Background: miR-21 is overexpressed in many human cancers, including glioblastoma.
Results: Insulin-like growth factor (IGF)-binding protein-3 (IGFBP3) is a novel miR-21 target gene and inhibits gliomagenesis in vitro and in vivo.
Conclusion: miR-21 down-regulates IGFBP3, which acts as a tumor suppressor in human glioblastoma.
Significance: IGFBP3 may have promise as a therapeutic target and prognostic marker for glioblastoma.
Keywords: Brain Tumor, Glioblastoma, Insulin-like Growth Factor (IGF), MicroRNA (MiRNA), Tumor Suppressor Gene, IGFBP3
Abstract
Despite advances in surgery, imaging, chemotherapy, and radiation, patients with glioblastoma multiforme (GBM), the most common histological subtype of glioma, have an especially dismal prognosis; >70% of GBM patients die within 2 years of diagnosis. In many human cancers, the microRNA miR-21 is overexpressed, and accumulating evidence indicates that it functions as an oncogene. Here, we report that miR-21 is overexpressed in human GBM cell lines and tumor tissue. Moreover, miR-21 expression in GBM patient samples is inversely correlated with patient survival. Knockdown of miR-21 in GBM cells inhibited cell proliferation in vitro and markedly inhibited tumor formation in vivo. A number of known miR-21 targets have been identified previously. By microarray analysis, we identified and validated insulin-like growth factor (IGF)-binding protein-3 (IGFBP3) as a novel miR-21 target gene. Overexpression of IGFBP3 in glioma cells inhibited cell proliferation in vitro and inhibited tumor formation of glioma xenografts in vivo. The critical role that IGFBP3 plays in miR-21-mediated actions was demonstrated by a rescue experiment, in which IGFBP3 knockdown in miR-21KD glioblastoma cells restored tumorigenesis. Examination of tumors from GBM patients showed that there was an inverse relationship between IGFBP3 and miR-21 expression and that increased IGFBP3 expression correlated with better patient survival. Our results identify IGFBP3 as a novel miR-21 target gene in glioblastoma and suggest that the oncogenic miRNA miR-21 down-regulates the expression of IGFBP3, which acts as a tumor suppressor in human glioblastoma.
Introduction
Brain tumors represent an important cause of cancer-related morbidity and mortality in the United States, with malignant gliomas being highly aggressive and difficult to treat (1). Although they rarely metastasize, malignant gliomas are invasive, vascular tumors with extensive areas of necrosis and hypoxia. Glioblastoma multiforme (GBM)2 is the most common histological subtype of glioma in adults. The median survival for GBM patients is ∼12 to 15 months and has remained this dismal for decades despite treatment advances (1). The primary treatment for GBM is surgical resection, followed by chemotherapy and radiation therapy, but this treatment provides little improvement in the disease course and outcome for the patients, and >90% of patients die within 3 years of diagnosis (2). Patients with recurrent GBM have an even bleaker prognosis (3). Thus, treatment of patients with GBM is a significant clinical problem requiring novel therapeutic approaches.
MicroRNAs (miRNAs) are an abundant family of small RNAs (∼22 nucleotides) that fine-tune the expression of genes implicated in fundamental biological processes such as differentiation, proliferation, and apoptosis (4, 5). miRNAs regulate the expression of multiple targets by binding to the 3′-untranslated regions of target mRNAs to promote mRNA degradation at a post-transcriptional level or by inhibiting the initiation of translation. miR-21 (also denoted as miR-21-5p) is frequently overexpressed in various human tumors and appears to play an important role in the oncogenic process as indicated by its association with high proliferation, low apoptosis, high invasion, and metastatic potential (6–12). Moreover, a number of validated miR-21 targets play an important role in the oncogenic process (13). We recently found that miR-21 regulates tumorigenesis in melanoma and prostate cancer models (14, 15).
In the present study, we examined the expression of miR-21 in human glioma cell lines and in human glioma tissue and found that miR-21 was overexpressed. Analysis of human glioma data in The Cancer Genome Atlas (TCGA) showed that miR-21 expression inversely correlated with patient survival. Knockdown of miR-21 in GBM cells markedly inhibited tumor formation when injected subcutaneously as compared with control GBM cells. Although a number of known miR-21 targets were identified by microarray analysis, IGFBP3 was identified and validated as a novel miR-21 target gene. IGFBP3 is an insulin-like growth factor (IGF) binding protein. The IGF signaling pathway plays an important role in growth, development, and maintenance of homeostasis in normal cells. Accumulating evidence suggests that disruption of the IGF system has major implications for growth retardation, atherosclerosis, insulin resistance, and cancer, including malignant glioma (16). The involvement of IGFBPs in cancer varies depending on the type of malignancy. For example, IGFBP2, IGFBP3, and IGFBP5 are associated with GBM more commonly than other brain tumors, suggesting they may play roles in glioma progression (17, 18). Overexpression of IGFBP3 in glioma cells inhibited cell proliferation and inhibited tumor formation of glioma xenografts. Examination of RNA expression from GBM patients showed that there was an inverse relationship between IGFBP3 and miR-21 expression and that IGFBP3 expression correlated with patient survival. Our results identify IGFBP3 as a novel miR-21 target gene in glioma and suggest that high expression of miR-21 down-regulates the expression of IGFBP3 that acts as a tumor suppressor in human glioma.
