Background: Although parasites commonly hijack the actin cytoskeleton, the Chlamydia pneumoniae CopN protein interferes with microtubules.
Results: CopN targets the β-tubulin longitudinal interface and inhibits microtubule assembly through a dual mechanism.
Conclusion: In addition to regulating type III secretion, C. pneumoniae CopN has evolved microtubule-related specific functions.
Significance: A framework for understanding the CopN role in the chlamydial infectious cycle is provided.
Keywords: Chlamydia, Microtubule, Protein-Protein Interaction, Structural Biology, Tubulin
Abstract
Although the actin network is commonly hijacked by pathogens, there are few reports of parasites targeting microtubules. The proposed member of the LcrE protein family from some Chlamydia species (e.g. pCopN from C. pneumoniae) binds tubulin and inhibits microtubule assembly in vitro. From the pCopN structure and its similarity with that of MxiC from Shigella, we definitively confirm CopN as the Chlamydia homolog of the LcrE family of bacterial proteins involved in the regulation of type III secretion. We have also investigated the molecular basis for the pCopN effect on microtubules. We show that pCopN delays microtubule nucleation and acts as a pure tubulin-sequestering protein at steady state. It targets the β subunit interface involved in the tubulin longitudinal self-association in a way that inhibits nucleotide exchange. pCopN contains three repetitions of a helical motif flanked by disordered N- and C-terminal extensions. We have identified the pCopN minimal tubulin-binding region within the second and third repeats. Together with the intriguing observation that C. trachomatis CopN does not bind tubulin, our data support the notion that, in addition to the shared function of type III secretion regulation, these proteins have evolved different functions in the host cytosol. Our results provide a mechanistic framework for understanding the C. pneumoniae CopN-specific inhibition of microtubule assembly.
Introduction
Microtubules are dynamic cytoskeletal assemblies involved in multiple cellular functions (1). They form tracks along which molecular motors convey cargos and organelles through the cytoplasm. In dividing cells, they reorganize spectacularly to form the architecture of the mitotic spindle, the structure that segregates chromosomes between daughter cells. Microtubules are made of αβ-tubulin heterodimers that assemble longitudinally, head-to-tail, to form protofilaments; these protofilaments interact laterally to shape the microtubule hollow cylinder (2). Both tubulin subunits bind a GTP molecule. The one on α-tubulin is structural; it is non-exchangeable and not hydrolyzed. In contrast, the β-tubulin GTP is exchangeable in unassembled tubulin. Upon microtubule assembly, this nucleotide becomes hydrolyzed, and its exchange is inhibited until disassembly.
The cytoskeleton is the target of intracellular parasites, most of which hijack the actin network (3). There are also some reports of microtubule destabilization by parasites, including Escherichia coli and Shigella through their related protein EspG (4) and VirA (5), respectively. But the mechanism of action is not known, and the microtubule-destabilizing activity has even been called into question (6). CopN3 from Chlamydia pneumonia (pCopN) has been recently identified as a microtubule-destabilizing protein (7). The development cycle of Chlamydia differentiates them from most other prokaryotes in that they are obligate intracellular pathogens (8). In consequence, methods to manipulate these bacteria genetically emerged only very recently (9, 10). To circumvent these limitations, chlamydial proteins have been studied in heterologous systems. In the case of pCopN, it has been shown that its expression in yeast and mammalian cells leads to inhibition of growth and to defects in cell division that are accompanied by the disruption of the mitotic spindle (11). This property is shared by CopN from Chlamydia abortus but not by the more distant Chlamydia trachomatis CopN (tCopN) (Fig. 1). Consistently, biochemical experiments have shown that pCopN, but not tCopN, interacts with tubulin directly and inhibits microtubule assembly in a dose-dependent manner (7). However, the underlying mechanism remains to be determined.
FIGURE 1.
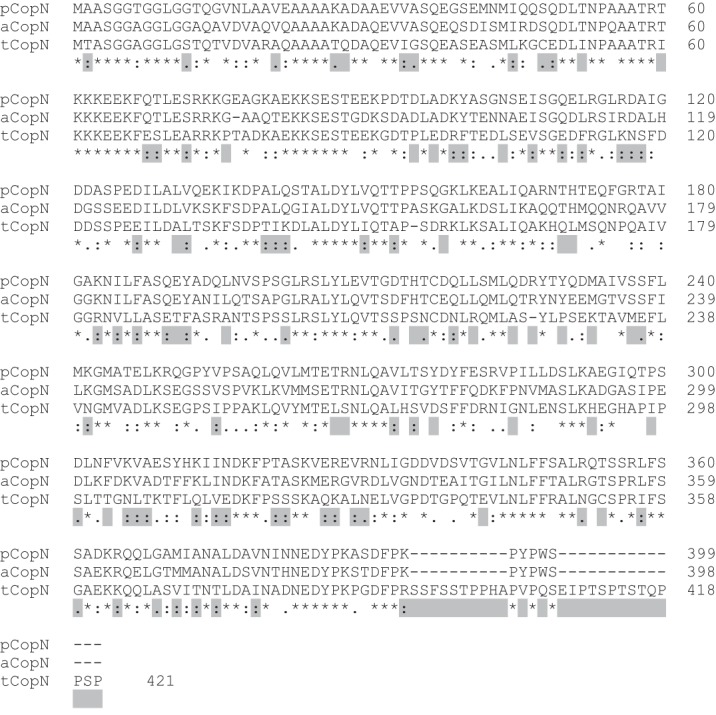
Sequence alignment of pCopN, C. abortus CopN (aCopN), and tCopN. Highlighted positions are those with identical features in pCopN and C. abortus CopN, both proteins interacting with tubulin, but that differ in tCopN, which does not bind tubulin (Ref. 7 and data not shown).
The purpose of this work was to gain further insight into pCopN function at the molecular level. We demonstrate that pCopN is a tubulin-sequestering protein that targets the β subunit longitudinal interface and binds tubulin with different affinities according to the nucleotide state. In addition, pCopN interacts directly with the microtubule nucleus and delays nucleation. We have also determined the 1.2 Å resolution structure of pCopN and mapped the surface interacting with tubulin. The structure confirms CopN as the Chlamydia homolog of the Yersinia YopN protein, as proposed (12, 13); as such, it seems likely that it serves as a plug of the type III secretion system of this bacteria (14) and is secreted during infection, albeit in small amounts (15, 16). Taken together, the natural abundance of tubulin (17) and its affinity for pCopN mean that, when secreted in the cytoplasm, pCopN will be bound to tubulin. Our observations provide a framework for understanding the way secreted pCopN plays a role in the chlamydial infectious cycle.
EXPERIMENTAL PROCEDURES
Cloning and Protein Preparation of pCopN
Full-length CopN and N-terminal truncation constructs were cloned by standard PCR methods from C. pneumonia and C. trachomatis genomic DNA into a pETM-11 plasmid, thus incorporating an N-terminal hexahistidine tag. C-terminal truncation and substitution mutants were obtained using PCR-based mutagenesis. The pCopN constructs used in this study are schematized in Fig. 2A. The pCopN mutations were introduced in the D29 construct, using either a pETM-11 plasmid (R268H mutant) or a pET28b plasmid (S238E/K242E, Q261E/T265E, and Q271E/T275E mutants). Proteins were expressed in BL21(DE3) E. coli cells grown in LB medium at 37 °C and induced with 0.1 mm isopropyl β-d-thiogalactopyranoside for 16 h at 18 °C. Bacteria were harvested by centrifugation and lysed by sonication. Proteins were purified by HisTrap affinity chromatography followed by gel filtration on Superdex75 column (GE Healthcare). All the constructs were correctly folded as judged by circular dichroism (data not shown). Proteins were snap frozen in liquid nitrogen and stored at −80 °C until use.
FIGURE 2.
