veterans Affairs Merit Review Program (to R. V. F.).
- aPKC
- atypical PKC
- PIP3
- phosphatidylinositol 3,4,5-(PO4)3
- PIP2
- phosphatidylinositol-4,5-(PO4)2
- PI
- phosphatidylinositol
- PH
- pleckstrin homology
- DAG
- diacylglycerol
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Fields A. P., Frederick L. A., Regala R. P. (2007) Targeting the oncogenic protein kinase C ι for the treatment of cancer. Biochem. Soc. Trans. 23, 1996–2000 [DOI] [PubMed] [Google Scholar]
- 2. Beeson M., Sajan M. P., Dizon M., Grebenev D., Gomez-Daspet J., Miura A., Kanoh Y., Powe J., Bandyopadhyay G., Standaert M. L., Farese R. V. (2003) Activation of protein kinase C-ζ by insulin and phosphatidylinositol-3,4,5-(PO4)3 is defective in muscle in type 2 diabetes and impaired glucose tolerance: amelioration by rosiglitazone and exercise. Diabetes 52, 1926–1934 [DOI] [PubMed] [Google Scholar]
- 3. Kim Y.-B., Kotani K., Ciaraldi T. P., Henry R. R., Kahn B. B. (2003) Insulin-stimulated protein kinase C-λ/ζ activity is reduced in skeletal muscle of humans with obesity and type 2 diabetes: reversal with weight reduction. Diabetes 52, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 4. Sajan M. P., Standaert M. L., Miura A., Bandyopadhyay G., Vollenweider P., Franklin D. M, Lea-Currie R., Farese R. V. (2004) Impaired activation of protein kinase C-ζ by insulin and phosphatidylinositol-3,4,5-(PO4)3 in cultured pre-adipocyte-derived adipocytes and myotubes of obese subjects. J. Clin. Endocrinol. Metab. 89, 3994–3998 [DOI] [PubMed] [Google Scholar]
- 5. Sajan M. P., Farese R. V. (2012) Insulin signalling in hepatocytes of type 2 diabetic humans: excessive expression and activity of PKC-ι and dependent processes and reversal by PKC-ι inhibitors. Diabetologia 55, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Standaert M. L., Galloway L., Karnam P., Bandyopadhyay G., Moscat J., Farese R. V. (1997) PKC-ζ as a downstream effector of PI 3-kinase during insulin stimulation in rat adipocytes: potential role in glucose transport. J. Biol. Chem. 272, 30075–30082 [DOI] [PubMed] [Google Scholar]
- 7. Liu Q., Ning W., Dantzer R., Freund G. G., Kelley K. W. (1998) Activation of protein kinase C-ζ and phosphatidylinositol 3-kinase and promotion of macrophage differentiation by insulin-like growth factor-1. J. Immunol. 160, 1393–13401 [PubMed] [Google Scholar]
- 8. Stokoe D., Stephens L. R, Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277, 567–570 [DOI] [PubMed] [Google Scholar]
- 9. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylate and activates protein kinase B α. Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 10. Hresko R. C., Mueckler M. (2005) mTOR.RICTOR is the serine473 kinase for Akt/protein kinase B in 3T3/L1 adipocytes. J. Biol. Chem. 280, 40406–40416 [DOI] [PubMed] [Google Scholar]
- 11. Standaert M. L., Bandyopadhyay G., Kanoh Y., Sajan M. P., Farese R.V. (2001) Insulin and PIP3 activate PKC-ζ by mechanisms that are both dependent and independent of phosphorylation of activation loop (T410) and autophosphorylation (T560) Sites. Biochemistry 40, 249–255 [DOI] [PubMed] [Google Scholar]
- 12. Standaert M. L., Bandyopadhyay G., Perez L., Price D., Galloway L., Poklepovic A., Sajan M. P., Cenni V., Sirri A., Moscat J., Toker A., Farese R. V. (1999) Insulin activates PKC-ζ and PKC-λ by an autophosphorylation-dependent mechanism and stimulates their translocation to GLUT4 vesicles and other membrane fractions in rat adipocytes. J. Biol. Chem. 274, 25308–25316 [DOI] [PubMed] [Google Scholar]
- 13. Pu Y., Peach M. L., Garfield S. H., Wincovitch S., Marquez V. E., Blumberg P. M. (2006) Effects on ligand interaction and membrane translocation of the positively charged arginine residues situated along the C1 domain binding cleft in the atypical PKC isoforms. J. Biol. Chem. 281, 33773–33788 [DOI] [PubMed] [Google Scholar]