MATERIALS AND METHODS
Biological Reagents and Cell Cultures
The biological activity of recombinant human IFNα (IFNcon1, InterMune) was expressed in terms of international reference units/ml using the human NIH reference standard (19). Antibodies against the following proteins were used: ANP32A, SPROUTY2, PTEN, and actin (Santa Cruz Biotechnology, Santa Cruz, CA); PDCD4, BTG2, and IGFBP3 (Abcam, Cambridge, MA); and AKT, IGF-1R, phospho-AKT, and phospho-IGF-1R (Cell Signaling Technology, Boston, MA). U87 (American Type Culture Collection), MT330 (UTHSC Department of Neurosurgery), and SJ-G2 (St. Jude Children's Research Hospital) GBM cell lines were grown in DMEM containing 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) supplemented with penicillin (100 international units/ml) and streptomycin (100 μg/ml) at 37 °C with 5% CO2.
Gene Expression Analysis
Total RNA was isolated using RNeasy Mini kit (Qiagen) from empty vector and miR-21 knockdown (KD) cells, and miRNA expression profiling was conducted on the nCounter Analysis System (NanoString Technologies) using the human V2 miRNA assay kit, containing ∼800 human miRNA probe sets derived from the miRNA database miRNA assay kit. In addition, 3–5 (1-μm) curls were cut from brain biopsy specimens from ∼40 GBM patients, RNA isolated using the RecoverAllTM Total Nucleic Acid Isolation kit (Ambion Inc.), and the expression of a panel of ∼230 cancer-related human genes was determined. In brief, total RNA was mixed with pairs of capture and reporter probes and hybridized on the nCounter Prep Station, and purified complexes were measured on the nCounter digital analyzer. To account for differences in hybridization and purification, data were normalized to the average counts for all control spikes in each sample and analyzed with nSolver software. Gene expression was further quantified by quantitative real time PCR (qPCR) as described previously (14). For miRNA expression, total RNA (5 μg) was reverse-transcribed into first-strand cDNA, and 40 ng of cDNA was used as a template for the PCR reaction with a forward primer specific to the mature miRNA sequence as described previously (14). SYBR Green-based real-time PCR was performed on a Bio-Rad iCycler, and gene expression was normalized relative to U6 or β-actin expression for miRNA or mRNA, respectively.
Lentiviral Knockdown of miR-21 and IGFBP3 Expression and IGFBP3 Overexpression
To knockdown miR-21 expression, oligonucleotides against the mature sequence of miR-21 gene were cloned into the lentiviral vector pLenti-U6-pgk-puro and antagomiR-21 lentivirus produced as described previously (14). To knock down IGFBP3 expression, miR-21KD GBM cells were transduced with the IGFBP33 short hairpin RNA (shRNA)-miR lentiviral vector pGIPZ, which contains a human miR-30 hairpin-based shRNA-miR structure against IGFBP3 (OPEN Biosystems). To overexpress IGFBP3, we purchased the IGFBP3 expression clone pReceiver-lv105, which contains a full open reading frame of the human IGFBP3 gene (Genecopoeia). GBM cells transduced with antagomiR-21 or IGFBP3 were selected with puromycin, and after selection stable pools with expression levels knocked down by >75% (see Fig. 2A) were maintained in growth medium without puromycin.
FIGURE 2.
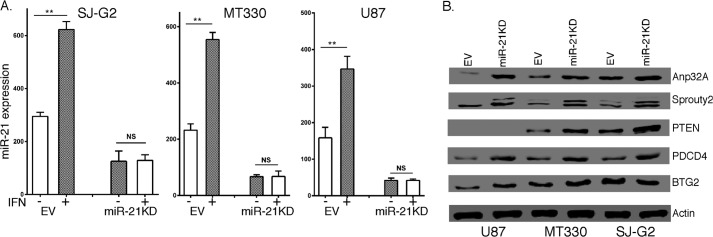
Effects of miR-21KD on IFN-inducibility and miR-21 target gene expression. Human glioma cells were transduced with antagomiR-21 lentivirus, and pools of stably KD cells (A) were treated with IFN (1000 international units/ml, for 5 h), total RNA was prepared, and miR-21 expression was determined by qPCR. B, cell lysates were prepared and immunoblotted as indicated for the expression of several known miR-21 targets. NS, not significant.
Immunoblot Analysis
Total cell lysates (25 μg) were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and immunoblotted with the indicated antibodies, followed by IRDye800CW goat anti-mouse IgG or IRDye680 goat anti-rabbit IgG (LI-COR Biosciences). Blots were visualized on an Odyssey Infrared Imaging System (LI-COR Biosciences).
Construction of Luciferase Reporter Gene Plasmids and Reporter Assays
The 3′-untranslated region (UTR) of IGFBP3 was amplified by PCR from genomic DNA of human 293T cells. After digestion with XhoI and BamHI, the PCR product was purified and cloned into pcDNA3.1-luc, resulting in the wild-type IGFBP3 reporter plasmid, pcDNA3.1-Luc-wtUTR. The mutant IGFBP3 reporter plasmid pcDNA3.1-Luc-muUTR was constructed by mutating the binding sites of miR-21 in the 3′-UTR of IGFBP3 using PCR based site-directed mutagenesis according to the manufacturer's instructions (Stratagene). The primers for amplifying the wild-type 3′-UTR were 5′-GATACTCGAGGGGGTAGGAGGGACAGAGAG-3′ and 5′-GCGGATCCAGCCATTCCTCCTTCCTGTT-3, and the primers for mutant construct were 5′-CAACTCAAGACGAAGCTTATTTCTGAGGAATTCCTCTTTAAAGGCAAAGCTTTATTTTCA-3 and 5′-GATGAAAATAAAGCTTTGCCTTTAAAGAGGAATTCCTCAGAAATAAGCTTCGTCTTGAGT-3′. Reporter gene binding assays were performed by co-transfection of 293T cells using wild-type and mutant reporter plasmids pcDNA3.1-Luc-wtUTR and pcDNA3.1-Luc-muUTR with miR-21 overexpressing (or knockdown) plasmid, respectively. pSV40-Renilla plasmid was co-transfected as an internal control. The ratio of luciferase and Renilla activities was determined at 24 h post-transfection using the Dual-Luciferase reporter gene kit (Promega).