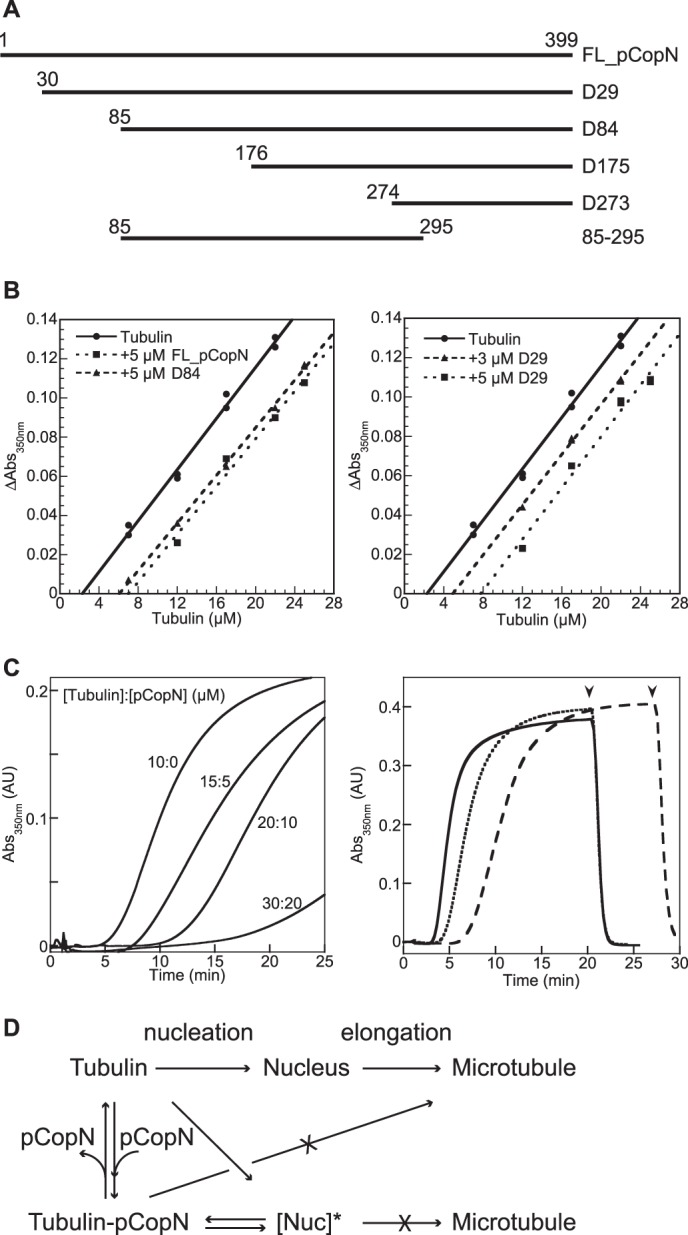
pCopN inhibits microtubule assembly by sequestering tubulin and by delaying nucleation. A, the pCopN constructs used in this study. The effect of mutations was studied in the framework of the D29 construct. B, critical concentration plots for microtubule assembly in the absence (●) and in the presence of 5 μm FL_pCopN (■) or D84 (▴) (left) or in the presence of 3 μm (▴) or 5 μm (■) D29 (right). C, time course of tubulin assembly in the presence of increasing amounts of tubulin-FL_pCopN complex. Left, the assembly of 10 μm tubulin or of tubulin/pCopN mixtures at the indicated concentrations was assayed turbidimetrically. The temperature was switched from 4 to 37 °C after 1 min of recording time in each case. Right, the same experiments but with 15 μm tubulin (solid line) or with mixtures of tubulin/pCopN at 20 μm/5 μm (dotted line) or 30 μm/15 μm (dashed line) concentrations. Arrowheads, time of the reverse temperature switch (same time for the 15:0 and the 20:5 tubulin/pCopN samples). The 1:1 tubulin/pCopN stoichiometry ensured that “free” tubulin was at the same concentration at the beginning of each set of experiments (either 10 μm (left) or 15 μm (right)). D, dual mechanism of pCopN on microtubule assembly. pCopN inhibits microtubule elongation by sequestering tubulin (B). The inhibition of microtubule nucleation is not solely explained by tubulin sequestration because increasing the tubulin-pCopN complex concentration for a given free tubulin concentration delays assembly (C). This implies that pCopN or its complex with tubulin interacts with the nucleus to yield a non-productive (as schematized here) or less productive nucleus for microtubule assembly ([Nuc]*). AU, absorbance units.
Tubulin Preparation
Tubulin was purified from ovine brain by two cycles of assembly-disassembly in a high molarity Pipes buffer (18). Before use, an additional cycle of assembly-disassembly was performed to remove inactive protein. GTP-tubulin and GDP-tubulin (i.e. with the β subunit loaded with GTP or GDP, respectively) were prepared by incubation with 1 mm nucleotide followed by a desalting step to remove the excess nucleotide.
Size Exclusion Chromatography
Samples were analyzed by gel filtration at a 0.5 ml/min flow rate on a Superdex S200 column (GE Healthcare) equilibrated with 20 mm Pipes-K, pH 6.8, 1 mm EGTA, and 2 mm MgCl2.
Microtubule Assembly
Microtubule assembly was monitored turbidimetrically in duplicate recordings in a Flexstation 3 plate reader (Molecular Devices) in 96-well plates. Plates with wells containing variable amounts of tubulin for a fixed concentration of pCopN constructs in a buffer consisting of 20 mm Pipes-K, pH 6.8, 6 mm MgCl2, 1 mm EGTA, 32% (v/v) glycerol, and 0.5 mm GTP in a total volume of 200 μl were prepared on ice and then transferred to the plate reader maintained at 37 °C. Microtubule assembly was monitored by recording the absorbance at 350 nm for 45 min at 30-s intervals, with the plates being shaken for 2 s before each measurement. We checked that the steady state had been reached after the 45-min recording time for each condition tested (data not shown). To ensure that the signal was due to microtubule assembly and not to cold stable tubulin aggregates, the absorbance was corrected for the residual signal after an additional incubation of the plate for 20 min on ice. The data were also corrected for optical path length using the “PathCheck” option of the Flexstation. The plot of the corrected signal as a function of the total tubulin concentration yields a linear curve, the abscissa intercept of which represents the critical concentration for tubulin assembly.
Additional experiments, in particular to evaluate the pCopN effect on microtubule nucleation and the effect on microtubule assembly of pCopN mutants and of some constructs, were performed in 50 mm Mes-K, pH 6.8, 6 mm MgCl2, 1 mm EGTA, 30% (v/v) glycerol, and 0.5 mm GTP and monitored at 350 nm with a Cary 50 spectrophotometer (Agilent Technologies), using a 0.7-cm path length cuvette.
Affinity Measurement by Fluorescence Spectroscopy
pCopN was labeled with acrylodan following established protocols (19). In short, the natural cysteine of pCopN (Cys-217) was reduced and then reacted with acrylodan, following which the excess dye was removed by desalting on a PD-10 column (GE Healthcare). This acrylodan-labeled pCopN was used for affinity measurements with GDP-tubulin and GTP-tubulin in a buffer consisting of 20 mm Pipes-K, pH 6.8, 1 mm MgCl2, 1 mm EGTA, and a 10 μm concentration of the relevant nucleotide. The interaction was followed by fluorescence spectrometry (λex, 280 nm; λem, 510 nm) at 20 °C in a Cary Eclipse spectrofluorometer (Agilent Technologies). Increasing amounts of tubulin were added every 2 min to a fixed (50 nm) concentration of labeled pCopN. The data were fitted to a 1:1 binding isotherm with Equation 1,
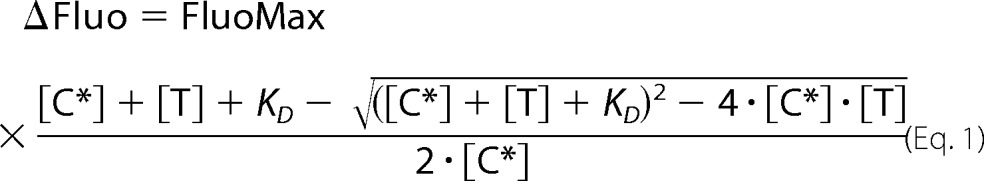 |
where ΔFluo is the variation of the fluorescence signal, FluoMax is the fluorescence at saturating concentration of tubulin, [T] and [C*] are the concentrations of tubulin and labeled pCopN, respectively, and KD is the equilibrium dissociation constant.
