Background: The knockout of the protein UNC-23, a cochaperone of Hsc70, leads to severe motility dysfunction in the model organism C. elegans.
Results: UNC-23 interacts with Hsc70, whose uncoupling from DNJ-13 cures the muscular phenotype.
Conclusion: The dystrophic muscle phenotype is caused by Hsc70 cycle deregulation.
Significance: Hsc70 and its cofactors might be potent targets for therapies against human myopathies.
Keywords: Caenorhabditis elegans (C. elegans), Heat Shock Protein (HSP), Molecular Chaperone, Muscular Dystrophy, Shear Stress, Bag2
Abstract
The molecular chaperone Hsc70 assists in the folding of non-native proteins together with its J domain- and BAG domain-containing cofactors. In Caenorhabditis elegans, two BAG domain-containing proteins can be identified, one of them being UNC-23, whose mutation induces severe motility dysfunctions. Using reporter strains, we find that the full-length UNC-23, in contrast to C-terminal fragments, localizes specifically to the muscular attachment sites. C-terminal fragments of UNC-23 instead perform all Hsc70-related functions, like ATPase stimulation and regulation of folding activity, albeit with lower affinity than BAG-1. Interestingly, overexpression of CFP-Hsc70 can induce muscular defects in wild-type nematodes that phenocopy the knockout of its cofactor UNC-23. Strikingly, the motility dysfunction in the unc-23 mutated strain can be cured specifically by down-regulation of the antagonistic Hsc70 cochaperone DNJ-13, implying that the severe phenotype is caused by misregulation of the Hsc70 cycle. These findings point out that the balanced action of cofactors in the ATP-driven cycle of Hsc70 is crucial for the contribution of Hsc70 to muscle functionality.
Introduction
The chaperone Hsc70 is involved in a variety of cellular processes like de novo protein folding, prevention of aggregation, and protein translocation through membranes (1–3). Hsc70 is composed of an N-terminal ATP/ADP-binding domain, a substrate-binding domain, and a C-terminal lid domain, which covers the substrate binding groove (4, 5). It acts via a regulated ATP-dependent cycle that is influenced by different cofactors, like Hsp40s/J domain-containing proteins and nucleotide exchange factors (NEFs).2 J domain-containing proteins bind to the ATP-bound state of Hsc70 and stimulate the hydrolysis activity of Hsc70. NEFs, like BAG domain-containing proteins, compete with J domain proteins and trigger nucleotide release (3, 6, 7). If both cofactors are present, the ATP turnover is activated strongly. Cells encode highly diverse J domain-containing proteins and NEFs for Hsc70, with different domain compositions and cellular functions (2, 3, 8–10).
In mammalian cells, several cytosolic Hsc70 isoforms and stress-inducible Hsp70s exist, and numerous cofactors increase the complexity of the Hsc70 system. In Caenorhabditis elegans, instead only the Hsc70 ortholog HSP-1 (F26D10.3) is present, and three heat-shock regulated Hsp70 isoforms (C12C8.1, F44E4.4, and F44E4.5) can be induced (11, 12). Nematode Hsc70 is present in most tissues under normal growth conditions, and knockdown via RNA interference (RNAi) leads to enhanced protein aggregation, induction of the heat shock response, and developmental alterations culminating in early larval arrest (11, 13–15).
Mutations at the unc-23 locus had been identified based on motility defects (16). The group of Donald Moerman showed that unc-23 encodes a BAG domain-containing protein, which represents a homolog of human Bag2 (17).3 In nematodes, unc-23 is linked to motility defects (16–18).3 In genome-wide investigations of muscular proteins and their localization, UNC-23 was found to be required for establishing the myosin ultrastructure (18, 19). Mutations in unc-23 result in motility dysfunctions in all developmental stages, including a head-bent phenotype and weakness in the attachment structures connecting hypodermis and muscle cells (16, 20). Here, we investigate the functional interaction of the nematode protein UNC-23 with Hsc70. Using a combination of in vitro and in vivo assays, we report a connection between UNC-23 and the Hsp40 protein DNJ-13, establishing the involvement of the wider Hsc70 machinery in the maintenance of muscle cell functionality and attachment.
EXPERIMENTAL PROCEDURES
Nematode Growth and Cultivation
All nematodes were treated and maintained according to standard procedures (21, 22). The N2 (wild type) and RB1301 (unc-23(ok1408)) strains were provided by the Caenorhabditis Genetics Center (Minneapolis, MN). RB1301 strains were outcrossed three times before performing the described experiments and are referred to as unc-23(ok1408) throughout this work.
RNA Interference
RNA interference experiments were performed by feeding nematodes with dsRNA-expressing Escherichia coli HT115 (DE3) strains as described (15). The RNAi constructs directed against dnj-12, dnj-13, dnj-19, unc-23, hsp-1 and the empty control vector L4440 were sequenced prior to transformation. With the exception of the unc-23 RNAi construct, which was acquired from Open Biosystems (Thermo Scientific, Darmstadt, Germany), all RNAi constructs were obtained from the genome-wide RNAi library (15). Synchronized L1 larvae were placed on the RNAi-expressing bacteria. RNAi motility phenotypes were scored using a Zeiss Stemi stereo microscope (Zeiss Microimaging, Jena, Germany) equipped with a SCHOTT KL1500 LCD unit (Mainz, Germany).
Life Span and Motility Assay
To determine the average life span of wild-type and unc-23(ok1408) nematodes, synchronized L1 larvae were cultivated on NGM/RNAi plates. Early adult nematodes were picked and transferred to new RNAi plates. Life span assays were monitored for adult nematodes and performed as described before (23). To test the motility of nematodes, 3-day-old young adults were placed in a droplet of M9 solution, and the number of lateral swimming movements/min was determined.
Generation of Transgenic Nematodes
An expression reporter construct for unc-23 was generated by cloning 1000 base pairs upstream the unc-23 (H14N18.1c) coding sequence containing the start codon of unc-23 in frame to the YFP gene (Clontech) in the PD95.79 vector. The following primers were used: unc-23 promoter, AGT CCA GCG CGG CCG CCA CTT TGA AAA GTA G (forward) and CGG ACT GCT AGC GCT GAA TAT TAG GAT GG (reverse). In order to investigate protein localization, the coding sequence of H14N18.1, obtained from Open Biosystems (Thermo Scientific) was amplified and inserted into the reporter construct in frame to YFP with the following primers: UNC-23-YFP fusion, ATT GCA GCT AGC ATG TTT CAG AAC ATA CCA ATC AAA ATA C (forward) and CAG CCT GCT AGC TTC GCT TTG ATC ATC CAT C (reverse). The construct for expression of C. elegans Hsc70 (referred to as Hsc70 throughout this work) was generated based on the endogenous hsp-1 (F26D10.3) promoter, which controls a CFP-Hsc70 construct. The following primers were used: Hsc70 promoter, GCA TGC GCG GCC GCT CGT CAC CAA CCA AAA GC (forward) and GGT TTT GCT AGC CTT ACT CAT TTT TAC TGT AAA AAA TAA TTT AAA AAT CAA G (reverse). The CFP-Hsc70 construct was described previously (24) and modified by exchanging the promoter. Further primers were used to generate the dnj-12, dnj-13, and dnj-19 constructs: dnj-12, GGT TTT GCG GCC GCC CGA AGT CTG ACG GCC AAA TGT GTA ATT CC (forward) and GGT TTT TCT AGA AGA CTG TTG GCA TTG AAC GCC TTG TGG C (reverse); dnj-13, GGT TTT GCG GCC GCG TCT CCG TGG CAG GAC CAT ACT GTG CGC CG (forward) and GGT TTT GCT AGC GAA GTT CCT CAG AAT CAC TTC CCG TTG AGT T (reverse); dnj-19, GGT TTT GCG GCC GCG GCA AGC GAA GTT GCG GCT GAC GCG TTA TTG G (forward) and GGT TTT GCT AGC TTG GTG TTG GCA TCT GAC GCC TTG TCC G (reverse). All PCR products with the exception of Hsc70 were inserted via NheI and NotI restriction sites in a modified PD95.79 vector containing either enhanced YFP or enhanced CFP. Microinjection was performed using standard procedures as described (25).