Tumor Formation in Mice
All animal experiments were performed in accordance with a study protocol approved by the Institutional Animal Care and Use Committee of the University of Tennessee Health Science Center. GBM xenografts were established in 5-week-old male NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) by injection of cells (1 × 106) directly into the flanks (15). Tumors were measured weekly with a handheld caliper. At the end of treatment the animals were sacrificed, and the tumors were removed, weighed, and subjected to analysis by immunofluorescent staining on Zeiss LSM700 laser scanning confocal microscope or gene expression by qPCR.
TCGA Data Query
To examine the relationship between miR-21 and IGFBP3 expression in human GBM brain tissue, we queried the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaCancerDetails.jsp?diseaseType=GBM&diseaseName=Glioblastoma multiforme) for all GBM samples with level 3 miRNA (UNC_H-miRNA_8 × 15K) and gene (BI_HT_HG-U133A Array Data Set) expression data available, as well as the accompanying clinical data. The data set was filtered for samples having expression data for miR-21, IGFBP3, and clinical data, yielding a final set of 418 independent patient samples. Statistical analyses were performed using Graphpad Prism.
Statistical Analyses
At least three independent experiments were performed in duplicate, and data are presented as means ± S.D. Analysis of variance and post hoc least significant difference analysis or Student's t tests were performed. p values < 0.05 (*), 0.01 (**), and 0.001 (***) were considered statistically significant.
RESULTS
miR-21 Expression in GBM Cell Lines and Tumor Tissue
To examine miR-21 expression in various human tumor cell lines, total RNA was isolated from human cell lines representing GBM (U87, MT330, and SJ-G2), prostate cancer (DU145 and PC-3), and melanoma (SK-MEL188 and WM164) as well as from normal human skin fibroblasts. Expression of miR-21 was determined by qPCR. As shown in Fig. 1A, although basal miR-21 expression varied among the different tumor cell lines, miR-21 was more highly expressed in GBM lines as compared with other human cancer cell lines and normal fibroblasts. Furthermore, using a platelet-derived growth factor-driven mouse glioblastoma model (20), we found miR-21 was markedly overexpressed in tumor tissue as compared with normal brain tissue (Fig. 1B). We next used bioinformatics to analyze The Cancer Genome Atlas (TCGA) database to determine the relationship between glioma grade and miR-21 expression in patient samples (Fig. 1C). Statistically significant higher miR-21 expression was found in tumor tissue from GBM patients as compared with low-grade glioma. To determine whether there was a relationship between patient survival and GBM diagnosis, brain biopsy specimens from 36 GBM patients representing longer term (>2 years) and shorter term (<1 year) survival after diagnosis were obtained from the UTHSC Tissue Services Core, RNA was isolated, and miR-21 expression was determined. Despite the expected patient-to-patient variability, higher miR-21 expression was found in patients that survived <1 year (Fig. 1D). Taken together, these results indicate that miR-21 is overexpressed in GBM and that high miR-21 expression is associated with poor patient survival.
FIGURE 1.
High miR-21 expression is observed in GBM cell lines, and in vivo is associated with poor patient survival. miR-21 expression was determined by qPCR on total RNA extracted from human U87, MT330, SJ-G2 glioma, SKMEL188 and WM164 melanoma, DU145 and PC3 prostate cancer cell lines, and normal skin fibroblasts (HF) (A) or tumor and normal (norm) brain tissue obtained from a mouse model of human glioma (B) (20). Expression was normalized to U6 RNA expression, and the data represent the mean ± S.D. of at least three experiments performed in duplicate. C, miR-21 expression in the TCGA database of GBM and low-grade glioma (LGG) patient samples was compared. D, RNA extracted from GBM patient biopsies was assayed for miR-21 gene expression by qPCR (n = 3), and expression plotted as a function of patient survival after diagnosis (long term, >2 years; short term, <1 year).
Characterization of IGFBP3 as a miR-21 Target Gene
To study the biological function of miR-21 in GBM, GBM cell lines were transduced with antagomiR-21, and stable pools of cells were isolated in which miR-21 expression was knocked down by >75% (Fig. 2A). We previously showed that the cytokine IFN induced miR-21 expression in a variety of cell lines (14). IFN treatment (1000 international units/ml for 5 h) induced miR-21 expression in the human GBM cell lines, and miR-21KD abrogated IFN-induced miR-21 expression (Fig. 2A). In contrast, although IFN also induces miR-100 and miR-125 expression in GBM cells, miR-21KD had no effect on IFN induction of these miRNAs, demonstrating the specificity and selectivity of miR-21KD (data not shown). To define the effect of miR-21KD on known target genes in GBM, whole cell lysates were prepared from control and antagomiR-21-transduced GBM cells, and protein expression was determined by immunoblotting. As shown in Fig. 2B, although PTEN expression was higher in MT330 and SJ-G2 miR-21KD cells as compared with empty vector-transduced cells, PTEN expression was absent in U87 cells due to a mutation in both PTEN alleles that is common in GBM (21). In contrast, Anp32A, Sprouty2, and PDCD4 expression was higher in all miR-21KD GBM cells. Thus, consistent with our previous studies, miR-21 regulates target gene expression in a highly cell line-dependent manner (14, 15).