Competition for Tubulin Binding Experiments
The competition for tubulin binding between FL_pCopN and designed ankyrin repeat proteins (DARPins) was monitored using the fluorescence assay used for the affinity measurements. To the preformed acrylodan-labeled FL_pCopN-tubulin complex we added an excess of the D2 DARPin (19), and the fluorescence decrease was recorded. As a positive chase control, unlabeled FL_pCopN was added to the fluorescent complex.
Tubulin Nucleotide Exchange
The fluorescence decrease associated with the fluorescence resonance energy transfer from tubulin tryptophans to S6-GDP (20, 21) was used to examine the kinetics of GDP release from 1 μm tubulin in the presence of a 5-fold excess of CopN. GDP-tubulin was equilibrated in 20 mm Pipes-K, pH 6.8, 1 mm MgCl2, 1 mm EGTA through a PD-10 desalting column before being mixed with CopN constructs. The exchange kinetics were monitored at 20 °C using a Cary Eclipse spectrofluorometer (λex, 295 nm; λem, 330 nm). After the addition of 10 μm S6-GDP, the signal decreases at the rate of GDP release from tubulin as it is replaced by S6-GDP.
Vinblastine-induced Tubulin Helical Assembly
Tubulin (10 μm) with or without 10 μm pCopN constructs was incubated with 10–100 μm vinblastine for 20 min on ice in 20 mm Pipes-K, pH 6.8, 0.2 mm MgCl2, 0.1 mm EGTA, and 10 μm GDP. After high speed centrifugation (300,000 × g for 10 min at 4 °C), the supernatant and pellet were analyzed by SDS-PAGE on Coomassie Blue-stained gels.
Crystallization and Structure Determination
The lysines of FL_pCopN and of the D29 construct were methylated prior to crystallization (22). The methylation reaction was performed in 20 mm Tris-HCl, pH 7.5, 100 mm NaCl at a 2 mg/ml protein concentration. To 0.5-ml aliquots, 20 μl of freshly prepared 1 m dimethylamine-borane complex (ABC) (Fluka) and 40 μl of 1 m formaldehyde were added twice, separated by an incubation time of 2 h at 4 °C. After a final 10-μl ABC addition, the reaction mixture was incubated overnight at 4 °C. The methylated proteins were purified on a desalting column preequilibrated with 20 mm Tris-HCl, pH 7.5, 100 mm NaCl and then concentrated to 30 mg/ml for crystallization. Crystals were obtained at 293 K by vapor diffusion in the following crystallization buffers: 0.1 m Tris-HCl, pH 8.5, 0.1 m lithium sulfate, 30% (w/v) polyethylene glycol 4000 for FL_pCopN and 0.1 m Hepes, pH 7.5, 25 mm NaCl, 1.6 m ammonium sulfate for D29. Before being flash-cooled in liquid nitrogen for data collection, crystals were transferred in the crystallization buffer supplemented with 20% glycerol. Both proteins crystallized in the P212121 space group with very similar cell dimensions (Table 1). Complete data sets were collected at 100 K on the Proxima 1 beamline (SOLEIL synchrotron, Saint Aubin, France). In each case, data were collected from a single crystal and processed with XDS (23). The structure of D29 was solved from a single-wavelength anomalous dispersion (SAD) experiment at the 1.9-Å wavelength exploiting the sulfur-SAD signal. The positions of 11 anomalous scatterers (corresponding to 7 of 12 Met + Cys residues expected in the asymmetric unit and to four ions from the crystallization buffer) were determined with SHELXD (24), and phasing was performed using Phaser (25). After density modification with Parrot (26), automatic building in the electron density was performed with Buccaneer (27) followed by manual building in Coot (28). The structure of FL_pCopN was solved by molecular replacement using Phaser (25) with the structure of D29 as a search model. The structures were refined with Phenix (29) and Buster (30), alternating refinement with model building in Coot. Structure validation, including a step of refinement, was performed using PDB_REDO (31). Statistics for data processing and refinement are reported in Table 1. Coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 4P40 (FL_pCopN) and 4P3Z (D29). Figures of structural models were generated with PyMOL (32). The electrostatic potential at the pCopN surface was calculated using APBS (33) and rendered in PyMOL.
TABLE 1.
Data collection, phasing, and refinement statistics
| D29 native | D29 Sulfur-SAD | FL_pCopN | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 67.4 67.5 85.19 | 67.2 67.2 85.7 | 66.8 68.0 85.0 |
| Wavelength (Å) | 0.9801 | 1.9074 | 0.9801 |
| Resolution (Å)a | 50-1.77 (1.88-1.77) | 100-2.62 (2.73-2.62) | 42.52-1.20 (1.28-1.20) |
| Rsym (%)a | 4.6 (64.9) | 3.2 (8.4) | 5.9 (57.0) |
| 〈I/σI〉a | 19.50 (2.02) | 57.8 (21.45) | 10.72 (2.01) |
| Completeness (%)a | 99.3 (97.9) | 99.5 (97.8) | 97.5 (95.6) |
| Multiplicitya | 4.3 ( 4.3) | 10.6 (6.8) | 3.53 (2.5) |
| Phasing | |||
| No. of sites | 11 | ||
| Figure of merit (before/after density modification) | 0.35/0.685 | ||
| Refinement | |||
| Resolution (Å) | 16.29-1.77 | 41.6-1.2 | |
| No. of reflections | 33431 | 118160 | |
| Rwork/Rfree | 0.1830 / 0.2353 | 0.1433/0.1624 | |
| No. of atoms | |||
| Protein | 4566 | 4596 | |
| Water | 151 | 449 | |
| Total | 4770 | 5089 | |
| B factors | |||
| Protein | 37.59 | 22.76 | |
| Water | 41.89 | 37.21 | |
| r.m.s deviations | |||
| Bond lengths (Å) | 0.016 | 0.013 | |
| Bond angles (degrees) | 1.28 | 1.435 | |
| Ramachandran | |||
| Favored region (%) | 98.98 | 99.02 | |
| Allowed region (%) | 1.02 | 0.98 | |
| Outliers (%) | 0 | 0 | |
a Values in parentheses are for the highest resolution shell.
RESULTS
pCopN Sequesters Tubulin in a 1:1 Complex and Delays Microtubule Nucleation
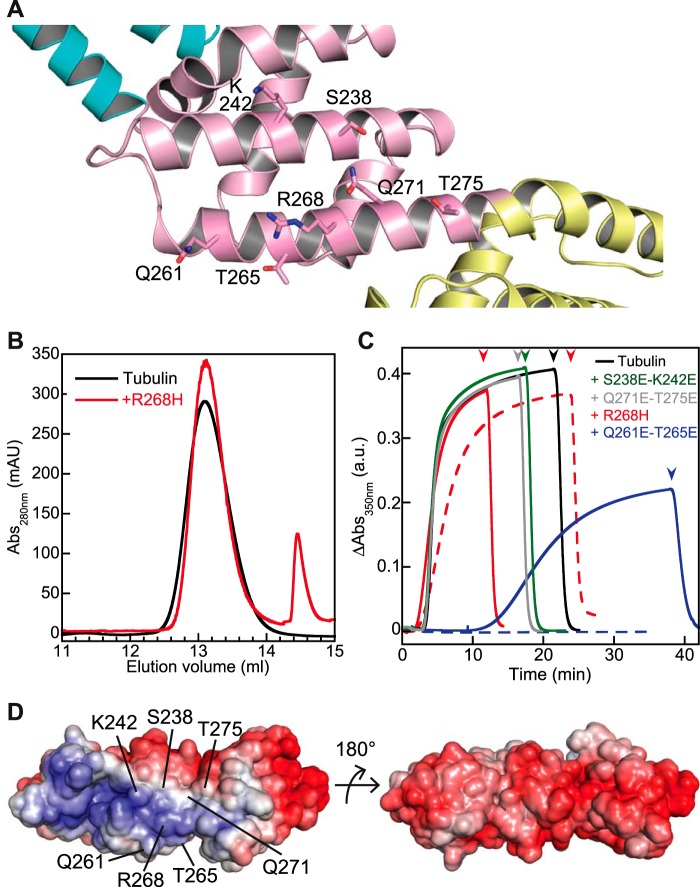
pCopN has been reported to interact with tubulin and to inhibit microtubule assembly in a dose-dependent manner (7). This could be achieved in at least two ways: by a sequestering mechanism (34, 35) or by capping microtubule ends (36). In order to distinguish between these two possibilities, we quantified the effect of pCopN on tubulin assembly at steady state using critical concentration plots derived from turbidimetric measurements. The critical concentration of tubulin alone for microtubule assembly was 2.3 μm. Full-length pCopN (FL_pCopN) as well as D29 and D84, the two N-terminally truncated constructs previously studied by Archuleta and co-workers (7) (Fig. 2A), indeed inhibited tubulin assembly in a dose-dependent manner. The critical concentration plots obtained were shifted while remaining parallel to the control straight line observed with tubulin alone, two characteristics of a sequestration mechanism (34, 35) (Fig. 2B). The amount of sequestered tubulin in complex with pCopN constructs can be derived from the shift of the plot. In the presence of 5 μm constructs, the apparent critical concentration varied from 6 to 7.7 μm, which corresponds to a 3.7–5.4 μm shift. We also tested the effect of D29 at a 3 μm concentration. In this case, the curve was shifted by 2.6 μm as compared with the control with tubulin alone (Fig. 2B). These values are consistent with a 1:1 tubulin/pCopN stoichiometry. They establish that pCopN is a tubulin-sequestering protein that makes a binary 1:1 tubulin-pCopN complex that is not incorporated in microtubules.
