Abstract
A clinical strain of Escherichia coli isolated from pleural liquid with high levels of resistance to cefotaxime, ceftazidime, and aztreonam harbors a novel CTX-M gene (blaCTX-M-32) whose amino acid sequence differs from that of CTX-M-1 by a single Asp240-Gly substitution. Moreover, by site-directed mutagenesis we demonstrated that this replacement is a key event in ceftazidime hydrolysis
The emergence of plasmid-mediated extended-spectrum β-lactamases in members of the family Enterobacteriaceae has become a worldwide problem (3, 4, 6, 7, 11-13, 16).
Most extended-spectrum β-lactamases are derivatives of TEM-1, TEM-2, or SHV-1 enzymes; however, there are an increasing number of reports that describe the worldwide emergence of β-lactamases belonging to other families, such as CTX-M and/or OXA derivatives (8).
The family of CTX-M enzymes is grouped on the basis of similarities in amino acid sequences into four major phylogenetic trees (6): the CTX-M-1 group (CTX-M-1 or MEN-1, CTX-M-3, CTX-M-10, CTX-M-12, CTX-M-15, and now CTX-M-32), the CTX-M-2 group (CTX-M-2, CTX-M-4, CTX-M-5, CTX-M-6, CTX-M-7, CTX-20, and Toho-1), the CTX-M-8 group, and the CTX-M-9 group (CTX-M-9, CTX-M-13, CTX-M-14, CTX-M-16, CTX-M-18, CTX-M-19, CTX-M-21, and Toho-2). The designation CTX-M refers to a potent activity against cefotaxime and having only a remnant of activity toward ceftazidime.
Here we report the molecular characterization of a new CTX-M β-lactamase derived from CTX-M-1 through a single Asp240-Gly substitution, CTX-M-32. In addition, we report experimental data showing that substitution of this amino acid is itself sufficient to confer hydrolytic activity against ceftazidime.
Patterns of antibiotic susceptibility shown by the clinical strain E. coli JC19325, as well as its transconjugant and transformants, are shown in Table 1. The MICs were determined by E-test and interpreted according to the method of the National Committee for Clinical Laboratory Standards (18). The clinical strain JC19325 showed a high level of resistance to cefotaxime, ceftazidime, and aztreonam (MICs of >256 μg/ml), cefoxitin (MIC of >256 μg/ml), and cefepime (MIC of 64 μg/ml). Moreover, clavulanic acid acted synergistically with amoxicillin, cefotaxime, and ceftazidime (E-test; ABBiodisk, Solna, Sweden), thus indicating the presence of a class A β-lactamase (9). An Escherichia coli TG1 transformant harboring the pMC-2 plasmid showed higher MICs of the affected antibiotics, probably due to more copies of the bla gene.
TABLE 1.
MICs of β-lactams for the JC19325 clinical strain, E. coli XL1(pMC-1), E. coli TG1, E. coli TG1(pMC-2), and E. coli TG1(pMC-3)
| Antibioticsa | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| JC19325 (produces CTX-M-32) | XL1(pMC-1b) (produces CTX-M-32) | TG1 | TG1(pMC-2c) (produces CTX-M-32) | TG1(pMC-3d) (produces CTX-M-1) | |
| Amoxicillin | >256 | >256 | 3 | >256 | >256 |
| Amoxicillin + clavulanate | 12 | 6 | 2 | 4 | 8 |
| Piperacillin | >256 | >256 | 0.38 | >256 | >256 |
| Cephalothin | >256 | >256 | 3 | >256 | >256 |
| Cefuroxime | >256 | >256 | 1.5 | >256 | >256 |
| Cefoxitin | >256 | 3 | 2 | 2 | 2 |
| Cefotaxime | >256 | >256 | 0.02 | >256 | >256 |
| Cefotaxime + clavulanate | >1 | 0.03 | 0.02 | 0.03 | 0.06 |
| Ceftazidime | 128 | 96 | 0.06 | >256 | 6 |
| Ceftazidime + clavulanate | >4 | 0.25 | 0.06 | 0.19 | 0.25 |
| Cefepime | 64 | 16 | 0.02 | 64 | 48 |
| Aztreonam | >256 | >256 | 0.03 | >256 | 48 |
| Imipenem | 0.19 | 0.25 | 0.12 | 0.19 | 0.19 |
| Meropenem | 0.047 | 0.03 | 0.008 | 0.02 | 0.02 |
Clavulanate was used at 4 μg/ml.