An important mechanism whereby miRNAs regulate gene expression is by targeting mRNAs for degradation. To identify novel miR-21 target genes in GBM, the expression of ∼230 cancer-related genes was examined in RNA prepared from empty vector and miR-21KD GBM cells. As shown in the heat map displayed in Fig. 3A, the expression of several mRNAs was consistently up-regulated in miR-21KD GBM cells, suggesting that they may be direct miR-21 targets. For example, consistent with our immunoblotting data shown in Fig. 2C, PTEN was up-regulated in miR-21KD MT330 and SJ-G2 GBM cell lines but not in U87 cells. Most interestingly, IGFBP3 was up-regulated in all three GBM lines examined. Thus, we next performed immunoblotting of cell extracts and found that IGFBP3 expression was higher in miR-21KD GBM cell lines (Fig. 3B). Similarly, immunostaining of miR-21KD cells showed enhanced IGFBP3 expression (data not shown). miRNAs target mRNAs for degradation by the binding of their 5–8 nucleotide seed sequence to the 3′-UTR of target genes. Thus, we examined the 3′-UTR of IGFBP3 and found that a miR-21 binding sequence was indeed present (Fig. 3C). To determine whether IGFBP3 was a direct miR-21 target, the 3′-UTR of IGFBP3 mRNA containing the predicted miR-21 target sequence as well as a mutated sequence were linked to luciferase, and a Dual-Luciferase (pcDNA3.1-Luc) reporter system was employed to evaluate miRNA-mRNA interactions. Overexpression of miR-21 in HEK293T cells down-regulated luciferase activity of the IGFBP3 reporter constructs, whereas miR-21KD enhanced reporter activity (Fig. 3C). In contrast, luciferase constructs with mutated miR-21 seed target sequences in IGFBP3 were unaffected by miR-21KD or overexpression. These results show that IGFBP3 is a direct miR-21 target gene in GBM.
FIGURE 3.
The effects of miR-21KD on target gene expression and the identification of IGFBP3 as a target gene. A, total RNA was prepared from EV and miR-21KD human glioma cells, and mRNA expression profiling was conducted on the nCounter Analysis System (Nanostring Technologies) using a human cancer-related gene panel. B, lysates were prepared from EV and miR-21KD human glioma cells and immunoblotted for IGFBP3 and actin. C, 293T cells were transiently cotransfected with wild-type (pcDNA3.1-Luc-wtUTR) or mutant (pcDNA3.1-Luc-muUTR) IGFBP3 reporter plasmids, and with EV, overexpressing or miR-21KD plasmid. pSV40-Renilla plasmid was cotransfected as an internal control. The ratio of luciferase and Renilla activities was determined at 24 h post-transfection using the Dual-Luciferase reporter gene kit (Promega).
IGFBP3 Negatively Regulates IGF Signaling in Glioma Cells
Because IGFBP3 has been shown to suppress cell proliferation by binding IGF and thereby inhibiting its growth promoting activity (16), we next examined IGF activity in conditioned medium prepared from control, IGFBP3-overexpressing, and miR-21KD SJ-G2 cells and determined its ability to stimulate the proliferation of serum-starved SJ-G2 cells. As shown in Fig. 4A, medium collected from EV-transduced cells stimulated the growth of serum-starved GBM cells. In contrast, conditioned medium prepared from miR-21KD and IGFBP3 overexpressing cells was markedly less effective in stimulating cell growth. We next added recombinant human IGF-1 to serum-starved glioma cells and measured IGF-1 receptor activation by phospho-IGF-R1 immunoblotting. As shown in Fig. 4B, IGF-1 (50 ng/ml) induced a time-dependent increase in IGF-1R activation with activation detectable at 5 min and peaking within 30 min. In contrast, IGF-R1 activation was only detectable at later times after IGF-1 addition to IGFBP3-overexpressing or miR-21KD cells. BMS-754807 is a potent small molecule inhibitor of the insulin-like growth factor 1 receptor and is currently in clinical trials for the treatment of a variety of human cancers (22). We examined the effect of BMS-754807 on IGF-1-induced phosphorylation of IGF-1R and its downstream target Akt (Fig. 4C) and found a dose-dependent inhibition of these components of the IGF-1R pathway similar to what had been reported previously (22). Based on these findings, we treated SJ-G2 cells with this IGF-1R inhibitor and determined whether it exhibited a growth suppressive effect. Most interestingly, although BMS-754807 inhibited the growth of SJ-G2 glioma cells (Fig. 4D), it had no further growth suppressive effect on cells overexpressing IGFBP3 or miR-21KD cells. These results suggest that the miR-21/IGFBP3 pathway plays a critical role in regulating glioma cell proliferation driven by IGF.
FIGURE 4.