Fluorescence and Confocal Microscopy
For fluorescence microscopy, either a Leica MZ16-FA (Leica, Wetzlar, Germany) stereo microscope or a Zeiss Axiovert 200 microscope equipped with a Hamamatsu C4742-95 camera (Hamamatsu, Herrsching, Germany) was used. For high resolution microscopy of individual muscle cells, confocal microscopy was performed on a Leica SP5 laser-scanning microscope as described previously (22). Images were cropped using ImageJ and Adobe Illustrator. No non-linear modifications of the image data were performed.
Cloning and Protein Expression
Expression plasmids for UNC-23 (H14N18.1), BAG-1 (F57B10.11), DNJ-12 (F39B2.10), and DNJ-13 (F54D5.8) were generated by PCR on the cDNA-containing plasmids obtained from Open Biosystems (Thermo Scientific). PCR products were inserted into the pET28b vector (Merck) and sequenced (GATC Biotech AG, Konstanz, Germany). Expression and purification were performed as described previously (7). Hsc70 was produced in a BioStat C fermenter culture of 7 liters (B. Braun Biotech, Melsungen, Germany). Induction was started at an A600 of 20 under constant monitoring of glucose levels and pH. Harvested bacteria were frozen at −80 °C, and 50 g of the bacterial pellet was used for each purification. The purification of Hsc70 was carried out as described previously (7).
Disulfo-succinimidyl Glutarate (DSSG) Cross-link
Cross-linking of proteins was used to analyze protein complex formation as described previously with a slightly different protocol (26). For cross-linking, we used the compound DSSG-D6/H6 (Creative Molecules Inc., Victoria, Canada), which is characterized by two functional amine-reactive groups. Further, it carries either six protons or six deuteriums to enable mass spectrometric identification of cross-linked peptides for future applications. The cross-linker was used in a 50-fold molar excess as described in the manufacturer's protocol (26–29). A 50 mm stock of the cross-linker DSSG-H6/D6 was prepared and added to a 10 μm protein solution containing either UNC-23 or Hsc70 or a combination of both proteins. The reaction was performed in 40 mm HEPES/KOH, 20 mm KCl, pH 7.5, and stopped with 1 μl of 1 m Tris/HCl, pH 8.0, after 1 and 5 min. Subsequently the samples were analyzed on a 12% SDS-PAGE.
Circular Dichroism Spectroscopy
Measurements were carried out in a 0.1-cm quartz cuvette in a thermostated cuvette holder on a Jasco J-710 spectropolarimeter (Jasco, Gross-Umstadt, Germany) equipped with a PFD-350S Peltier-type attachment unit (Jasco, Gross-Umstadt, Germany). Proteins were dialyzed in 40 mm potassium phosphate, pH 7.5, in concentrations of 0.2 mg/ml and measured as described previously (7). Spectra were measured with 10 accumulations, and the molar ellipticity (ΘMRW) was calculated. For comparability, the temperature transitions were normalized and evaluated to obtain the melting temperature of the purified proteins.
ATPase Measurements
The ATPase activity of Hsc70 was determined using an enzyme-coupled assay described previously (7, 30). For the activity determination without J domain-containing proteins 3 μm Hsc70 and 5 μm NEF (BAG-1 or UNC-23 and the corresponding fragments) were used. The titration assays were performed with 3.2 μm Hsc70 and 0.8 μm DNJ-12 or DNJ-13. ATPase assays were measured at a wavelength of 340 nm and 25 °C. The influence of BAG-1 and various UNC-23 fragments was analyzed by adding different concentrations of these proteins. Data analysis was performed using the OrginPro 8.6 G software package.
Refolding of Luciferase
Luciferase refolding was performed as described before (7). Different UNC-23 constructs were added to 3.2 μm Hsc70 and 0.8 μm DNJ-13. Plateau values of luciferase activities were obtained to evaluate refolding yields and plotted against the UNC-23 concentrations.
Analytical Ultracentrifugation
Analytical ultracentrifugation with fluorescence detection was used to determine the interaction of BAG-1 or UNC-23 with other proteins. To this end, Δ258-UNC-23 was labeled randomly at its lysine residues with 5- (and 6-)carboxyfluorescein succinimidyl ester (Invitrogen). BAG-1 was labeled at its cysteines as described previously (7). Proteins were added to the labeled cofactors, as indicated in the figure legends. To directly quantify the affinity of BAG-1 or Δ258-UNC-23 for Hsc70, the sedimentation properties of the labeled proteins were determined in the presence of different Hsc70 concentrations. The weight average sedimentation coefficient was used as a measure to indicate binding. This parameter, which is calculated from the sedimentation coefficients and concentrations of each species in the solution, can be utilized for direct KD calculations (31, 32). The weight-averaged sedimentation coefficient was obtained from full analysis of the sedimentation velocity experiments using the C(s) module of UltraScanII. For better comparison between the two very different cofactors, the parameter was normalized. Normalized data were then fit to binding isotherms using the concentration Mtot of the labeled protein, the concentration Ltot of Hsc70, and the dissociation constant KD as parameters.
Oligomerization of unlabeled UNC-23 fragments was investigated in an XL-A analytical ultracentrifuge. Data acquisition with the UV detection system was performed at 280 nm. Data analysis of sedimentation velocity experiments was performed using the C(s) analysis tool of the UltraScanII software package (33). Sedimentation coefficients (s20,w), diffusion coefficients (D20,w), and molar masses were obtained from this analysis.
Statistical Analysis
For statistical analysis of the phenotypes, the Mann-Whitney test was used, and the p values were calculated with the OriginPro 8.6 G software package. The p values are given in the figure legends.
RESULTS
UNC-23 Contains a Disordered N-terminal Domain with PXXP Motifs
In C. elegans, two BAG domain-containing proteins can be identified. BAG-1 consists of a C-terminal BAG domain and an N-terminal ubiquitin-like domain, and BAG-1 is the homolog of the human Bag1 with an overall amino acid identity of 30%. UNC-23 is the assigned homolog of human Bag2 (17). Employing HMMER3-based search algorithms (24, 34) with known human and nematodal BAG domains as a starting set, no further BAG domain proteins in the C. elegans proteome could be found (data not shown). The similarity between UNC-23 and Bag2 is strongest in the BAG domain with about 40% amino acid identity (Fig. 1A). Based on the sequence of the obtained cDNA, the N-terminal part of UNC-23 contains two poly(Q)-rich regions, several phenylalanine/proline (F/P) repeats, and three PXXP motifs, making UNC-23 about 200 amino acids longer than its assigned human homolog, which does not contain these domains. PXXP repeats, which are thought to interact with Src homology 3 domains (35), instead can be found in other human BAG proteins, like Bag3 and Bag4 (Fig. 1A), but the homology of their BAG domain to UNC-23 is much lower. Thus, nematode UNC-23 possibly represents a protein that combines characteristics of human Bag2, Bag3, and Bag4 (Fig. 1A).
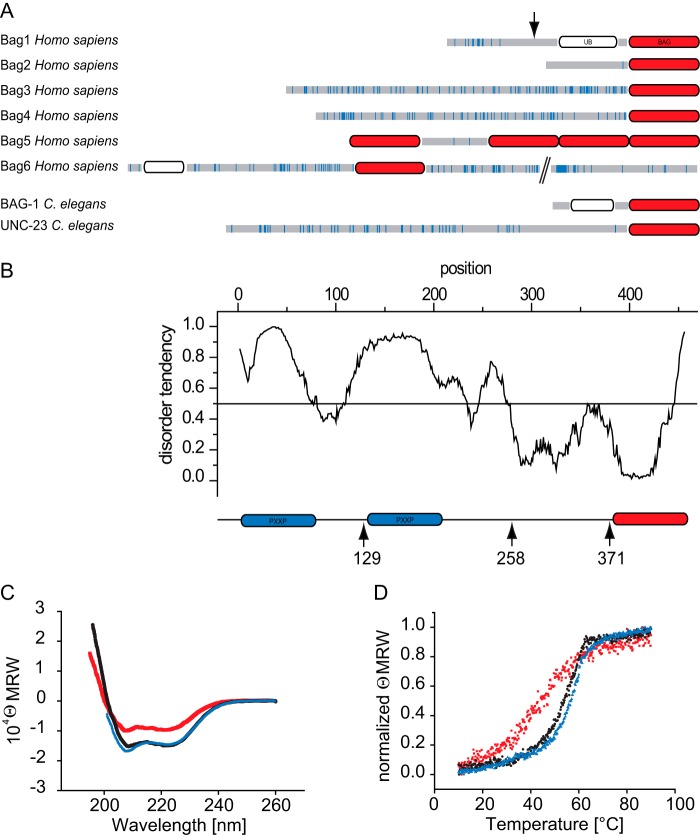
FIGURE 1.