In the course of these experiments, we noticed that pCopN delayed microtubule assembly, and it did so in a dose-dependent manner (Fig. 2C). Microtubule assembly is a two-step process, in which a nucleus forms and subsequently elongates (Fig. 2D); the turbidity increases mainly along with microtubule elongation. We showed that raising the tubulin-pCopN complex concentration for a given free tubulin concentration correlated with a longer lag time preceding the turbidity increase (Fig. 2C). Nevertheless, and as expected for a sequestering mechanism, a similar turbidity level was observed at steady state (Fig. 2C, right). This behavior is typical of the inhibition of the nucleation step (37). Therefore, our data point to a dual interference of pCopN with microtubule assembly (Fig. 2D).
pCopN Prevents the Self-assembly of Tubulin by Targeting Its β Longitudinal Interface
To gain further insight into the dual mechanism of the pCopN inhibition of microtubule assembly, we mapped the pCopN footprint on tubulin by studying its interference with well characterized tubulin properties. First, we recorded the effect of pCopN on vinblastine-induced tubulin self-assembly. Vinblastine induces the formation of helical assemblies (38) by promoting tubulin-tubulin longitudinal associations (39). The size of the oligomers is large enough to be analyzed in a high speed centrifugation assay. Using a 10 μm tubulin concentration, we found that almost all of the tubulin went to the pellet after centrifugation in the presence of 100 μm vinblastine but that, in the presence of 10 μm pCopN constructs, tubulin remained in the supernatant (Fig. 3A). This result indicated that pCopN prevents tubulin longitudinal self-assembly.
FIGURE 3.
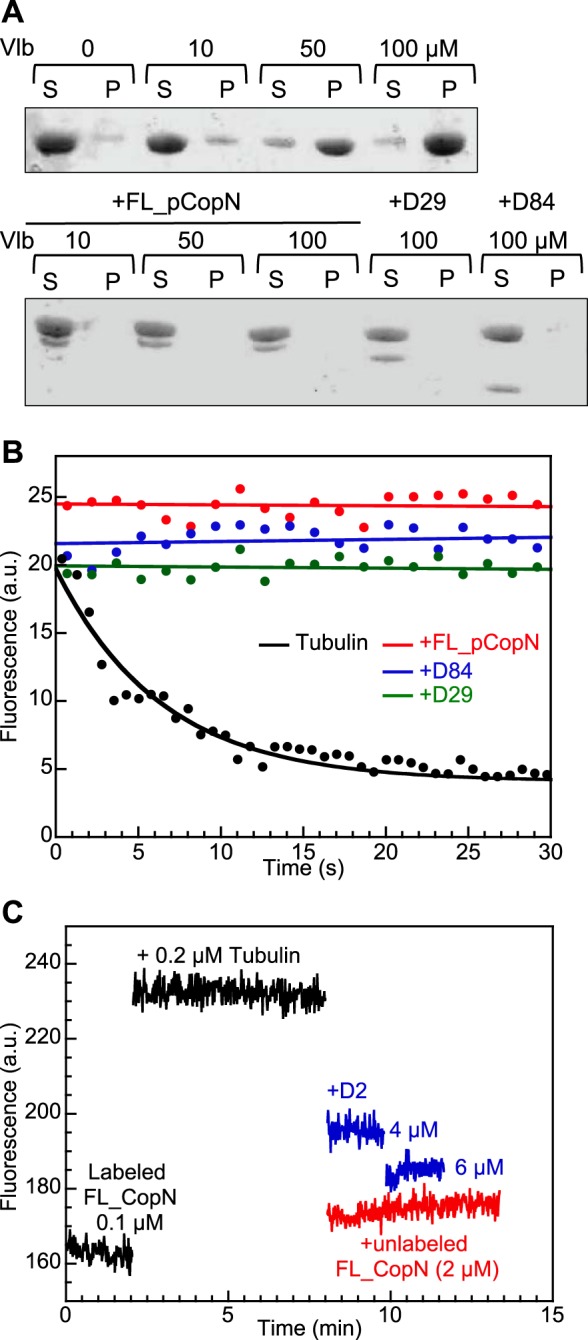
pCopN prevents tubulin assembly by targeting the β subunit longitudinal interface. A, inhibition of the vinblastine-induced tubulin helical assembly by pCopN. Vinblastine-dependent tubulin self-association was assayed under the conditions described under “Experimental Procedures” by analyzing the tubulin content in the supernatant (S) and pellet (P) after centrifugation of mixtures of the relevant proteins with vinblastine (Vlb) at the indicated concentrations. The vinblastine-induced assembly (top) is inhibited by pCopN constructs (bottom). B, pCopN inhibits the tubulin nucleotide exchange. Nucleotide exchange was assayed by recording the fluorescence decrease associated with the substitution of GDP by S6-GDP in the β subunit nucleotide site. For tubulin alone, the experimental data points (black dots, 20% of which are represented) were fitted with a monoexponential decay function (black curve). In the presence of FL_pCopN (red), D29 (green), or D84 (blue), the fluorescence signal does not vary appreciably with time; the curves are linear fits of the experimental data (10% of which are represented). C, FL_pCopN competes with the D2 DARPin for tubulin binding. The fluorescence signal of 0.1 μm acrylodan-labeled FL_pCopN is enhanced upon the addition of 0.2 μm tubulin. It decreases upon the addition of D2 at the indicated concentrations (blue curves) or of 2 μm unlabeled FL_pCopN (red) used as a positive control. The somewhat lower efficiency of D2 as compared with that of FL_pCopN suggests that the former has a slightly lower affinity for tubulin. a.u., arbitrary units.