The effects of IGFBP3 overexpression on growth promotion, IGF-1 induced receptor activation and sensitivity to an IGF-1R inhibitor. A, conditioned medium was collected from EV, miR-21KD, and IGFBP3-overexpressing SJ-G2 (or MT330) cells, added to SJ-G2 cells that were grown in low serum (0.1%) containing medium for 18 h, and cell growth rate was determined. B, serum-deprived EV-, IGFBP3-overexpressing, or miR-21KD SJ-G2 cells transduced with scrambled or shIGFBP3 were treated with recombinant human (rh) IGF-1 (50 ng/ml) for the indicated time, lysed, and immunoblotted for phospho-IGF-1R (pIGF-1R), IGF-1R, or actin. C, serum-deprived SJ-G2 cells were pretreated with varying concentrations of BMS-754807 (BMS) for 2 h and then stimulated with IGF-1 (50 ng/ml) for 1 h. Cells were lysed and immunoblotted for phospho-IGF-1R, IGF-1R, phospho-Akt (pAkt), Akt, or actin. D, EV, IGFBP3-overexpressing, and miR-21KD SJ-G2 cells were grown in the presence or absence of BMS-754807 (2.5 μm), and cell proliferation was determined at daily intervals.
IGFBP3 Inhibits GBM Tumor Formation, and Its Knockdown Enhances the Tumorigenicity of miR-21KD GBM Cells
We then sought to determine whether overexpression of IGFBP3 could affect the tumorigenicity of GBM cells in vivo. NSG mice were injected subcutaneously with MT330 GBM cells, and tumor volume was determined by caliper measurement. As shown in Fig. 5A, the formation of tumors was markedly suppressed by IGFBP3 overexpression, and this reduction in GBM tumorigenesis was statistically significant. Moreover, when tumors were weighed at 7 weeks after injection, there was ∼65% reduction in tumor weight in mice injected with IGFBP3-overexpressing GBM cells. Histopathological analysis of tumor tissue indicated that EV and IGFBP3-overexpressing tumors were both high-grade gliomas and were histologically similar.
FIGURE 5.
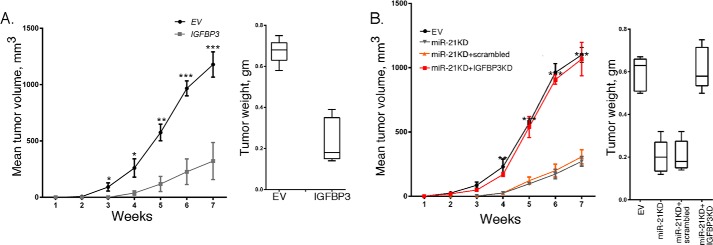
The effects of IGFBP3 overexpression on GBM tumor formation. A, NSG mice were injected subcutaneously with 106 EV or IGFBP3-overexpressing MT330 cells, and tumor growth was determined twice per week by caliper measurements, and at necropsy, tumors were weighed. B, NSG mice were injected subcutaneously with 106 EV-, IGFBP3-overexpressing, or miR-21KD SJ-G2 cells transduced with scrambled or shIGFBP3, and tumor growth was determined twice per week by caliper measurements, and at necropsy, tumors were weighed.
Moreover, we found that miR-21KD markedly inhibited GBM tumor formation (Fig. 5B), which was not surprising as we previously showed that miR-21KD inhibited tumorigenicity of prostate cancer cells and melanoma (14, 15). As IGFBP3 expression is up-regulated by miR-21KD, we next examined whether IGFBP3 plays a direct role in the action of miR-21 on GBM tumorigenesis. Thus, IGFBP3 expression in miR-21KD MT330 cells was knocked down by ∼80% (data not shown), and the cells were injected into the flanks of NSG mice. As shown in Fig. 5B, transduction of miR-21KD cells with IGFBP3 shRNA restored the tumorigenicity to that of EV-transduced MT330 cells. In contrast, transduction of cells with scrambled shRNA had no effect on the tumorigenicity of miR-21KD cells. These results demonstrate that IGFBP3KD restores GBM tumorigenesis, and thus IGFBP3 is an important miR-21 target in GBM. Moreover, as anticipated, IGFBP3KD rescued the effects of miR-21 on the IGF-1 signaling pathway as detected by IGF-1R phosphorylation (Fig. 4B). Consistent with the data from MT330 cells grown in vitro, whereas IGFBP3 expression was increased in miR-21KD MT330 tumors, IGFBP3 expression was knocked down in GBM tumors from miR-21KD MT330 cells transduced with IGFBP3 shRNA (data not shown).
IGFBP3 Expression Correlates with Longer Patient Survival in GBM and Inversely Correlates with miR-21 Expression
As shown in Fig. 1D, higher miR-21 expression correlates with short term survival of GBM patients from analysis of the TCGA database. We next sought to determine the relationship of IGFBP3 expression to patient survival after diagnosis with GBM. As shown in Fig. 6A, higher IGFBP3 expression was found in patients that survived more than 2 years after GBM diagnosis as compared with patients that survived for shorter times, and this difference was statistically significant. To directly determine the relationship between miR-21 and IGFBP3 expression, brain biopsy specimens from 36 GBM patients that represented long term and short term survival after diagnosis were obtained from the UTHSC Tissue Services Core, RNA was isolated, and expression of miR-21 and IGFBP3 was determined. As shown in Fig. 6B, although there was expected patient-to-patient variability, there was a statistically significant inverse correlation between miR-21 and IGFBP3 expression. Taken together, the results from the TCGA database and our own analysis of GBM specimens showed that IGFBP3 is a positive indicator of GBM survival and that miR-21 and IGFBP3 expression are negatively correlated.