- 14. Sajan M. P., Rivas J., Li P., Standaert M. L., Farese R. V. (2006) Repletion of atypical protein kinase C following RNAi-mediated depletion restores insulin-stimulated glucose transport. J. Biol. Chem. 281, 17466–17473 [DOI] [PubMed] [Google Scholar]
- 15. Sajan M. P., Standaert M. L., Rivas J., Miura A., Kanoh Y., Soto J., Taniguchi C. M., Kahn C. R., Farese R. V. (2009) Role of atypical protein kinase C in activation of sterol regulatory element binding protein-1c and nuclear factor κ B (NFκB) in liver of rodents used as model of diabetes, and relationships to hyperlipidaemia and insulin resistance. Diabetologia 52, 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Standaert M. L, Sajan M. P., Miura A., Kanoh Y., Chen H. C., Farese R. V., Jr., Farese R. V. (2004) Insulin-induced activation of atypical protein kinase C, but not protein kinase B, is maintained in diabetic (ob/ob and Goto-Kakazaki) liver: contrasting insulin signaling patterns in liver versus muscle define phenotypes of type 2 diabetic and high-fat-induced insulin-resistant states. J. Biol. Chem. 279, 24929–24934 [DOI] [PubMed] [Google Scholar]
- 17. Bandyopadhyay G., Standaert M. L., Zhao L., Yu B., Avignon A., Galloway L., Karnam P., Moscat J., Farese R. V. (1997) Activation of Protein Kinase C (α, β and ζ) by insulin in 3T3/L1 cells. J. Biol. Chem. 272, 2551–2598 [DOI] [PubMed] [Google Scholar]
- 18. Hannun Y. A., Loomis C. R., Bell R. M. (1985) Activation of protein kinase C by Triton-X mixed micelles containing diacylglycerol and phosphatidylserine. J. Biol. Chem. 260, 10039–10043 [PubMed] [Google Scholar]
- 19. Hannun Y. A., Loomis C. R., Bell R. M. (1986) Protein kinase C activation in mixed micelles. J. Biol. Chem. 261, 7184–7190 [PubMed] [Google Scholar]
- 20. Bandyopadhyay G., Standaert M. L., Kikkawa U., Ono Y., Moscat J., Farese R. V. (1999) Effects of transiently expressed atypical (ζ,λ), conventional (α,β) and novel (δ,ϵ) protein kinase C isoforms on insulin-stimulated translocation of epitope-tagged GLUT4 glucose transporters in rat adipocytes: specific interchangeable effects of protein kinases C-ζ and C-l. Biochem. J. 337, 461–470 [PMC free article] [PubMed] [Google Scholar]
- 21. Standaert M. L., Ortmeyer H. K., Sajan M. P., Kanoh Y., Bandyopadhyay G., Hansen B. C., Farese R. V. (2002) Skeletal muscle insulin resistance in obesity-associated type 2 diabetes in monkeys is linked to a defect in insulin activation of protein kinase C-ζ/λ/ι. Diabetes 51, 2936–2943 [DOI] [PubMed] [Google Scholar]
- 22. Auguin D., Barthe P., Augé-Sénégas M.-T., Stern M.-H., Noguchi M., Roumestand C. (2004) Solution structure and backbone dynamics of the pleckstrin homology domain of the human protein kinase B (PKB/Akt): interaction with inositol phosphates. J. Biomol. NMR 28, 137–155 [DOI] [PubMed] [Google Scholar]
- 23. Komander D., Fairservice A., Deak M., Kular G. S., Prescott A. R., Peter Downes C., Safrany S. T., Alessi D. R., van Aalten D. M. (2004) Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 23, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez-Garcia L. A., Schulze J. O., Fröhner W., Zhang H., Süss E., Weber N., Navratil J., Amon S., Hindie V., Zeuzem S., Jørgensen T. J., Alzari P. M., Neimanis S., Engel M., Biondi R. M. (2011) Allosteric regulation of protein kinase PKC ζ by the N-terminal C1 domain and small compounds to the PIF-pocket. Chem. Biol. 18, 1463–1473 [DOI] [PubMed] [Google Scholar]
- 25. Graybill C., Wee B., Atwood S. X., Prehoda K. E. (2012) Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J. Biol. Chem. 287, 21003–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Müller G., Ayoub M., Storz P., Rennecke J., Fabbro D., Pfizenmaier K. (1995) PKC ζ is a molecular switch in signal transduction of TNF-α, bifunctionally regulated by ceramide and arachidonic acid. EMBO J. 14, 1961–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]