Transconjugant harboring CTX-M-32.
Transformant harboring CTX-M-32 β-lactamase gene.
Transformant harboring CTX-M-32mut or CTX-M-1 β-lactamase gene.
Isoelectric focusing was performed using polyacrylamide gels containing Ampholine, within a pH range of 3.5 to 9.5, as previously described (17). The clinical isolate produced one enzyme with a pI of 9.0.
In the present study the E. coli XL1-Blue MRF′Kan strain (Stratagene Europe, Amsterdam, The Netherlands) was used in the conjugation experiments.
The clinical strain JC19325 had one plasmid which harbored a β-lactamase with a pI of 9.0 that was transferred by conjugation into E. coli XL1-Blue MRF′Kan using kanamycin (25 μg/ml) and cefotaxime (2 μg/ml) as selective antibiotics. A few of the transconjugants which grew harbored an identical plasmid of approximately 15 kb, which was named pMC-1.
Plasmid DNA was isolated by the alkaline lysis method (23) from the transconjugant that produced a single β-lactamase with a pI of 9.0. Plasmid DNA was digested with KpnI and ligated to the plasmid vector pBGS18− (25); afterwards, the ligation mixture was introduced into E. coli TG1 cells by transformation with CaCl2, and transformants were detected on Luria-Bertani agar plates with cefotaxime (2 μg/ml) and kanamycin (25 μg/ml). The resulting plasmid, designated pMC-2, carried a bla-producing insert of size circa 4 kb. Double-stranded templates were subjected to nucleotide sequencing by using the method of Sanger et al. (23, 23a).
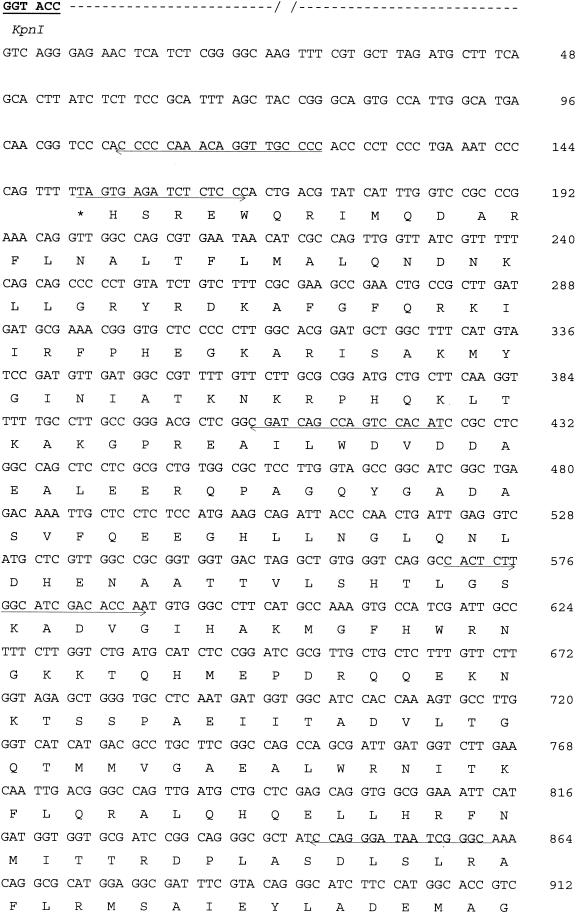
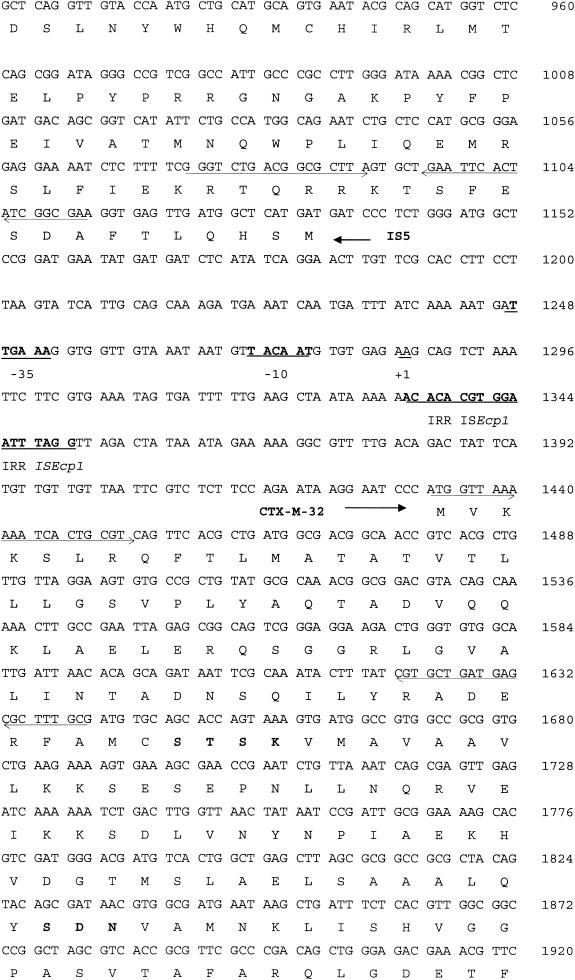