FIGURE 6.

High IGFBP3 expression in vivo is associated with better patient survival. A, IGFBP3 expression in the TCGA database of GBM patient samples was related to patient survival after diagnosis as indicated. B, RNA was extracted from 36 GBM patient biopsies, representing long term and short term survival after diagnosis. The expression of miR-21 and IGFBP3 was determined by qPCR (n = 3) and normalized to the expression of U6A and actin, respectively (14).
DISCUSSION
GBM is the most aggressive and deadly form of glioma. Despite improved molecular characterization and aggressive surgery, radiation, and chemotherapy, the median survival of GBM patients remains only 12 to 15 months. Thus, identification of new molecular targets in GBM may lead to improved therapeutic approaches. In the present study, we analyzed the relationship between miR-21 expression in GBM and patient prognosis. We found that miR-21 was expressed at higher levels in GBM cell lines as compared with normal cells (fibroblasts and astrocytes) and that miR-21 expression was elevated in tumor tissue in a mouse model of human glioma. We then examined the relationship between miR-21 expression and glioma tumor grade and patient survival based on samples from the UTHSC Tissue Services Core and information in the TCGA database. High miR-21 was found to be associated with shorter term survival. This is not surprising based on the previous findings that relatively high miR-21 levels were found in various human tumors and miR-21 appears to play an important role in the oncogenic process as indicated by its association with high cell proliferation, low apoptosis, high invasion, and metastatic potential (6–12). miR-21 is believed to play an important role in cancer development as well as in the resistance of cancers to chemotherapy and radiation. For example, we recently showed that miR-21KD sensitized cells to the apoptotic actions of chemotherapeutic agents and inhibited the metastatic potential of melanoma cells (14, 15, 23).
A number of miR-21 target genes have been described previously, including PTEN, PDCD4, etc. We found that miR-21KD in several glioma cell lines up-regulated the expression of these miR-21 target genes in a cell line-dependent manner, demonstrating that these genes are truly miR-21 targets. We then performed gene expression analysis of a cancer-related gene panel on RNA samples derived from control and miR-21KD GBM cell lines to identify new miR-21 target genes. By this approach, we identified that IGFBP3 was up-regulated upon miR-21KD in several GBM cell lines, indicating that it was a potential miR-21 target gene. By luciferase reporter assays driven by wild-type and mutant 3′-UTR of IGFBP3, we showed that IGFBP3 expression was directly regulated by miR-21 expression. The present study is the first to show that IGFBP3 is a miR-21 target gene. The family of IGFBPs is comprised of six members (IFGBP1–6), which bind to and regulate the functions of IGFs. By modulating the bioavailability of IGFs, IGFBPs regulate tumor growth and invasion (24). Overexpression of soluble and its receptor IGF-1R has been detected in several cancers, including prostate cancer, melanoma, and GBM (25–28). IGFR1 signaling has also been found to mediate resistance to chemotherapy and radiation (29–32). IGF-1 has been implicated in tumor resistance to radiation and chemotherapy (31, 33) and is associated with poor prognosis in GBM (25). Our studies showing that IGFBP3 negatively regulates IGF-1R signaling and is associated with better prognosis (better patient function and longer survival) are especially important, as IGFBP3 is the major binding protein and regulator of IGF-1 ligand bioavailability. Moreover, IGFBP3 reportedly can inhibit or enhance the activity of IGF-1R signaling in various cancers (34, 35).
IGFBP3 plays a critical role in many cancers, including GBM. For example, IGFBP3 was found to be among a group of serum markers in GBM associated with prolonged survival (15 months) after tumor resection in a cohort of 23 patients (36). IGFBP3 reportedly can have antiproliferative or growth promoting actions on tumor cells (34). In our study, we found that IGFBP3 overexpression inhibited cell proliferation. Moreover, we found that high IGFBP3 expression in GBM was associated with better patient prognosis as indicated by increased survival postoperatively. Consistent with our findings are the studies showing that IGFBP3 has tumor-suppressive properties (37) and that increased serum levels of IGFBP3 are associated with better prognosis in prostate cancer (38). Low IGFBP3 expression in patients with esophageal tumors correlates with higher tumor grade, advanced stage, and poor survival (39). Moreover, in patients with colorectal cancer, higher circulating IGFBP3 levels are associated with a greater response to chemotherapy and better overall survival (40). Due to its central role in cancer cell signaling, IGF-1R has become an attractive therapeutic target, and various strategies targeting IGF-1R are being presently tested (41). We found that both miR-21KD and IGFBP3 overexpression are inhibitory to IGF-1R phosphorylation and IGF-1 signaling. In addition, the IGF-1R antagonist BMS-754807 inhibited glioma cell proliferation but had little effect on the proliferation of either miR-21KD or IGFBP3-overexpressing glioma cells. Therefore, future studies should be directed to test whether small molecule inhibitors of IGF-1R synergize with cytotoxic and targeted agents in GBM. Finally, we showed in GBM tumor biopsies that IGFBP3 and miR-21 expression was inversely correlated and that low miR-21 and high IGFBP3 expression was associated with improved patient performance and longer survival. Thus, IGFBP3 and miR-21 may have diagnostic and prognostic utility in GBM. In summary, we identified IGFBP3 as a novel miR-21 target gene in GBM, and characterized the role of the miR-21/IGFBP3 axis in glioma, which suggests that miR-21 promotes tumorigenesis in GBM through suppressing the expression of the tumor suppressor IGFBP3.