To demonstrate that pCopN targets the β subunit longitudinal interface, we studied the effect of pCopN on the tubulin nucleotide exchange as well as its competition with a DARPin that binds to that surface. We studied the interference of pCopN with tubulin nucleotide exchange by recording the quenching of the fluorescence of tubulin's tryptophan residues upon substitution of GDP by S6-GDP (20, 21). Fitting the data with a monoexponential decay function, we found a rate constant of 0.15 ± 0.02 s−1 for tubulin alone, in agreement with previous results (21). In the presence of a 5-fold excess of pCopN constructs over tubulin, the fluorescence signal remained constant over time, demonstrating that pCopN prevented nucleotide release from β-tubulin (Fig. 3B). This result strongly suggests that pCopN interacts with the β-tubulin longitudinal interface in the vicinity of the exchangeable nucleotide binding site. To confirm the identity of the tubulin surface contacted by pCopN, we performed a competition experiment for tubulin binding between FL_pCopN and the D2 DARPin, which binds to the longitudinal interface of the tubulin β subunit (19). To this end, we labeled the natural cysteine of FL_pCopN with acrylodan. The binding of tubulin to acrylodan-labeled FL_pCopN led to an increase of the fluorescence signal (Fig. 3C). We then performed chase experiments by adding to the fluorescent complex D2 in excess or, as a control, unlabeled FL_pCopN. We observed a fluorescence decrease, back to a level close to that of labeled FL_pCopN alone (Fig. 3C). We conclude that pCopN and D2 compete for the same binding site at the β-tubulin longitudinal interface.
pCopN Discriminates between Distinct Nucleotide-bound States of Tubulin
Because pCopN interferes with nucleotide exchange, it might also discriminate tubulin according to its nucleotide state. To evaluate this possibility and, more generally, to quantify the strength of the interaction, we determined the affinity of pCopN for GTP- and GDP-tubulin by recording the fluorescence change upon the stepwise addition of tubulin to acrylodan-labeled pCopN (Fig. 4). Fitting the data with Equation 1 (see “Experimental Procedures”) yielded the value of the equilibrium dissociation constant. We found that FL_pCopN had a higher affinity for GDP-tubulin than for GTP-tubulin (KD = 135 ± 15 and 300 ± 20 nm, respectively, averaged from three independent experiments). The difference in affinity according to the tubulin nucleotide state is consistent with pCopN interacting with the longitudinal interface of the β subunit, where the structural differences between GTP- and GDP-tubulin are found (40).
FIGURE 4.
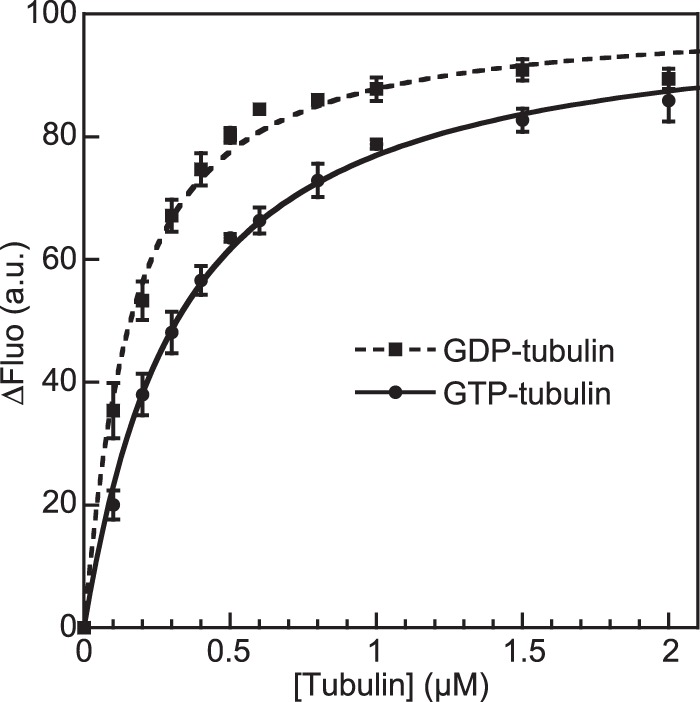
pCopN discriminates between distinct nucleotide-bound states of tubulin. The fluorescence variations of 50 nm acrylodan-labeled FL_pCopN upon the addition of GTP-tubulin (●) or GDP-tubulin (■) are shown. The curves are the fits of the experimental data points with Equation 1 from which the KD is deduced. Error bars, S.D. values calculated using data from three experiments. a.u., arbitrary units.
pCopN Belongs to the LcrE Family of Proteins
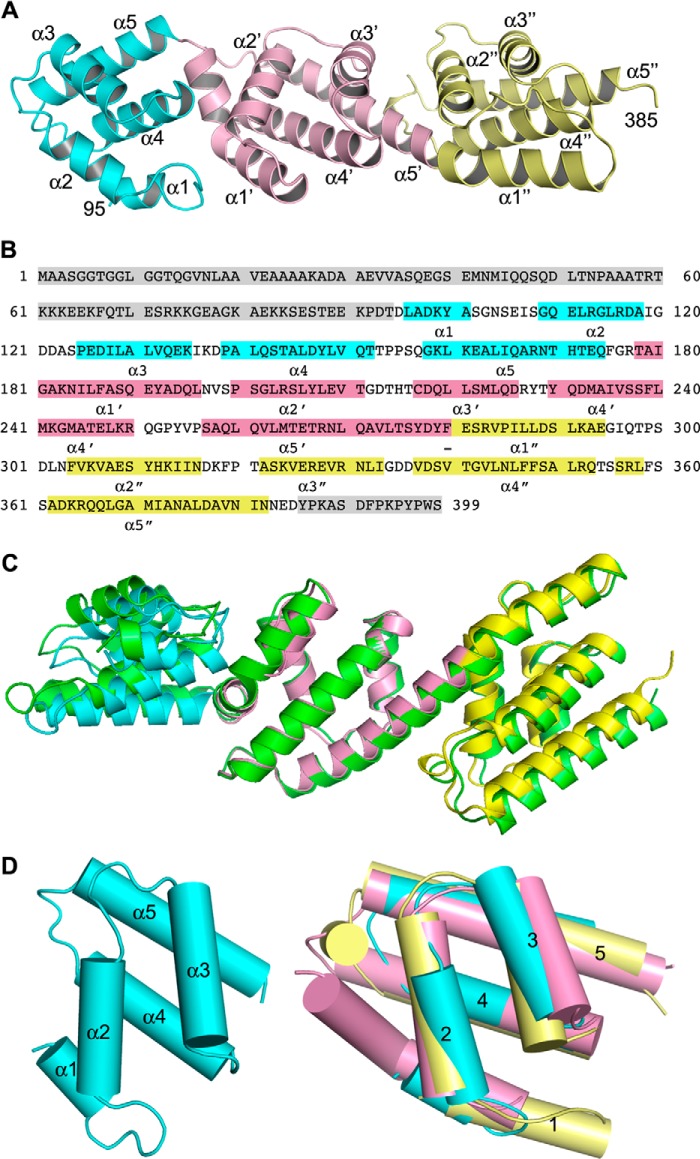
Having established that pCopN targets the β-tubulin longitudinal interface, we undertook to narrow down the tubulin-interacting surface on the pCopN side. To this end, we first determined the pCopN structure. After reductive methylation of lysines, we obtained crystals of the D29 construct and determined its structure using the anomalous signal of sulfur atoms. We also obtained crystals of methylated FL_pCopN that diffracted to 1.2 Å and determined the structure of the full-length protein by molecular replacement, taking the D29 structure as a model (Table 1). pCopN is an elongated protein comprising three repeated helical motifs flanked by disordered N-terminal and C-terminal extensions (Fig. 5). The N-terminal region, corresponding to the first ∼95 residues in FL_pCopN, and the last ∼15 C-terminal residues were disordered both in FL_pCopN and in D29. We defined the N-terminal helical motif (R1) as residues 96–175, the second one (R2) as residues 176–278, and the C-terminal motif (R3) as residues 279–385. There is some flexibility in the definition of the boundaries, in particular between R2 and R3, which share a long, kinked helix. The relative orientation of the domains differs slightly between the FL_pCopN and D29 structures, probably reflecting the plasticity of the protein (Fig. 5C). The helical motifs superposed quite well (Fig. 5D); taking R2 as a reference, the r.m.s. deviation after superposition (using SSM (41)) was 2.2 Å in the case of R1 (77 Cαs compared) and 1.7 Å in the case of R3 (81 Cαs compared). Interestingly, this is despite a very low similarity at the amino acid sequence level (data not shown).
FIGURE 5.
The pCopN structure. A, overview of the D29 structure. The helical motifs are colored cyan, pink, and yellow from the N-terminal residue (Asp-95) to the C-terminal residue (Asp-385) of the structured region. The helices are labeled according to B. B, secondary structure of pCopN. The disordered N- and C-terminal regions are highlighted in gray. The helices are colored by motif in connection with A; loop residues are not highlighted. The N-terminal moiety of the long helix from Ser-257 to Glu-294 constitutes the last R2 helix (α5′), whereas its C-terminal moiety corresponds to the first R3 helix (α1″). The region equivalent to this long helix but at the R1-R2 junction comprises two distinct helices, α5 and α1′ (which is kinked (see D)), separated by a 3-residue-long loop. The R3 motif also comprises a short 3/10 helix (residues 356–358). C, superposition of FL_pCopN and D29 structures on the second helical motif. D29 is colored as in A, and FL_pCopN is shown in green. D, the five-helix motif. Left, the R1 repeat; right, superposition of R1 and R3 on the second (R2) repeat.