Structure and stability of UNC-23. A, scheme of C. elegans and human BAG domain-containing proteins. The BAG domain (BAG) is depicted in red, and the ubiquitin domain (UB) is shown in white. Proline residues are highlighted by blue bars. The arrow indicates the length of the human Bag-1S splicing isoform. B, IUPred intrinsically unstructured domain search and correspondingly designed UNC-23 protein fragments. Numbers mark the truncation points starting from the N terminus. PXXP motifs are depicted in blue, and the BAG domain is shown in red. C, circular dichroism spectra of the three truncation fragments Δ371-UNC-23 (red), Δ258-UNC-23 (black), and Δ129-UNC-23 (blue). MRW, mean residue weight. D, thermal transitions of Δ371-UNC-23 (red square), Δ258-UNC-23 (black circle), and Δ129-UNC-23 (blue triangle), as measured by circular dichroism at 222 nm. Starting values were normalized for better comparability.
The unusual N-terminal domain of UNC-23 is thought to be regulated by alternative splicing, with three different splice variants (H14N18.1a, H14N18.1b, and H14N18.1c), encoding proteins of 458, 399, and 457 amino acids, respectively (see the WormBase Web site). The cDNA we obtained from Open Biosystems encodes variant H14N18.1c, implying that this splicing form is relevant in vivo.
We were interested in the structural organization of the N-terminal domain. To this end, we performed an unstructured domain search with the algorithm IUPred, which determines the probability of individual amino acids to form stable interactions with their neighboring amino acids (36). This algorithm revealed a very high probability of the N terminus to be disordered, whereas the C terminus containing the BAG domain is predicted to be folded (Fig. 1B). To address this experimentally, we expressed variants of UNC-23, omitting stretches from the N-terminal part (Fig. 1B). Although the fragment corresponding to human Bag2 (Δ258-UNC-23) expressed stably, the presence of PXXP regions at the N terminus strongly reduced the expression and solubility of UNC-23 fragments. The full-length UNC-23 could not be expressed in sufficient amounts, but a UNC-23 variant encoding about half of the N-terminal region (Δ129-UNC-23) was soluble. We investigated the structure content of the purified proteins with CD spectroscopy. Here both proteins showed a similar extent of α-helical structure (Fig. 1C), implying that the N terminus between amino acids 128 and 259 additionally contains some secondary structures.
We further investigated the stability of the proteins by recording thermal transitions using CD spectroscopy. The transitions showed unfolding of both fragments with midpoints at 55 °C for Δ258-UNC-23 and 56 °C for Δ129-UNC-23 (Fig. 1D and Table 1), indicating that the presence of the extended N-terminal domain does not influence the overall stability of the protein. We then deleted the entire region N-terminal of the BAG domain to express the isolated C-terminal BAG domain of UNC-23 (Δ371-UNC-23). This fragment showed a reduced α-helical content (Fig. 1C) and also a reduced thermal stability (Fig. 1D), revealing that amino acids 258–371 contribute to structure and stability of the BAG domain and hence may form an interdependent structure. Nevertheless, all fragments are stable at the experimental temperatures of 20–25 °C.
TABLE 1.
Characteristics of the different UNC-23 truncation fragments
Values were obtained by protein expression, CD thermal transitions, analytical ultracentrifugation, and evaluation of the ATPase assays. Data were analyzed as described under “Experimental Procedures.” S.E. values are shown. Values for BAG-1 were obtained from Sun et al. (7). UZ, analytical ultracentrifugation; ND, not determined.
| Expression | Tm | s20,w | D20,w | Mr(UZ) | Mr(MS) | KD(app) ATPase Hsc70/DNJ-12 [μm] | KD(app) ATPase Hsc70/DNJ-13 [μm] | KD UZ | |
|---|---|---|---|---|---|---|---|---|---|
| °C | S | m2 s−1 | kDa | kDa | μm | ||||
| UNC-23 | − | ||||||||
| Δ129-UNC-23 | + | 56 | 3.17 ± 0.19 | 4.04 ± 0.5 × 10−7 | 68.8 ± 9.9 | 39.438 | 6.84 ± 3.9 | ND | ND |
| Δ258-UNC-23 | +++ | 55 | 3.01 ± 0.12 | 4.5 ± 0.6 × 10−7 | 57.9 ± 7.3 | 24.921 | 15.06 ± 6.0 | ND | 0.5 ± 0.4 |
| Δ371-UNC-23 | + | 46 | 2.92 ± 0.1 | 11.0 ± 0.6 × 10−7 | 23.2 ± 1.45 | 11.964 | 138 ± 23.4 | ND | ND |
| BAG-1 | +++ | ND | 2.1 ± 0.4 (7) | 7.36 ± 1.5 × 10−7 (7) | 23.5 (7) | 24.010 | 0.39 ± 0.12 | 1.35 ± 0.48 | 0.02 ± 0.02 |
Finally, we performed analytical ultracentrifugation to obtain information about the quaternary structure of the purified fragments. In sedimentation velocity experiments, we obtained molecular masses that are roughly twice as high as the calculated molecular weight of the purified fragments (Table 1), implying that the BAG domain with the very C-terminal amino acids is capable of forming dimers. Based on this analysis, we conclude that the extended N-terminal region does not contribute to structure and stability of the more C-terminal parts of the UNC-23 protein.
unc-23(ok1408) Nematodes Show Severe Muscular Defects
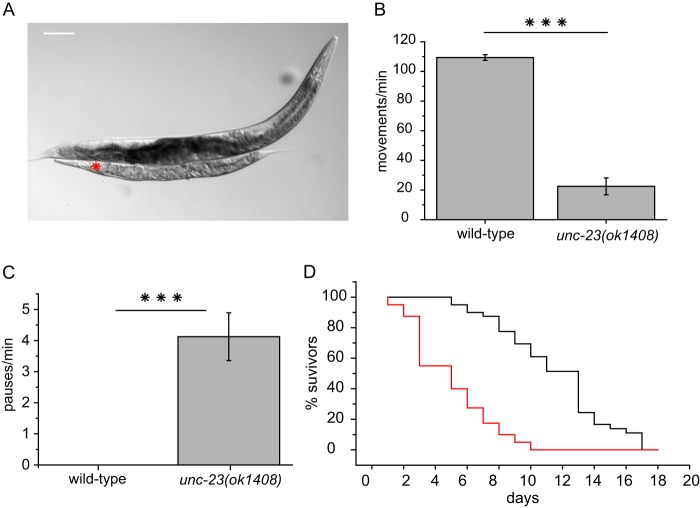
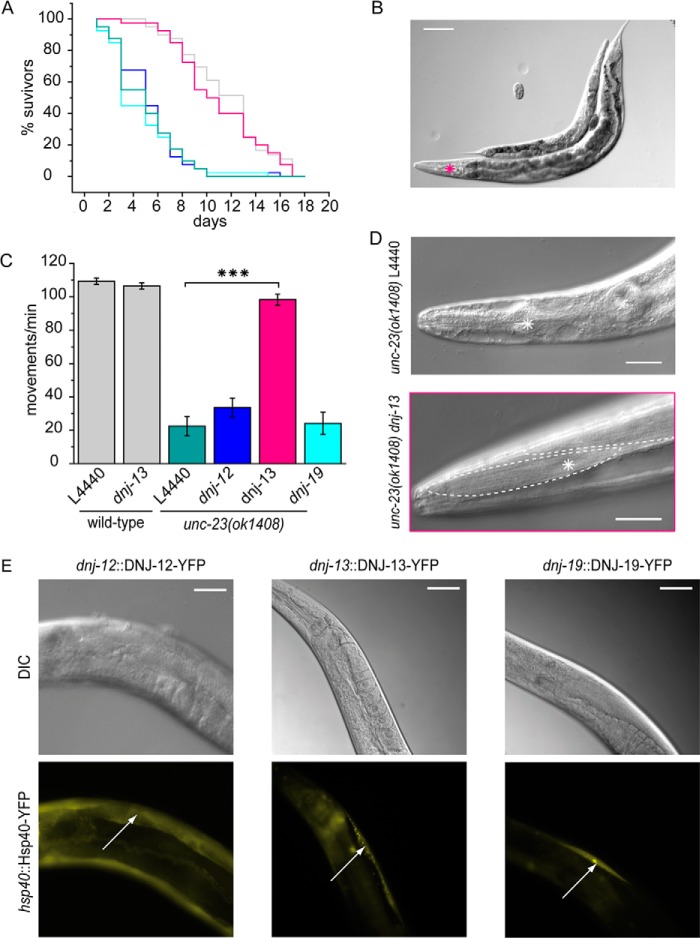
Different mutations in the unc-23 gene had been identified decades ago, and a detailed phenotypic characterization of those strains is available (16). Mutations in unc-23 lead to a reduced overall size of the worms and a head-bent phenotype during forward movements due to failed muscle attachment (16, 20). A knock-out strain of unc-23, containing a large deletion in the gene (strain RB1301), is available from the OMRF Knock-out Group. unc-23(ok1408) worms are smaller than wild-type animals, thinner and more translucent (Fig. 2A). unc-23(ok1408) nematodes show severe motility disorders worsening during development, including the very prominent head-bent phenotype known from other unc-23 alleles (16, 17).3 To quantify the differences in general motility, a thrashing assay was performed with young adult animals. Motility of unc-23(ok1408) nematodes at that developmental stage had decreased significantly with only 23 ± 6 swimming strokes/min compared with 109 ± 2 of the wild-type worm (Fig. 2B). Furthermore, we observed irregularities in pharyngeal pumping, measureable as frequent stops during the normally regular swallowing movements (Fig. 2C). Strikingly, the average life span of the nematodes is decreased to only 5 days compared with 12 days for wild-type nematodes (Fig. 2D).