This work was supported by National Institute of Health Grants CA133322 (to L. M. P. and A. M. D.), Department of Defense Award W81XWH-11-1-0533 (to L. M. P.), Cancer Center Support Grant 21766 from NCI, National Institutes of Health (to A. M. D.), and by the Muirhead Chair Endowment (to L. M. P.) at the University of Tennessee Health Science Center.
- GBM
- glioblastoma multiforme
- EV
- empty vector
- IGFBP3
- insulin-like growth factor (IGF)-binding protein-3
- IGF-1R
- IGF-1 receptor
- miRNA
- microRNA
- TCGA
- The Cancer Genome Atlas
- UTHSC
- University of Tennessee Health Science Center
- qPCR
- quantitative real time PCR
- KD
- knockdown.
REFERENCES
- 1. Ostrom Q. T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N. E., Kruchko C., Barnholtz-Sloan J. S. (2013) CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro. Oncol. 15, ii1–ii56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 [DOI] [PubMed] [Google Scholar]
- 3. Wong E. T., Hess K. R., Gleason M. J., Jaeckle K. A., Kyritsis A. P., Prados M. D., Levin V. A., Yung W. K. (1999) Outcomes and prognostic factors in recurrent glioma patients enrolled onto phase II clinical trials. J. Clin. Oncol. 17, 2572–2578 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 5. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 6. Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007) miR-21-mediated tumor growth. Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 7. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Folini M., Gandellini P., Longoni N., Profumo V., Callari M., Pennati M., Colecchia M., Supino R., Veneroni S., Salvioni R., Valdagni R., Daidone M. G., Zaffaroni N. (2010) miR-21: an oncomir on strike in prostate cancer. Mol. Cancer 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Löffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermüller J., Kretzschmar A. K., Burger R., Gramatzki M., Blumert C., Bauer K., Cvijic H., Ullmann A. K., Stadler P. F., Horn F. (2007) Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 10. Chan J. A., Krichevsky A. M., Kosik K. S. (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 [DOI] [PubMed] [Google Scholar]
- 11. Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359 [DOI] [PubMed] [Google Scholar]
- 12. Wang P., Zou F., Zhang X., Li H., Dulak A., Tomko R. J., Jr., Lazo J. S., Wang Z., Zhang L., Yu J. (2009) microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 69, 8157–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buscaglia L. E., Li Y. (2011) Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 30, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C. H., Yue J., Fan M., Pfeffer L. M. (2010) IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 70, 8108–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C. H., Yue J., Pfeffer S. R., Handorf C. R., Pfeffer L. M. (2011) MicroRNA miR-21 Regulates the Metastatic Behavior of B16 Melanoma Cells. J. Biol. Chem. 286, 39172–39178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trojan J., Cloix J. F., Ardourel M. Y., Chatel M., Anthony D. D. (2007) Insulin-like growth factor type I biology and targeting in malignant gliomas. Neuroscience 145, 795–811 [DOI] [PubMed] [Google Scholar]
- 17. Santosh V., Arivazhagan A., Sreekanthreddy P., Srinivasan H., Thota B., Srividya M. R., Vrinda M., Sridevi S., Shailaja B. C., Samuel C., Prasanna K. V., Thennarasu K., Balasubramaniam A., Chandramouli B. A., Hegde A. S., Somasundaram K., Kondaiah P., Rao M. R. (2010) Grade-specific expression of insulin-like growth factor-binding proteins-2, -3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol. Biomarkers Prev. 19, 1399–1408 [DOI] [PubMed] [Google Scholar]
- 18. Wang H., Wang H., Zhang W., Fuller G. N. (2006) Overexpression of IGFBP5, but not IGFBP3, correlates with the histologic grade of human diffuse glioma: a tissue microarray and immunohistochemical study. Technol. Cancer Res. Treat. 5, 195–199 [DOI] [PubMed] [Google Scholar]
- 19. Pfeffer L. M., Mullersman J. E., Pfeffer S. R., Murti A., Shi W., Yang C. H. (1997) STAT3 as an adapter to couple phosphatidylinositol-3 kinase to the IFNAR-1 chain of the type I IFN receptor. Science 276, 1418–1420 [DOI] [PubMed] [Google Scholar]
- 20. Hambardzumyan D., Amankulor N. M., Helmy K. Y., Becher O. J., Holland E. C. (2009) Modeling adult gliomas using RCAS/t-va technology. Transl. Oncol. 2, 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wen S., Stolarov J., Myers M. P., Su J. D., Wigler M. H., Tonks N. K., Durden D. L. (2001) PTEN controls tumor-induced angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 4622–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carboni J. M., Wittman M., Yang Z., Lee F., Greer A., Hurlburt W., Hillerman S., Cao C., Cantor G. H., Dell-John J., Chen C., Discenza L., Menard K., Li A., Trainor G., Vyas D., Kramer R., Attar R. M., Gottardis M. M. (2009) BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol. Cancer Ther. 8, 3341–3349 [DOI] [PubMed] [Google Scholar]