The structure of pCopN closely resembles that of the MxiC protein from Shigella (42); they share both the disordered N-terminal region and the following three repeated helical motifs (Fig. 6). Whereas the repeated motif was described as a four-helix X-bundle fold (43) in the case of MxiC (42), in the case of pCopN, it was best described as a five-helix motif, two roughly parallel helices (α2 and α3) lying on the other three, also approximately parallel, helices (α1, α4, and α5) (Fig. 5D). Based on genomes and sequence analysis, it has been suggested that CopN belongs to the YopN/TyeA/InvE/MxiC (or LcrE) family of proteins (13, 15). These proteins are either composed of a single polypeptide chain, as is the case of CopN and MxiC, or of two chains, the YopN-TyeA complex from Yersinia being one example (44). In this complex, TyeA is structurally equivalent to the R3 repeat (42). The structure of pCopN therefore confirms that it belongs to this family of proteins.
FIGURE 6.
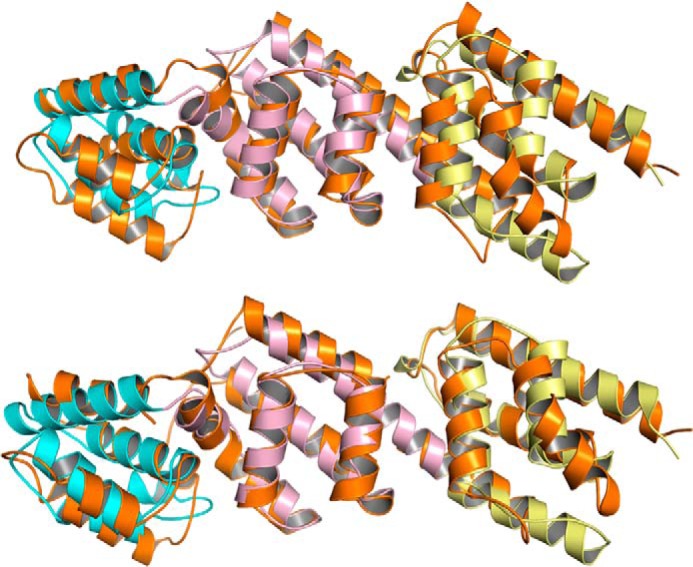
Superposition of MxiC (colored in orange, Protein Data Bank code 2VJ4) on pCopN (colored as in Fig. 5A). The r.m.s. deviation after an overall superposition is 3.9 Å (239 Cαs compared; top), whereas the r.m.s. deviation after a motif-based superposition is 2.8 Å for the first helical repeat (86 Cαs compared), 1.8 Å for the second (90 Cαs compared), and 2.3 Å for the third (91 Cαs compared) (bottom). The lower r.m.s. deviation as compared with that after superposition of the structures as a whole reflects the flexibility at the junction between the repeated motifs.
The Second and Third pCopN Helical Motifs Are Needed for the Interaction with Tubulin
Because the D84 construct inhibits microtubule assembly (Ref. 7 and Fig. 2) and tubulin nucleotide exchange (Fig. 3B) to the same extent as the full-length protein, the pCopN N-terminal disordered region is unlikely to play a role in the interaction with tubulin. To further delineate the pCopN tubulin-interacting site and to evaluate the respective contribution of the repeated motifs, we produced three additional constructs that were defined based on the structure. One of them (the 85–295 construct) included the R1 and R2 motifs and terminated after the long R2-R3 connecting helix. The D175 construct comprised R2 and R3, and the last one (D273) was composed mainly of the R3 motif (Fig. 2A). Among these three additional shorter constructs, only D175 behaved similarly to FL_pCopN in terms of its interaction with tubulin (Fig. 7A) and its microtubule assembly (Fig. 7B) and nucleotide exchange (Fig. 7C) inhibitions. The other constructs, lacking either the R2 motif (D273) or R3 (85–295 construct), had essentially no effect (Fig. 7, A–C).
FIGURE 7.
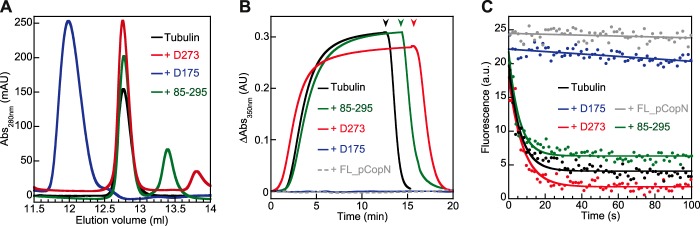
pCopN interacts with tubulin through its second and third helical motifs. The interference with tubulin of the D175, D273, and 85–295 constructs (as defined in Fig. 2A) was assayed by three methods. A, size exclusion chromatography of 20 μm tubulin either alone or in the presence of 20 μm constructs. B, microtubule assembly measured by turbidimetry of 20 μm tubulin either alone or with 20 μm constructs. Arrowheads, temperature switch from 37 to 4 °C. C, tubulin nucleotide exchange (for experimental details, see “Experimental Procedures” and Fig. 3B (from which data for FL_pCopN are taken)). In each case, 10% of the experimental data points are displayed. In the case of tubulin alone or in the presence of D273 or 85–295, the curve is the fit of the data with a monoexponential decay function; in the presence of D175 and of FL_pCopN, the curve is a linear fit of the experimental points. Black, tubulin alone; green, tubulin with the 85–295 construct; blue, tubulin with D175; red, tubulin with D273. The control with FL_pCopN in B and C is displayed in gray. AU, absorbance units; mAU, milliabsorbance units; a.u., arbitrary units.
A screen for pCopN mutants that reverted the growth inhibition phenotype in yeast had identified a R268H substitution (11). Arg-268 is located in the N-terminal part of the R2-R3 connecting helix (Figs. 5B and 8A), exposed to the surface, which would argue for a direct role of this residue in the interaction with tubulin. To characterize the R268H substitution further, we introduced it into the pCopN D29 construct and evaluated its effect in vitro. We found that the R268H mutation strongly reduced the strength of the tubulin-pCopN interference; no interaction with tubulin was detected in our gel filtration assay (Fig. 8B), and R268H retained only a very weak inhibitory effect on microtubule assembly (Fig. 8C). We also tested the effect of the substitution by glutamates of nearby residues (Fig. 8A). The S238E/K242E and the Q271E/T275E double mutants led to proteins that do not inhibit microtubule assembly even at a 40 μm pCopN mutant concentration (Fig. 8C). The Q261E/T265E double mutant displayed an intermediate effect (Fig. 8C), further delineating the pCopN tubulin-interacting surface. Therefore, introducing acidic residues in this overall basic surface of pCopN (Fig. 8D) reduces or annihilates the inhibitory effect on microtubule assembly. To summarize, taken together and as far as the interaction with tubulin is concerned, our results narrow the pCopN minimal functional domain down to the R2-R3 region. Furthermore, because the tubulin surface is mostly electronegative, it is not surprising that the R2-R3 surface interacting with it comprises a basic patch.
FIGURE 8.
Delineating the pCopN tubulin-interacting surface. A, close-up of the pCopN structure showing the location of the residues mutated in this study. B, the pCopN R268H mutation leads to a protein that does not interact with tubulin, as analyzed by gel filtration. Samples of tubulin either alone (black line) or in the presence of R268H (red line) were loaded on the column. C, microtubule assembly of 20 μm tubulin alone or in the presence of R268H (20 μm (solid line) or 30 μm (dashed line)), of S238E/K242E (40 μm), of Q261E/T265E (20 μm (solid line) or 30 μm (dashed line)), or of Q271E/T275E (40 μm). Arrowheads, temperature switch from 37 to 4 °C. The color code is as shown. D, views of the pCopN molecular surface colored by electrostatic potential (red, negative; blue, positive, ranging from −7 to 7 kBT). Left, view of the surface comprising the mutated residues. Right, view along the same orientation as in Fig. 5A. a.u., arbitrary units; mAU, milliabsorbance units.