During isoelectric focusing, the pI 9.0 β-lactamase activity band from the E. coli transformants cofocused with the β-lactamase activity band from the clinical strain JC19325. Nucleotide sequencing of the KpnI insert revealed some interesting features, including (i) a new bla gene. This new bla gene was 876 bp long, initiated with an ATG codon, and ended with a TGA codon (291 amino acids long). The initiation codon was preceded by a Shine-Dalgarno ribosome-binding sequence, AAGGAA. The EMBL and Swiss-Prot database searches for this open reading frame revealed similarities to CTX-M β-lactamases. The deduced amino acid sequence had the closest homology (99%) with the CTX-M-1 enzyme (2, 3), from which it differed by the single amino acid substitution Asp240-Gly (Ambler numbering) (1). (ii) The second interesting feature was the inverted repeat right (IRR) sequence of ISEcp1B 80 bp upstream of the ATG start codon of CTX-M-32. No putative promoter sequences were found in the 80-bp sequence that separated the IRR of ISEcp1B from the ATG site of the blaCTX-M-32 gene; moreover, this IRR provided −35 and −10 promoter sequences, thus probably contributing to the expression of the blaCTX-M-32 gene. (iii) Third, this IRR was downstream of a tnpA gene that encoded the transposase of IS5. Figure 1 shows the 2,326-bp sequence of the original 4-kpb KpnI fragment.
FIG. 1.
Nucleotide sequence of a 2,326-bp DNA fragment of the pMC-2 plasmid. The deduced amino acid sequence is indicated in single-letter code below the nucleotide sequence. Stop codons are indicated by asterisks. The −35 and −10 promoter sequences of the blaCTX-M-32 gene and the IRR sequence of ISEcp1 are underlined and indicated by bold letters, as is the +1 position of the transcriptional start of the blaCTX-M-32 gene (10). The CTX-M-32 and transposase of IS5 proteins are indicated by arrows. Bold amino acids are those conserved in class A β-lactamases (15). Oligonucleotides used for sequencing are indicated by arrows, and KpnI restriction sites delimiting the 4-kbp insert are also underlined.