- 23. Yang C. H., Yue J., Sims M., Pfeffer L. M. (2013) The curcumin analog EF24 targets NF-kappaB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PLoS One 8, e71130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vincent A. M., Feldman E. L. (2002) Control of cell survival by IGF signaling pathways. Growth Horm. IGF Res. 12, 193–197 [DOI] [PubMed] [Google Scholar]
- 25. Trojan J., Blossey B. K., Johnson T. R., Rudin S. D., Tykocinski M., Ilan J., Ilan J. (1992) Loss of tumorigenicity of rat glioblastoma directed by episome-based antisense cDNA transcription of insulin-like growth factor I. Proc. Natl. Acad. Sci. U.S.A. 89, 4874–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hellawell G. O., Turner G. D., Davies D. R., Poulsom R., Brewster S. F., Macaulay V. M. (2002) Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 62, 2942–2950 [PubMed] [Google Scholar]
- 27. All-Ericsson C., Girnita L., Seregard S., Bartolazzi A., Jager M. J., Larsson O. (2002) Insulin-like growth factor-1 receptor in uveal melanoma: a predictor for metastatic disease and a potential therapeutic target. Invest. Ophthalmol. Vis. Sci. 43, 1–8 [PubMed] [Google Scholar]
- 28. Nickerson T., Chang F., Lorimer D., Smeekens S. P., Sawyers C. L., Pollak M. (2001) In vivo progression of LAPC-9 and LNCaP prostate cancer models to androgen independence is associated with increased expression of insulin-like growth factor I (IGF-I) and IGF-I receptor (IGF-IR). Cancer Res. 61, 6276–6280 [PubMed] [Google Scholar]
- 29. Dallas N. A., Xia L., Fan F., Gray M. J., Gaur P., van Buren G., 2nd, Samuel S., Kim M. P., Lim S. J., Ellis L. M. (2009) Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 69, 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eckstein N., Servan K., Hildebrandt B., Pölitz A., von Jonquières G., Wolf-Kümmeth S., Napierski I., Hamacher A., Kassack M. U., Budczies J., Beier M., Dietel M., Royer-Pokora B., Denkert C., Royer H. D. (2009) Hyperactivation of the insulin-like growth factor receptor I signaling pathway is an essential event for cisplatin resistance of ovarian cancer cells. Cancer Res. 69, 2996–3003 [DOI] [PubMed] [Google Scholar]
- 31. Turner B. C., Haffty B. G., Narayanan L., Yuan J., Havre P. A., Gumbs A. A., Kaplan L., Burgaud J. L., Carter D., Baserga R., Glazer P. M. (1997) Insulin-like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 57, 3079–3083 [PubMed] [Google Scholar]
- 32. Yu D., Watanabe H., Shibuya H., Miura M. (2003) Redundancy of radioresistant signaling pathways originating from insulin-like growth factor I receptor. J. Biol. Chem. 278, 6702–6709 [DOI] [PubMed] [Google Scholar]
- 33. Bartucci M., Morelli C., Mauro L., Andò S., Surmacz E. (2001) Differential insulin-like growth factor I receptor signaling and function in estrogen receptor (ER)-positive MCF-7 and ER-negative MDA-MB-231 breast cancer cells. Cancer Res. 61, 6747–6754 [PubMed] [Google Scholar]
- 34. Firth S. M., Baxter R. C. (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23, 824–854 [DOI] [PubMed] [Google Scholar]
- 35. Schedlich L. J., Graham L. D. (2002) Role of insulin-like growth factor binding protein-3 in breast cancer cell growth. Microsc. Res. Tech. 59, 12–22 [DOI] [PubMed] [Google Scholar]
- 36. Elstner A., Stockhammer F., Nguyen-Dobinsky T. N., Nguyen Q. L., Pilgermann I., Gill A., Guhr A., Zhang T., von Eckardstein K., Picht T., Veelken J., Martuza R. L., von Deimling A., Kurtz A. (2011) Identification of diagnostic serum protein profiles of glioblastoma patients. J. Neurooncol. 102, 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamada P. M., Lee K. W. (2009) Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am. J. Physiol. Cell Physiol. 296, C954–C976 [DOI] [PubMed] [Google Scholar]
- 38. Shariat S. F., Lamb D. J., Kattan M. W., Nguyen C., Kim J., Beck J., Wheeler T. M., Slawin K. M. (2002) Association of preoperative plasma levels of insulin-like growth factor I and insulin-like growth factor binding proteins-2 and -3 with prostate cancer invasion, progression, and metastasis. J. Clin. Oncol. 20, 833–841 [DOI] [PubMed] [Google Scholar]
- 39. Torng P. L., Lee Y. C., Huang C. Y., Ye J. H., Lin Y. S., Chu Y. W., Huang S. C., Cohen P., Wu C. W., Lin C. T. (2008) Insulin-like growth factor binding protein-3 (IGFBP-3) acts as an invasion-metastasis suppressor in ovarian endometrioid carcinoma. Oncogene 27, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 40. Fuchs C. S., Goldberg R. M., Sargent D. J., Meyerhardt J. A., Wolpin B. M., Green E. M., Pitot H. C., Pollak M. (2008) Plasma insulin-like growth factors, insulin-like binding protein-3, and outcome in metastatic colorectal cancer: results from intergroup trial N9741. Clin. Cancer Res. 14, 8263–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chitnis M. M., Yuen J. S., Protheroe A. S., Pollak M., Macaulay V. M. (2008) The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. 14, 6364–6370 [DOI] [PubMed] [Google Scholar]