FIGURE 2.
unc-23(ok1408) shows loss in motility. A, comparison between a young adult knock-out worm of the unc-23(ok1408) strain (red asterisk) and a young adult wild-type hermaphrodite. Scale bar, 100 μm. B, thrashing assay of young adult wild-type nematodes and unc-23(ok1408). Swimming movements/min were counted in M9 droplets. Means with S.E. (error bars) were plotted. For statistical analysis, the Mann-Whitney test was used: p = 2.83 × 10−6 < α = 0.001. Significance is indicated by three asterisks. C, irregularities in swallowing movements were counted in young adult wild-type and unc-23(ok1408) nematodes. For statistical analysis, the Mann-Whitney test was used: p = 3.22 × 10−6 < α = 0.001. Significance is indicated by three asterisks. D, life span analysis represented by a Kaplan-Meyer plot of unc-23(ok1408) (red) and wild type (black). All assays were performed with at least 10 animals.
Hsc70 and UNC-23 Are Expressed in Muscular Tissues
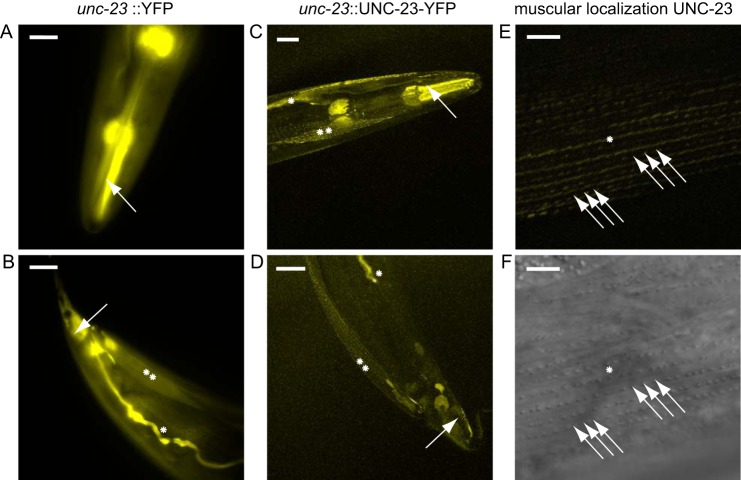
To address the function of UNC-23 in vivo, we determined its tissue-specific expression. We thus generated a YFP-fused promoter construct (unc-23::YFP) and injected it into gonads of wild-type nematodes. Under normal growth conditions, the promoter is active in the pharyngeal muscle cells (Fig. 3A) and the body wall muscle cells (Fig. 3B). Further strong expression can be observed in the excretory canal cell and the anal depressor muscle (Fig. 3B). We then generated a construct containing the full-length UNC-23 fused to YFP (unc-23::UNC-23-YFP). Upon injection, progeny also showed expression in the excretory canal and the body wall muscle cells (Fig. 3, C and D). Confocal microscopy on fluorescent nematodes was performed to investigate UNC-23 localization in the body wall muscle cells in detail. As a marker in the muscle cells, we utilized the dense body structures, which are clearly visible in the differential interference contrast microscopic images (16, 22). UNC-23 colocalized with dense bodies and is additionally found in the M-line of the muscular ultrastructure (Fig. 3E). Although no stable line could be obtained from the described construct, we consistently observed this pattern in all fluorescent nematode progeny (n = 14). Despite lacking information on the functionality of the YFP fusion construct, the localization of UNC-23-YFP in muscular cells was clearly specific.
FIGURE 3.
unc-23::YFP is ubiquitously expressed in C. elegans. A and B, transgenic wild-type nematodes containing the unc-23 promoter fused to YFP. Scale bar, 20 μm. A, the promoter is active in the pharyngeal muscle cells (arrow). B, the nematodal tail is shown with YFP fluorescence in the anal depressor cell (arrow), the excretory canal cell (asterisk) and the body wall muscles (two asterisk). C and D, wild-type nematode containing UNC-23 fused to YFP under the control of the endogenous unc-23 promoter. Scale bar, 20 μm. C, YFP fluorescence is visible in the pharynx (arrow). D, YFP fluorescence can be observed in the anal depressor cell (arrow), the excretory canal (asterisk), and the body wall muscles (two asterisks). Scale bar, 20 μm. E, detailed view of subcellular localization of UNC-23-YFP in the M-line (asterisk) and at the dense bodies (arrows) on the basal lamina side of the striated body wall muscle. F, corresponding differential interference contrast image used for comparison with the dense body localization. Scale bar, 5 μm.
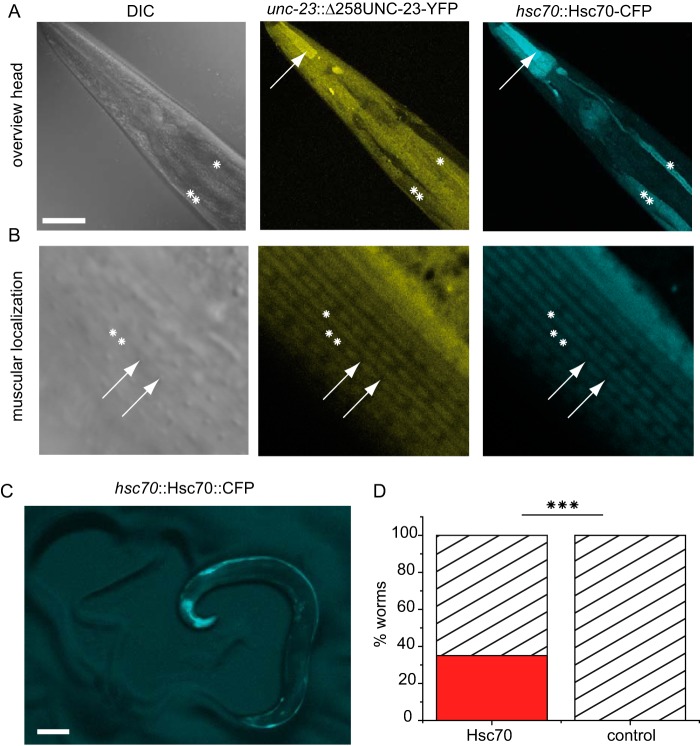
Further, a YFP-fused UNC-23 fragment lacking the N-terminal domain (unc-23::Δ258-UNC-23-YFP) together with the nematode Hsc70 (hsp-1::CFP-Hsc70) were injected. Like other unc-23 constructs, Δ258-UNC-23 showed expression in the pharynx, the excretory canal, and the body wall muscles (Fig. 4A). Strikingly, however, Δ258-UNC-23 localized differently compared with the full-length construct, being absent from the dense body superstructure but enriched in the space between dense bodies (Fig. 4B). Thus, the specific localization of UNC-23 to the dense bodies apparently requires the full-length protein. Hsc70 expression can be observed in muscle cells among other tissues (Fig. 4A). Within the muscular ultrastructure, Hsc70 was present in the M- and I-band, excluding the dense body structures, resulting in a localization overlapping with Δ258-UNC-23 (Fig. 4B).