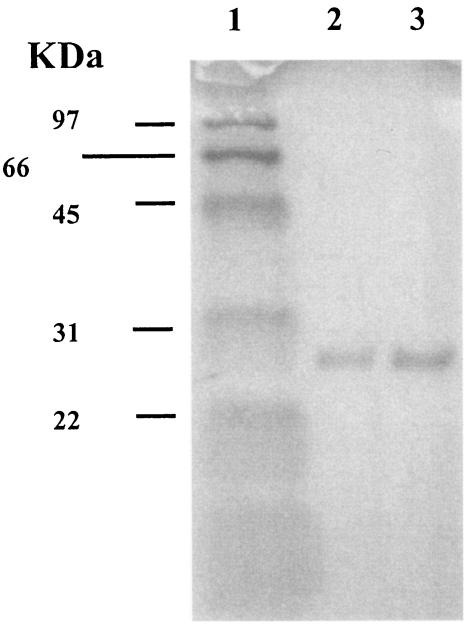
To purify the CTX-M enzyme, the blaCTX-M-32 gene was cloned in the pGEX-6P-1 vector, which allowed a fusion protein between glutathione S-transferase (GST) and the CTX-M enzyme. The β-lactamase was purified to homogeneity following the manufacturer's directions for the GST gene fusion system (Amersham Pharmacia Biotech, Europe GmbH). The purified protein appeared on sodium dodecyl sulfate-polyacrylamide gel electrophoresis as a band of 28 kDa (≥99% pure) (Fig. 2).
FIG. 2.
Electrophoresis analysis of CTX-M-32 and CTX-M-1 purified extracts in a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel stained with Coomassie brilliant blue R-250. Lanes: 1, protein molecular markers; 2, purified CTX-M-1 protein used in kinetic experiments; 3, purified CTX-M-32 protein used in kinetic experiments.
For kinetic experiments, CTX-M-32 β-lactamase was used at a 1,800 μM concentration. The β-lactamase showed a hydrolytic profile similar to that expected for a molecular class A CTX-M enzyme (6), with the Km for ampicillin lower than the Km for cefalothin, a Km for cefotaxime of <500 μM, and a clear hydrolytic activity towards cefotaxime. Moreover, moderate hydrolytic activity was detected against ceftazidime, as ceftazidime MICs suggested (Table 2).
TABLE 2.
Substrate profile of β-lactamase CTX-M-32
| Antibiotic | Km (μM) | Kcat (s−1) | Kcat/Km (μM−1) · (s−1) |
|---|---|---|---|
| Ampicillin | 8 ± 2.6 | 3 ± 1.1 | 0.4 |
| Cephalothin | 211 ± 0.5 | 928 ± 16.4 | 4.4 |
| Cefuroxime | 261 ± 0.2 | 162 ± 11.6 | 0.6 |
| Cefotaxime | 322 ± 2.7 | 320 ± 10.4 | 1 |
| Ceftazidime | 271 ± 0.2 | 0.91 ± 0.5 | 0.003 |
| Cefepime | 1,287 ± 4.1 | 43 ± 8.8 | 0.03 |
| Aztreonam | 31 ± 0.7 | 1 ± 0.4 | 0.03 |
The CTX-M-32 enzyme is derived from CTX-M-1 by a single amino acid replacement, Asp240-Gly. To confirm the importance of the Asp-Gly substitution in the hydrolysis of ceftazidime, we replaced the Gly240 with Asp in CTX-M-32 by using site-directed mutagenesis as previously described (14). The CTX-M-32mut or CTX-M-1 gene was then cloned into the pBGS18− vector, yielding the pMC-3 plasmid. The mutagenesis was confirmed by nucleotide sequencing. The MICs for E. coli TG1 harboring pMC-2 and pMC-3 are shown in Table 1. The MICs of ceftazidime corresponding to E. coli TG1 harboring CTX-M-1 β-lactamase were clearly lower than those corresponding to E. coli TG1 carrying CTX-M-32. To confirm this result, the substrate profile of the CTX-M-1 β-lactamase was determined with the enzyme purified as mentioned above for CTX-M-32 (GST gene fusion system) (Fig. 2). For kinetic experiments, CTX-M-1 β-lactamase was used at a 1,670 μM concentration. Kcat/Km (in micromolar per second) values for ceftazidime and cefotaxime were 0.0001 and 1.5; therefore, a lower catalytic efficiency with respect to ceftazidime was detected with CTX-M-1, according to the differences in ceftazidime MICs between CTX-M-32 and CTX-M-1 enzymes (Table 1).