DISCUSSION
In this study, we have biochemically characterized the interaction of the CopN protein from C. pneumoniae with tubulin. We have shown that pCopN inhibits microtubule assembly by a sequestering mechanism (Fig. 2B). So far, two families of proteins have been described to sequester tubulin, those of stathmin (34) and related proteins (45, 46) and of the PN2–3 domain of the CPAP protein (35). In both cases, detached from tubulin, these sequestering proteins are intrinsically unfolded (35, 47). In addition, several proteins, such as tubulin tyrosine ligase (48) and TOG domains (49), inhibit microtubule assembly in a dose-dependent manner. Both proteins are folded on their own. The pCopN structure comprises a folded core of three repeats of a five-helix motif and disordered N-terminal and C-terminal extensions (Fig. 5). We have shown that the repeated motifs 2 and 3 are both necessary and sufficient to interact with tubulin and to interfere with its assembly (Fig. 7), a pCopN basic surface including Arg-268 most likely being directly implicated in the interaction (Fig. 8). Therefore, pCopN belongs to the group of microtubule inhibitor proteins that bind tubulin via an autonomously structured region.
In addition to sequestering tubulin, pCopN also inhibits microtubule nucleation (Fig. 2C). Such a property has not been reported for the aforementioned sequestering proteins. However, the inhibition of microtubule nucleation is not very efficient because large amounts of pCopN are needed for a noticeable effect to be observed; moreover, pCopN only delays microtubule assembly (Fig. 2C). The fact that pCopN and/or the tubulin-pCopN complex interact directly with the microtubule nucleus but not appreciably with growing microtubules (Fig. 2D) indicates that pCopN targets a binding site present in the former assembly but not in the latter one. A tubulin structural change might be at the origin of this difference. Alternatively, a surface available in the nucleus might be masked in growing microtubules. If this is the case, the pCopN tubulin-binding site is expected to comprise surfaces in addition to the β subunit longitudinal interface described here (Fig. 3), which is accessible at the microtubule (+)-end (36). In agreement with this hypothesis, pCopN interferes with microtubule elongation by sequestering tubulin without any interaction with microtubules (Fig. 2B), whereas the D1 DARPin, which targets a tubulin site restricted to the β subunit longitudinal interface, interacts with the microtubule (+)-end and destabilizes it by a direct effect (36).
pCopN was originally identified as a putative homolog of Yersinia YopN within a locus of type III secretion-related genes, although the conservation at the sequence level was rather low (12). The YopN/TyeA/InvE/MxiC family of bacterial proteins prevents the premature release of effectors through the type III secretion system found in many Gram-negative bacteria (16). Bacterial plug proteins have also been found to be secreted, their secretion being regulated by chaperones (50). The structural relatedness of CopN with MxiC (Fig. 6), together with the observation that it is secreted by the Yersinia secretion system (15) in a way that is modulated by chaperones (16), definitively confirms that CopN is the chlamydial member of this family of plug proteins. Our structural data also give a rationale for the interaction with chaperones that modulate the pCopN secretion. The interaction with the Scc1 and Scc4 chaperones is abolished along with the removal of the first 100 pCopN residues (16), which correspond to the disordered N-terminal extension (Fig. 5B). pCopN also binds the Scc3 chaperone in a way that depends on the presence of the last 79 residues of pCopN (16) (i.e. encompassing the C-terminal disordered region). Regarding the time of CopN expression, proteomic data show that tCopN accumulates in the infectious elementary bodies, whereas it is not detected in the replicative reticulate bodies (51). The transcription profiles of copN also strongly argue for an accumulation of the protein late in elementary bodies (52, 53). tCopN and pCopN were observed associated with the inclusion membrane at mid-infection times (15, 54). Altogether, taking into account the known function of the LcrE family members, it is very likely that pCopN functions as a plug and is secreted upon elementary body contact with the host cell. It is possible that, once elementary bodies have converted to reticulate bodies, CopN is still being synthesized and secreted, never accumulating in the bacteria to a detectable level. In addition to its role in regulating type III secretion, CopN could thus act in the host cytosol at the early to mid-stage of the bacterial developmental cycle.
The next question is the relevance of the tubulin-pCopN interaction in the C. pneumoniae cycle. During the replicative phase of infection, pCopN appears to be of low abundance (54). A low cytoplasmic concentration of a tubulin-sequestering protein is unlikely to significantly alter microtubule dynamics. This is because the amount of trapped tubulin, equivalent to the amount of the sequestering protein, will remain low compared with the pool available for microtubule assembly, the soluble (non-microtubular) concentration of tubulin in the cell having been estimated to be 5–15 μm (17) for a total tubulin concentration of ∼25 μm (55). As discussed above, the anti-nucleation activity of pCopN is not likely to have a significant effect either. The physiological situation therefore contrasts with experiments where pCopN was overexpressed and had a marked effect on microtubules (11). Nevertheless, compounds alleviating growth inhibition by pCopN in yeast have been shown to also inhibit growth of C. pneumoniae in host cells (11). Because yeast growth inhibition is due to the interaction of pCopN with tubulin, this observation suggests that this same interaction is required for C. pneumoniae growth in its host. Moreover, these compounds are inactive on C. trachomatis replication in host cells (11), consistent with the fact that tCopN does not interact with tubulin (Ref. 7 and data not shown). These observations establish a link between the activity in a heterologous system and the function in the bacterial cycle and feed the hypothesis that the tubulin-pCopN interaction is essential for C. pneumoniae development. We formulate two hypotheses for the role of this interaction below.
One possibility is that secreted pCopN remains concentrated in a restricted area. In that case, although being low overall, the concentration of pCopN may be high enough locally to perturb the nucleation or the elongation of a subset of microtubules. Restriction of CopN labeling to the vicinity of the inclusion membrane is consistent with this scenario (15, 54). Another attractive hypothesis is that the physiological relevance of the association of pCopN with tubulin is to trap pCopN until an unknown activation signal comes in. This mechanism is reminiscent of that proposed for the myocardin-related transcription factor (MRTF) family of proteins, whose cellular localization is conditioned by G-actin binding (56). In that case, upon actin polymerization and concomitant G-actin depletion, the actin-MRTF complex dissociates, and the MRTF protein accumulates in the nucleus. In the case of pCopN, the submicromolar KD of the tubulin-pCopN complex (Fig. 4), together with a non-microtubular tubulin concentration at least about 20 times above the KD value (17), ensures that almost all pCopN molecules that are secreted in the cytoplasm bind tubulin. The observation that the R268H pCopN point mutation or the introduction of acidic residues is sufficient to eliminate the interference effects with tubulin (11) (Fig. 8) suggests that the regulation of the tubulin-pCopN interaction might be achieved through some pCopN modification. This would lead to the dissociation from tubulin and to the release of pCopN, making it available to fulfill a function that remains to be identified.
Acknowledgments
We are grateful to N. McDonald for critical reading of the manuscript. We thank I. Mignot for excellent assistance and D. Mauchand (Unité Commune d'Expérimentation Animale, Institut National de la Recherche Agronomique, Jouy en Josas, France) for providing the material from which tubulin was purified. This work has benefited from the facilities and expertise of the LEBS/IMAGIF crystallization and biophysics platforms (CNRS, Gif-sur-Yvette, France). Diffraction data were collected at the European Synchrotron Research Facility (ID14eh4) and at the SOLEIL synchrotron (PX1 and PX2 beamlines). We thank the machine and beam line groups for making these experiments possible.
This work was supported by grants from the Fondation ARC pour la Recherche sur le Cancer (to B. G.), by Agence Nationale de la Recherche Grant ANR-12-BSV8-0002-01 (to B. G.), and by CNRS.
The atomic coordinates and structure factors (codes 4P40 and 4P3Z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CopN
- chlamydial outer protein N
- DARPin
- designed ankyrin repeat protein
- pCopN
- CopN from C. pneumoniae
- tCopN
- CopN from C. trachomatis
- SAD
- single-wavelength anomalous dispersion
- MRTF
- myocardin-related transcription factor
- r.m.s.
- root mean square.