FIGURE 4.
Localization of Δ258-UNC-23-YFP. Transgenic wild-type nematode expressing CFP-Hsc70 under the control of its endogenous hsp-1 promoter, Δ258-UNC-23-YFP, under the control of the endogenous unc-23 promoter. A, Hsc70 and Δ258-UNC-23 are expressed in the pharynx (arrow), the excretory canal (asterisk), and the body wall muscles (two asterisks). Δ258-UNC-23 is additionally observable in the intestinum. Scale bar, 40 μm. B, detailed view on the basal lamina side of the striated body wall muscles. CFP-Hsc70 and Δ258-UNC-23 show a diffuse localization in the M-band (asterisk) and I-band (two asterisks). No localization at the dense bodies was detectable (arrows). Scale bar, 5 μm. C, transgenic hsp-1::CFP-Hsc70-expressing nematode showing the head-bent phenotype. Scale bar, 100 μm. D, quantification of the head-bent phenotype in the F1 progeny (red) compared with the control (C34B2.5::CFP). For statistical analysis, the Mann-Whitney test was used. p = 1.5 × 10−9 < α = 0.001. Statistical significance is indicated by three asterisks.
During these injections, it became evident that nematodes injected with the CFP-Hsc70 construct can obtain a phenotype similar to the head-bent phenotype of unc-23(ok1408) nematodes (Fig. 4C). This phenotype is observable in about 40% of the F1 progeny (Fig. 4D). This strongly suggests that the deregulation of Hsc70 can develop a harmful gain-of-function phenotype. These results indicate how sensitively the muscle attachment structures react to changes in the Hsc70-system and that deregulation leads to weakened attachment structures similarly to the unc-23 knockout.
UNC-23 Binds Hsc70 via Its BAG Domain with Rather Low Affinity
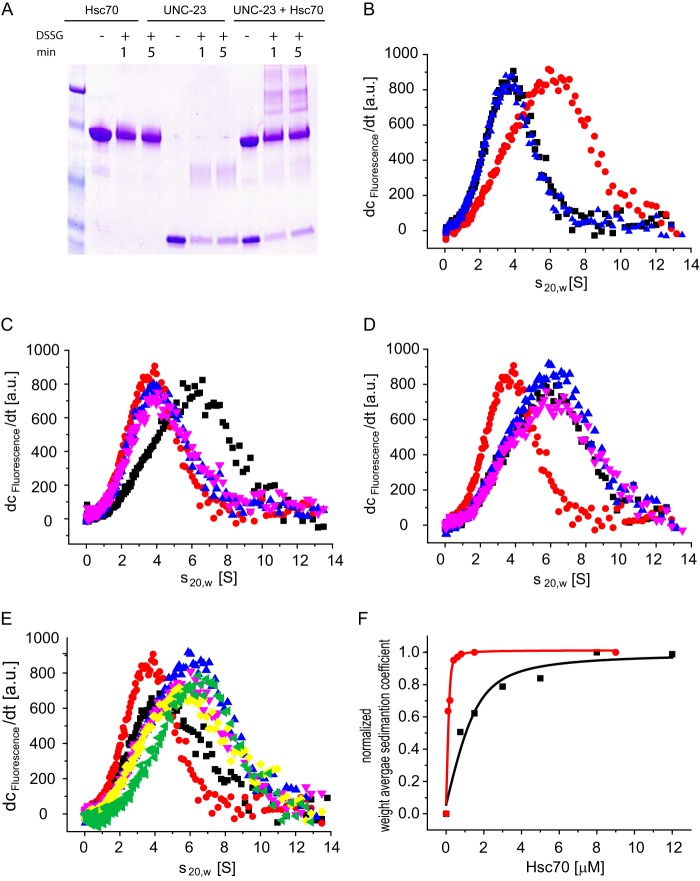
Having seen the localization differences between unc-23 constructs in muscle cells and the phenotypic similarities, we set out to characterize the interaction between Hsc70 and UNC-23 in vitro using purified proteins. We first analyzed the interaction of Δ258-UNC-23 and Hsc70 by cross-linking (Fig. 5A). Δ258-UNC-23 forms dimers upon the addition of the cross-link reagent. Heterooligomers of Δ258-UNC-23 and Hsc70 are formed in the presence of the cross-linker, implying that, in agreement with earlier two-hybrid data (17), the purified proteins can directly interact with each other. To further characterize this complex, analytical ultracentrifugation was performed. To this end, we labeled Δ258-UNC-23 by coupling a fluorescent dye to lysine residues (*Δ258-UNC-23). *Δ258-UNC-23, like the unlabeled protein, sedimented with a sedimentation coefficient of 3.3 S, confirming that the dimeric nature of the protein was maintained after the labeling reaction. The addition of 3 μm Hsc70 increased the s20,w to 6.1 S, proving complex formation between UNC-23 and Hsc70. In contrast, nematode Hsp110/C30C11.4, which is a homolog of Hsc70, did not increase the s20,w of *Δ258-UNC-23, implying that this interaction is specific to Hsc70 (Fig. 5B).
FIGURE 5.
UNC-23 specifically binds to Hsc70 in vitro. A, DSSG-cross-linking experiments were performed in 40 mm HEPES/KOH, 20 mm KCl, pH 7.5. Samples were taken after 1 and 5 min. Samples contained either Hsc70 alone (lanes 2–4), Δ258-UNC-23 alone (lanes 5–7), or a combination of both (lanes 8–10). Lane 1 contains the protein standard. B, dc/dt profiles of velocity sedimentation experiments of 3 μm *Δ258-UNC-23. The assay was performed in 40 mm HEPES/KOH, 150 mm KCl, pH 7.5. Shown are sedimentation profiles of *Δ258-UNC-23 alone (black squares) and in the presence of 5 μm Hsc70 (red circles) or 5 μm Hsp110 (blue triangles). C, sedimentation profiles of *Δ258-UNC-23 in the absence (red circles) or in the presence of 3 μm Hsc70 (black squares). Further additions of ATP (blue triangles) or ADP (pink triangles) were made. The experiment buffer was 40 mm HEPES/KOH, 150 mm KCl, pH 7.5, and 5 mm MgCl2. D, the binding of *Δ258-UNC-23 to Hsc70 in the presence of Hsp40 proteins. Shown are sedimentation profiles of *Δ258-UNC-23 alone (red circles), in the presence of Hsc70 (blue triangles), in the presence of Hsc70 and 5 μm DNJ-12 (black squares), or in the presence of Hsc70 and 5 μm DNJ-13 (pink triangles). E, competition binding of nucleotide exchange factors to the *Δ258-UNC-23·Hsc70 complex. *Δ258-UNC-23 was sedimented alone (red circles) or in the presence of Hsc70 (blue triangles), Hsc70 and 10 μm BAG-1 (black squares), Hsc70 and 10 μm Δ129-UNC-23 (pink triangles), Hsc70 and 10 μm Δ258-UNC-23 (yellow rhombi), or Hsc70 and 10 μm Δ371-UNC-23 (green triangle). F, the affinity of *Δ258-UNC-23 (black) and *BAG-1 (red) was quantified by the addition of increasing concentrations of Hsc70. Weight-averaged sedimentation coefficients were normalized and plotted against Hsc70 concentrations. The evaluations were performed as described under “Experimental Procedures” a.u., arbitrary units.