Three different enzymes, CTX-M-15, -16, -19 and, recently, CTX-M-27 have been reported to be associated with ceftazidime hydrolysis (4, 5, 20, 21). The amino acid changes associated with the phenotype of ceftazidime hydrolysis were a Pro-to-Ser substitution at position 167 in CTX-M-19 with respect to CTX-M-18 (20) and an Asp-to-Gly substitution at position 240 in CTX-M-16 with respect to CTX-M-9 (4) and in CTX-M-27 with respect to CTX-M-14 (5). In agreement with these previous results, we also report that the Asp240 substitution is a key factor in the evolution of CTX-M β-lactamases, as it increases their hydrolytic activity toward ceftazidime.
Regarding the CTX-M enzymes, to our knowledge only six different enzymes have been published in the group 1 CTX-M enzymes: CTX-M-1, -3, -10, -12, -15, and -32 (3, 16, 19, 20). Among these, only CTX-M-15 and -32 showed more efficient ceftazidime hydrolysis than their parental enzymes, CTX-M-3 and CTX-M-1, respectively. The two former enzymes share the same amino acid substitution, although CTX-M-15 differs from CTX-M-32 in four additional amino acid changes. In terms of evolution, CTX-M-32 is probably an ancestor between CTX-M-1 and CTX-M-15 and constitutes a step forward in the evolution of β-lactamase in broad-spectrum hydrolysis of antibiotics such as ceftazidime.
In summary, we report the genetic and biochemical characterization of a new CTX-M enzyme, CTX-M-32. This is the fourth report of a CTX-M β-lactamase isolation in Spain, as CTX-M-9, CTX-M-10, and CTX-M-14 have previously been isolated in this country (7, 19, 22, 24).
Nucleotide sequence accession number.
The GenBank accession number for the CTX-M-32 β-lactamase is AJ557142.
..
Acknowledgments
This work was supported by Direccion Xeral de I+D, Xunta de Galicia (PGIDT01PXR90101PR and PGIDIT02SAN91604PR) and Fondo de Investigaciones Sanitarias (PI021415).
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, et al. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthélémy, M., J. Péduzzi, H. Bernard, C. Tancrède, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R., C. Dutour, J. L. Samapaio, C. Chanal, D. Sirot, R. Labia, C. D Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240-Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., C. Recule, R. Baraduc, C. Chanal, D. Sirot, C. De Champs, and J. Sirot. 2003. Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52:29-35. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou, G., M. Cartelle, M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 beta-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistant threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanawong, A., F. H. M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coque, T. M., A. Oliver, J. C. Pérez-Díaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutour, C., R. Bonnet, H. Marchandin, M. Boye, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 15.Joris, B., P. Ledent, O. Dideberg, E. Fonze, J. Lamotte-Brasseur, J. A. Kelly, J. M. Ghuysen, and J. M. Frere. 1991. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob. Agents Chemother. 35:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kariruki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 21.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23a.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simarro, E., F. Navarro, J. Ruiz, E. Miro, J. Gomez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like beta-lactamase in Spain. J. Clin. Microbiol. 38:4676-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt, B., P. J. Hedge, T. S. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]