REFERENCES
- 1. Desai A., Mitchison T. J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 2. Nogales E., Whittaker M., Milligan R. A., Downing K. H. (1999) High-resolution model of the microtubule. Cell 96, 79–88 [DOI] [PubMed] [Google Scholar]
- 3. Carabeo R. (2011) Bacterial subversion of host actin dynamics at the plasma membrane. Cell Microbiol. 13, 1460–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hardwidge P. R., Deng W., Vallance B. A., Rodriguez-Escudero I., Cid V. J., Molina M., Finlay B. B. (2005) Modulation of host cytoskeleton function by the enteropathogenic Escherichia coli and Citrobacter rodentium effector protein EspG. Infect. Immun. 73, 2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshida S., Handa Y., Suzuki T., Ogawa M., Suzuki M., Tamai A., Abe A., Katayama E., Sasakawa C. (2006) Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science 314, 985–989 [DOI] [PubMed] [Google Scholar]
- 6. Germane K. L., Ohi R., Goldberg M. B., Spiller B. W. (2008) Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules. Biochemistry 47, 10241–10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Archuleta T. L., Du Y., English C. A., Lory S., Lesser C., Ohi M. D., Ohi R., Spiller B. W. (2011) The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J. Biol. Chem. 286, 33992–33998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dautry-Varsat A., Subtil A., Hackstadt T. (2005) Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 7, 1714–1722 [DOI] [PubMed] [Google Scholar]
- 9. Nguyen B. D., Valdivia R. H. (2012) Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc. Natl. Acad. Sci. U.S.A. 109, 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wood D. O., Wood R. R., Tucker A. M. (2014) Genetic systems for studying obligate intracellular pathogens: an update. Curr. Opin. Microbiol. 17, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang J., Lesser C. F., Lory S. (2008) The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456, 112–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsia R. C., Pannekoek Y., Ingerowski E., Bavoil P. M. (1997) Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25, 351–359 [DOI] [PubMed] [Google Scholar]
- 13. Pallen M. J., Beatson S. A., Bailey C. M. (2005) Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Betts-Hampikian H. J., Fields K. A. (2010) The Chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front. Microbiol. 1, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fields K. A., Hackstadt T. (2000) Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38, 1048–1060 [DOI] [PubMed] [Google Scholar]
- 16. Silva-Herzog E., Joseph S. S., Avery A. K., Coba J. A., Wolf K., Fields K. A., Plano G. V. (2011) Scc1 (CP0432) and Scc4 (CP0033) function as a type III secretion chaperone for CopN of Chlamydia pneumoniae. J. Bacteriol. 193, 3490–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gardner M. K., Charlebois B. D., Jánosi I. M., Howard J., Hunt A. J., Odde D. J. (2011) Rapid microtubule self-assembly kinetics. Cell 146, 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castoldi M., Popov A. V. (2003) Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32, 83–88 [DOI] [PubMed] [Google Scholar]
- 19. Mignot I., Pecqueur L., Dorléans A., Karuppasamy M., Ravelli R. B., Dreier B., Plückthun A., Knossow M., Gigant B. (2012) Design and characterization of modular scaffolds for tubulin assembly. J. Biol. Chem. 287, 31085–31094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yarbrough L. R., Fishback J. L. (1985) Kinetics of interaction of 2-amino-6-mercapto-9-β-ribofuranosylpurine 5′-triphosphate with bovine brain tubulin. Biochemistry 24, 1708–1714 [DOI] [PubMed] [Google Scholar]
- 21. Amayed P., Carlier M. F., Pantaloni D. (2000) Stathmin slows down guanosine diphosphate dissociation from tubulin in a phosphorylation-controlled fashion. Biochemistry 39, 12295–12302 [DOI] [PubMed] [Google Scholar]
- 22. Schubot F. D., Waugh D. S. (2004) A pivotal role for reductive methylation in the de novo crystallization of a ternary complex composed of Yersinia pestis virulence factors YopN, SycN and YscB. Acta Crystallogr D Biol. Crystallogr 60, 1981–1986 [DOI] [PubMed] [Google Scholar]
- 23. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, version 2.8.0, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 31. Joosten R. P., Joosten K., Murshudov G. N., Perrakis A. (2012) PDB_REDO: constructive validation, more than just looking for errors. Acta Crystallogr. D Biol. Crystallogr. 68, 484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 33. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jourdain L., Curmi P., Sobel A., Pantaloni D., Carlier M. F. (1997) Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry 36, 10817–10821 [DOI] [PubMed] [Google Scholar]
- 34. Cormier A., Clément M. J., Knossow M., Lachkar S., Savarin P., Toma F., Sobel A., Gigant B., Curmi P. A. (2009) The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J. Biol. Chem. 284, 6909–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pecqueur L., Duellberg C., Dreier B., Jiang Q., Wang C., Plückthun A., Surrey T., Gigant B., Knossow M. (2012) A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. U.S.A. 109, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carlier M. F., Didry D., Pantaloni D. (1997) Hydrolysis of GTP associated with the formation of tubulin oligomers is involved in microtubule nucleation. Biophys. J. 73, 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weisenberg R. C., Timasheff S. N. (1970) Aggregation of microtubule subunit protein. Effects of divalent cations, colchicine and vinblastine. Biochemistry 9, 4110–4116 [DOI] [PubMed] [Google Scholar]
- 38. Gigant B., Wang C., Ravelli R. B., Roussi F., Steinmetz M. O., Curmi P. A., Sobel A., Knossow M. (2005) Structural basis for the regulation of tubulin by vinblastine. Nature 435, 519–522 [DOI] [PubMed] [Google Scholar]
- 39. Nawrotek A., Knossow M., Gigant B. (2011) The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 412, 35–42 [DOI] [PubMed] [Google Scholar]
- 40. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 41. Deane J. E., Roversi P., King C., Johnson S., Lea S. M. (2008) Structures of the Shigella flexneri type 3 secretion system protein MxiC reveal conformational variability amongst homologues. J. Mol. Biol. 377, 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris N. L., Presnell S. R., Cohen F. E. (1994) Four helix bundle diversity in globular proteins. J. Mol. Biol. 236, 1356–1368 [DOI] [PubMed] [Google Scholar]
- 43. Schubot F. D., Jackson M. W., Penrose K. J., Cherry S., Tropea J. E., Plano G. V., Waugh D. S. (2005) Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346, 1147–1161 [DOI] [PubMed] [Google Scholar]
- 44. Charbaut E., Curmi P. A., Ozon S., Lachkar S., Redeker V., Sobel A. (2001) Stathmin family proteins display specific molecular and tubulin binding properties. J. Biol. Chem. 276, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 45. Ravelli R. B., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M. (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202 [DOI] [PubMed] [Google Scholar]
- 46. Steinmetz M. O., Kammerer R. A., Jahnke W., Goldie K. N., Lustig A., van Oostrum J. (2000) Op18/stathmin caps a kinked protofilament-like tubulin tetramer. EMBO J. 19, 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szyk A., Deaconescu A. M., Piszczek G., Roll-Mecak A. (2011) Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ayaz P., Ye X., Huddleston P., Brautigam C. A., Rice L. M. (2012) A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joseph S. S., Plano G. V. (2013) The SycN/YscB chaperone-binding domain of YopN is required for the calcium-dependent regulation of Yop secretion by Yersinia pestis. Front. Cell Infect. Microbiol. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saka H. A., Thompson J. W., Chen Y. S., Kumar Y., Dubois L. G., Moseley M. A., Valdivia R. H. (2011) Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol. Microbiol. 82, 1185–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Slepenkin A., Motin V., de la Maza L. M., Peterson E. M. (2003) Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect. Immun. 71, 2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Belland R. J., Zhong G., Crane D. D., Hogan D., Sturdevant D., Sharma J., Beatty W. L., Caldwell H. D. (2003) Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lugert R., Kuhns M., Polch T., Gross U. (2004) Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 193, 163–171 [DOI] [PubMed] [Google Scholar]
- 54. Gard D. L., Kirschner M. W. (1987) Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. J. Cell Biol. 105, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mouilleron S., Wiezlak M., O'Reilly N., Treisman R., McDonald N. Q. (2012) Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity. Structure 20, 1960–1970 [DOI] [PubMed] [Google Scholar]