We then tested the influence of nucleotides on the binding of UNC-23. To this end, ADP and ATP were added to these complexes. We indeed observed a competition between nucleotide binding to Hsc70 and UNC-23 interaction, resulting in disruption of the complexes and sedimentation of *Δ258-UNC-23 with 3.4 S even in the presence of Hsc70 (Fig. 5C). This competition could be confirmed in the absence of MgCl2, highlighting the ability of Hsc70 to bind nucleotides independently of MgCl2 (data not shown). We then tested the influence of Hsp40 proteins on the complex formation between *Δ258-UNC-23 and Hsc70. To this end, we added DNJ-12 or DNJ-13 to *Δ258-UNC-23 and Hsc70 in analytical ultracentrifugation experiments and determined whether complexes are altered. Neither of the Hsp40 proteins could displace *Δ258-UNC-23 from Hsc70 (Fig. 5D), which is supportive of previous studies that ATP presence is required for Hsp40 binding to Hsc70 (6).
We finally tested the competition between different BAG domain proteins during analytical ultracentrifugation. Here the displacement of *Δ258-UNC-23 from Hsc70 by BAG-1 was very efficient. Instead, the competition with equal amounts of either Δ129-UNC-23 or Δ258-UNC-23 did not lead to full displacement (Fig. 5E), implying a weaker affinity compared with BAG-1. We performed the same competition experiment with Δ371-UNC-23. Here, we found the ability to compete with *Δ258-UNC-23 severely compromised (Fig. 5E). Apparently, the shortest UNC-23 fragment is not able to bind with the same affinity as the longer UNC-23 fragments although it contains the BAG domain. To prove the weaker affinity of UNC-23 for Hsc70 in comparison with BAG-1, we analyzed the complex formation of the proteins upon the addition of increasing concentrations of Hsc70 (Fig. 5F and Table 1). We obtained weight-averaged sedimentation coefficients for each Hsc70 concentration and used the increase in this values to determine the binding affinities. For BAG-1, we obtained a binding constant of roughly 0.02 μm in the absence of nucleotides; for Δ258-UNC-23, we obtained 0.5 μm. These data confirm that UNC-23 represents a protein that binds in the absence of nucleotides to Hsc70, albeit with much lower affinity than BAG-1.
DNJ-12 and DNJ-13 Cooperate with UNC-23 during ATPase Stimulation and Protein Folding
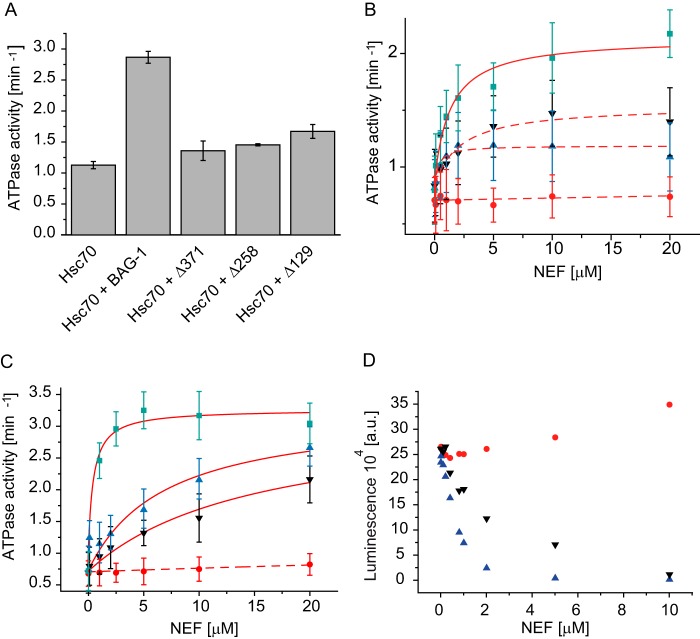
Having characterized the interaction between UNC-23 and Hsc70, we tested the influence of UNC-23 on the ATPase activity of Hsc70. The activation of Hsc70 by UNC-23 is very weak in particular when compared with the stimulation induced by BAG-1 (Fig. 6A). For BAG-1, a very strong activation can be seen, especially in cooperation with Hsp40 proteins (7). This cooperative stimulation is a general feature of BAG proteins and Hsp40 proteins as these two cofactors act on different reaction steps to enhance the ATPase of Hsc70 severalfold (37).
FIGURE 6.
Δ258-UNC-23 can regulate Hsc70 activity. The steady-state ATPase activity of Hsc70 in the presence of different cofactors was determined in 40 mm HEPES/KOH, 150 mm KCl, pH 7.5. A, the ATPase activity of Hsc70 after the addition of BAG-1, Δ129-UNC-23, Δ258-UNC-23, and Δ371-UNC-23. Means with S.E. were plotted. B, ATPase activity of the Hsc70/DNJ-13 system upon the addition of increasing amounts of BAG-1 (green squares), Δ129-UNC-23 (blue triangles), Δ258-UNC-23 (black triangles), or Δ371-UNC-23 (red circles). C, ATPase activity of the Hsc70/DNJ-12 system upon the addition of increasing amounts of BAG-1 (green squares), Δ129-UNC-23 (blue triangles), Δ258-UNC-23 (black triangles), or Δ371-UNC-23 (red circles). The activities were plotted against the protein concentration and evaluated. The assays were analyzed as described previously. Dotted lines were not evaluated further due to small signal changes relative to the error bars. Means and S.D. values (error bars) were plotted. D, refolding of firefly luciferase by the Hsc70/DNJ-13 system was investigated in standard buffer with increasing amounts of Δ129-UNC-23 (blue triangles), Δ258-UNC-23 (black triangles), or Δ371-UNC-23 (red circles).
We thus tested the effect of UNC-23 in the presence of DNJ-13 (Fig. 6B). Here we observed a stimulation with the longer UNC-23 fragments, resulting in an almost 2-fold increase of the ATPase (Table 1). The isolated BAG domain instead did not increase the turnover of the Hsc70 ATPase together with DNJ-13 (Fig. 6B). Likewise, the stimulation in the presence of DNJ-12 was analyzed (Fig. 6C and Table 1). Both UNC-23 fragments, Δ129-UNC-23 and Δ258-UNC-23, induced similar activities, implying that the unstructured N-terminal domain is not involved in this interaction (Fig. 6, B and C). The isolated BAG domain Δ371-UNC-23, however, showed much lower affinity and activity, confirming that the BAG domain alone is not sufficient to exert the Hsc70-related effects of UNC-23 (Fig. 6, B and C). In cases where the stimulation factors were high enough, we analyzed the affinities, which again highlight that the affinity of UNC-23 for Hsc70 under enzymatically relevant conditions is in the micromolar range and thus much weaker than the affinity to BAG-1 (Table 1).
We also evaluated whether UNC-23 can influence the refolding activity of Hsc70 in the presence of DNJ-13. Using urea-denatured luciferase, we added Hsc70, DNJ-13, and different concentrations of UNC-23 to refolding solutions and recorded the change in luciferase activity. Although no gain of luciferase activity was observed in the absence of the Hsp40 protein, the presence of DNJ-13 led to a marked increase of Hsc70 refolding activity (7). Δ129-UNC-23 reduced this activity, leading to lower refolding rates. This was also observed for Δ258-UNC-23. In contrast to the longer fragments, the isolated BAG domain barely influenced the refolding activity even at higher concentrations (Fig. 6D). These data show that UNC-23 is able to modulate Hsc70 reactions in vitro in combination with the Hsp40 protein DNJ-13.
Knockdown of DNJ-13 Rescues the Phenotype of unc-23(ok1408) Nematodes
The Hsc70 cofactor system comprising the Hsp40 proteins DNJ-12 and DNJ-13 cooperates with UNC-23 in vitro. Is this interaction with Hsc70 cofactors also evident in vivo? To address this question, we performed RNAi knockdown experiments with the Hsp40 proteins DNJ-12, DNJ-13, and DNJ-19. No obvious phenotype was observed upon knockdown of either dnj-12, dnj-13, or dnj-19 by feeding RNAi to wild-type nematodes (data not shown). Although RNAi against dnj-12 or dnj-19 also did not alter the life span of unc-23(ok1408) nematodes, RNAi against dnj-13 improved the health of the mutated strain. The life span of unc-23(ok1408) nematodes in the presence of RNAi against dnj-13 increased from roughly 5 days, as observed in untreated unc-23 deletion strains, to about 11 days (Fig. 7A). The debilitating growth phenotype was almost absent, with only a weak head-bent phenotype remaining (Fig. 7B). The motility as observed in thrashing assays also improved significantly (Fig. 7C). Further, the head muscle cells observable from the dense body structure in differential interference contrast images are apparent in RNAi-treated nematodes, whereas they are not visible in unc-23 mutated nematodes (Fig. 7D) (16). Thus, large parts of the phenotypes observed in unc-23 mutated nematodes originate only in the presence of DNJ-13, and the reduction of DNJ-13 levels by RNA interference restores the wild-type appearance. Interestingly, RNAi against dnj-12 or dnj-19 did not lead to similar effects. Based on this strong rescue, it is evident that the presence of DNJ-13 is in part responsible for the debilitating effects observed in unc-23 mutated worms. Because these two proteins behave antagonistically in the Hsc70 ATPase cycle, it is tempting to speculate that reduction in one of the cofactors can compensate for loss of the other. Thus, the Hsc70 cycle is apparently shifted out of balance in the absence of UNC-23, which is restored again by diminishing the levels of the competing cofactor DNJ-13.
FIGURE 7.
Knockdown of dnj-13 rescues unc-23(ok1408) phenotype. A, life span assay of wild-type nematodes (gray) and unc-23(ok1408) after treatment with RNAi against dnj-12 (blue), dnj-13 (pink), dnj-19 (cyan), and empty control (green). 20 individuals were subjected to analysis starting from synchronized L1 larvae on RNAi plates at 20 °C. The assay was monitored for adult worms. B, growth comparison between young adult unc-23(ok1408) worms fed on empty vector L4440 and dnj-13 RNAi (pink star). Synchronized L1 larvae were fed on RNAi plates for 3 days at 20 °C. Scale bar, 100 μm. C, thrashing assay of 3-day-old wild-type and unc-23(ok1408) nematodes fed on RNAi against the empty vector L4440, dnj-12, dnj-13, or dnj-19. Means with S.E. (error bars) were plotted. For statistical analysis, the Mann-Whitney test was used. p = 2.83 × 10−6 < α = 0.001. Statistical significance is indicated by three asterisks. The assay was performed with at least 10 animals. D, top, head of young unc-23(ok1408) nematode. Bottom, young adult unc-23(ok1408) nematode after 3 days of dnj-13 RNAi. Muscular tissue is present in the head region with dense bodies and visible striature. Boundaries of muscle cells are highlighted with dotted lines. Metacorpus of pharynx is indicated by an asterisk. Scale bar, 20 μm. E, transgenic wild-type nematodes containing the endogenous promoter of different Hsp40s expressing YFP-fused DNJ-12, DNJ-13, and DNJ-19. Top, differential interference contrast images; bottom, YFP fluorescent images. Arrows, body wall muscle expression of the corresponding protein. Scale bar, 50 μm.
To test whether the selectivity for dnj-13 compared with dnj-12 and dnj-19 is a result of lacking expression, we examined whether all three Hsp40s are expressed under these conditions. We generated nematodes by injecting full-length fusions of these Hsp40s under their own promoter sequences (dnj-12::DNJ-12-YFP, dnj-13::DNJ-13-YFP, and dnj-19::DNJ-19-YFP).All three Hsp40s were found to express in body wall muscle cells and several other cell types (Fig. 7E), implying that the participation of DNJ-13 in these processes may occur as a selective Hsp40 cofactor, whereas DNJ-12 and DNJ-19 may be involved in other tasks in the nematode.
DISCUSSION
The Bcl-2-associated athanogene (BAG) family proteins are conserved through evolution with homologs in mammals, invertebrates (C. elegans and Drosophila melanogaster), plants (Arabidopsis thaliana), and yeast (Saccharomyces cerevisiae) (3, 9, 38, 39). They are nucleotide exchange factors for Hsc70. They often contribute functionality to the Hsc70 machinery. As such, BAG proteins, like Bag1, bind interaction partners, including the protein kinase Raf1, ubiquitin ligases, growth factor receptors, and retinoic acid receptors. Thereby, nucleotide dissociation-dependent delivery of Hsc70-bound clients is connected with cellular processes (9, 37, 38, 40–43). However, it is unknown what role Hsc70 and the client processing activity play in these events. Here we investigate the interaction of the nematode BAG protein UNC-23 with the Hsc70 system. UNC-23 acts as NEF for Hsc70, although its affinity for Hsc70 is rather low. It is additionally required to maintain muscle functionality during development and aging. UNC-23 is one of the few genes known to be involved in muscle attachment, a phenotype commonly termed mua (20).
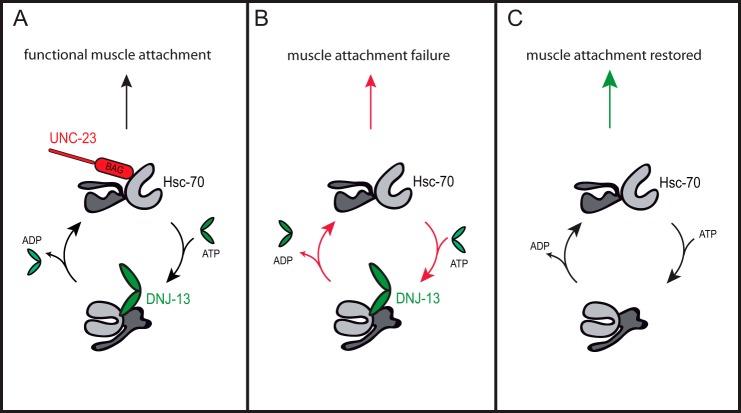
The characteristics of the UNC-23 protein and the in vivo data presented here suggest a potential mechanism for UNC-23 function in cooperation with Hsc70 and its cofactor DNJ-13 (Fig. 8). Besides other localizations, UNC-23 is prominent as part of the dense body structures. This, in principle, could be relevant for the stability of the muscle cells or the ultrastructure of the myofilaments, which is compromised if mutated or during unc-23 RNAi (16–18). We could observe a loss of this localization if the N terminus is not present. Probably, the full-length protein is required for the correct localization to the attachment sites, whereas the C terminus regulates all Hsc70-related functions.
FIGURE 8.
Role of UNC-23 in muscle maintenance. Schematic model of Hsc70 function in C. elegans muscle attachment. UNC-23 is depicted in red with the highlighted BAG domain. Hsc70 with its substrate binding domain, the nucleotide binding domain, and the helical lid is shown in gray. DNJ-13 is shown in green. A, under physiological conditions. B, the attachment fails if UNC-23 is not present. C, if the two cofactors UNC-23 and DNJ-13 are missing, the Hsc70 cycle can restore muscle attachment.
The rescue of the unc-23 mutant allele by the reduction of DNJ-13 levels and the imitation of the phenotype by overexpression of CFP-Hsc70 clearly shows that the involvement of the wider Hsc70 system is required to fulfill the important UNC-23 functions. In vitro DNJ-13 supports exactly the opposite Hsc70 conformational states in terms of ATP binding and client processing, locking Hsc70 in an ADP client-bound state, whereas BAG proteins support client and nucleotide release. Thus, in the absence of UNC-23, DNJ-13 may indeed be harmful, blocking nucleotide and client release. The Hsc70 system without both cofactors apparently is able to sufficiently support muscle attachment, making the knockdown of DNJ-13 beneficial under these conditions (Fig. 8).
It is interesting to see whether homolog functions are conserved in mammalian BAG proteins. Human Bag2 is known to be involved in the cellular stress response and proteosomal degradation pathways, regulating the degradation of Tau fragments and the cystic fibrosis transmembrane conductance regulator together with Hsc70 and CHIP (42, 43), but it is not known to be involved in muscle-related functions. Human Bag3 instead regulates the degradation of filamin in muscle cells (44). The wider processes are regulated by the chaperone-associated autophagy network to remodel the muscle cells and maintain the filamin balance in the muscle cells (44, 45). Bag3 in mice interacts with filamin, which, if damaged, is targeted for degradation via the Hsc70/HspB8/CHIP system and thus may represent a potential regeneration mechanism (44). Indeed, Bag3 and filamin C mutations are responsible for a large fraction of genetic myofibrillar myopathies in humans (46–48).
The homology to the mammalian system here is interesting, in particular because UNC-23 may combine sequence elements of the human Bag2, Bag3, and Bag4 proteins. It thus is possible that this involvement similarly is relevant for the muscular attachment functions of the BAG domain-containing UNC-23 in C. elegans. Dense bodies are integrin-based adhesion complexes and resemble vertebrate focal adhesion complexes (49), which contain filamin (50). Thus, it is tempting to speculate that UNC-23 fulfills similar tasks in C. elegans, although the direct interaction partners of UNC-23 remain to be identified.
Acknowledgments
We are grateful to Professor Donald Moerman for fruitful discussions and for sharing unpublished data. We also thank Professor Johannes Buchner for continuous support. We thank Professor Andreas Bausch for access to the confocal microscope and Carina Pelzl and Christoph Kaiser for introducing us to the instrument. We further thank Matthias Scherr for excellent practical assistance and Christopher Stratil and Alina Röhl for critically reading the manuscript. Some strains were provided by the Caenorhabditis Genetics Center. The license to UltraScanII was generously provided by Borries Demeler (University of San Antonio).
This work was supported by Deutsche Forschungsgemeinschaft Grant RI1873/1-3 (to K. R.) and the Graduate School of the Technische Universität München (to K. P.). The Caenorhabditis Genetics Center is supported by the National Institutes of Health Office of Research Infrastructure Programs (Grant P40 OD010440).
D. Moerman, personal communication.
- NEF
- nucleotide exchange factor
- DSSG
- disulfo-succinimidyl glutarate.
REFERENCES
- 1. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 2. Zhuravleva A., Clerico E. M., Gierasch L. M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Popp S., Packschies L., Radzwill N., Vogel K. P., Steinhoff H.-J., Reinstein J. (2005) Structural dynamics of the DnaK-peptide complex. J. Mol. Biol. 347, 1039–1052 [DOI] [PubMed] [Google Scholar]
- 5. Schlecht R., Erbse A. H., Bukau B., Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
- 6. Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. (1999) Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun L., Edelmann F. T., Kaiser C. J. O., Papsdorf K., Gaiser A. M., Richter K. (2012) The lid domain of Caenorhabditis elegans Hsc70 influences ATP turnover, cofactor binding and protein folding activity. PloS One 7, e33980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kabbage M., Dickman M. B. (2008) The BAG proteins: a ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 65, 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doong H., Vrailas A., Kohn E. C. (2002) What's in the “BAG”? a functional domain analysis of the BAG-family proteins. Cancer Lett. 188, 25–32 [DOI] [PubMed] [Google Scholar]
- 11. Guisbert E., Czyz D. M., Richter K., McMullen P. D., Morimoto R. I. (2013) Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 9, e1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. GuhaThakurta D., Palomar L., Stormo G. D., Tedesco P., Johnson T. E., Walker D. W., Lithgow G., Kim S., Link C. D. (2002) Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 12, 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaiser A. M., Brandt F., Richter K. (2009) The non-canonical Hop protein from Caenorhabditis elegans exerts essential functions and forms binary complexes with either Hsc70 or Hsp90. J. Mol. Biol. 391, 621–634 [DOI] [PubMed] [Google Scholar]
- 14. Nollen E. A., Garcia S. M., van Haaften G., Kim S., Chavez A., Morimoto R. I., Plasterk R. H. (2004) Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A. 101, 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 16. Waterston R. H., Thomson J. N., Brenner S. (1980) Mutants with altered muscle structure in Caenorhabditis elegans. Dev. Biol. 77, 271–302 [DOI] [PubMed] [Google Scholar]
- 17. Rahmani P. (2002) Characterization of the unc-23 gene, an HSP-1 chaperone regulator, in Caenorhabditis elegans. Ph.D. thesis, University of British Columbia, Vancouver, Canada [Google Scholar]
- 18. Meissner B., Warner A., Wong K., Dube N., Lorch A., McKay S. J., Khattra J., Rogalski T., Somasiri A., Chaudhry I., Fox R. M., Miller D. M., 3rd, Baillie D. L., Holt R. A., Jones S. J. M., Marra M. A., Moerman D. G. (2009) An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 5, e1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meissner B., Rogalski T., Viveiros R., Warner A., Plastino L., Lorch A., Granger L., Segalat L., Moerman D. G. (2011) Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PloS One 6, e19937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plenefisch J. D., Zhu X., Hedgecock E. M. (2000) Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development 127, 1197–1207 [DOI] [PubMed] [Google Scholar]
- 21. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaiser A. M., Kaiser C. J. O., Haslbeck V., Richter K. (2011) Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans. PloS One 6, e25485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolkow C. A., Kimura K. D., Lee M. S., Ruvkun G. (2000) Regulation of C. elegans Life-Span by insulinlike signaling in the nervous system. Science 290, 147–150 [DOI] [PubMed] [Google Scholar]
- 24. Haslbeck V., Eckl J. M., Kaiser C. J. O., Papsdorf K., Hessling M., Richter K. (2013) Chaperone-interacting TPR proteins in Caenorhabditis elegans. Journal of molecular biology 425, 2922–2939 [DOI] [PubMed] [Google Scholar]
- 25. Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richter K., Walter S., Buchner J. (2004) The co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 342, 1403–1413 [DOI] [PubMed] [Google Scholar]
- 27. Seebacher J., Mallick P., Zhang N., Eddes J. S., Aebersold R., Gelb M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing research articles. J. Proteome Res. 5, 2270–2282 [DOI] [PubMed] [Google Scholar]
- 28. Green N. S., Reisler E., Houk K. N. (2001) Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 10, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 30. Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1998) ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17, 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dam J., Schuck P. (2005) Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys. J. 89, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stafford W. F. I. (1994) Modern Analytical Ultracentrifugation Emerging Biochemical and Biophysical Techniques, pp. 119–137, Birkhäuser, Boston [Google Scholar]
- 33. Demeler B., Brookes E., Wang R., Schirf V., Kim C. A. (2010) Characterization of reversible associations by sedimentation velocity with UltraScan. Macromol. Biosci. 10, 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eddy S. R. (2009) A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211 [PubMed] [Google Scholar]
- 35. Alexandropoulos K., Cheng G., Baltimore D. (1995) Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc. Natl. Acad. Sci. U.S.A. 92, 3110–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dosztányi Z., Csizmok V., Tompa P., Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 37. Höhfeld J., Jentsch S. (1997) GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takayama S., Xie Z., Reed J. C. (1999) An evolutionarily conserved Family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274, 781–786 [DOI] [PubMed] [Google Scholar]
- 39. Sondermann H. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 40. Song J., Takeda M., Morimoto R. I. (2001) Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3, 276–282 [DOI] [PubMed] [Google Scholar]
- 41. Liu R. (1998) Interaction of BAG-1 with retinoic acid receptor and its inhibition of retinoic acid-induced apoptosis in cancer cells. J. Biol. Chem. 273, 16985–16992 [DOI] [PubMed] [Google Scholar]
- 42. Dai Q., Qian S.-B., Li H.-H., McDonough H., Borchers C., Huang D., Takayama S., Younger J. M., Ren H. Y., Cyr D. M., Patterson C. (2005) Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J. Biol. Chem. 280, 38673–38681 [DOI] [PubMed] [Google Scholar]
- 43. Arndt V., Daniel C., Nastainczyk W., Alberti S. (2005) BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol. Biol. Cell 16, 5891–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ulbricht A., Eppler F. J., Tapia V. E., van der Ven P. F. M., Hampe N., Hersch N., Vakeel P., Stadel D., Haas A., Saftig P., Behrends C., Fürst D. O., Volkmer R., Hoffmann B., Kolanus W., Höhfeld J. (2013) Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430–435 [DOI] [PubMed] [Google Scholar]
- 45. Ulbricht A., Höhfeld J. (2013) Tension-induced autophagy. Autophagy 9, 920–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Selcen D., Muntoni F., Burton B. K., Pegoraro E., Sewry C., Bite A. V., Engel A. G. (2009) Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann. Neurol. 65, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vorgerd M., van der Ven P. F. M., Bruchertseifer V., Löwe T., Kley R. A., Schröder R., Lochmüller H., Himmel M., Koehler K., Fürst D. O., Huebner A. (2005) A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet. 77, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harms M. B., Sommerville R. B., Allred P., Ma D., Ph D., Cooper P., Lopate G., Pestronk A., Weihl C. C., Baloh R. H. (2013) Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol. 71, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hynes R. O. (1992) Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 50. Tu Y., Wu S., Shi X., Chen K., Wu C. (2003) Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell 113, 37–47 [DOI] [PubMed] [Google Scholar]