Background: Intestinal expression of peptide absorption transporter (PepT1)/Slc15a1 exhibits circadian oscillation, but the mechanism is unknown.
Results: During the daily feeding cycle, bile acids accumulated in intestinal cells, thereby suppressing PPARα-mediated expression of PepT1/Slc15a1.
Conclusion: Time-dependent suppression of PPARα activity by bile acids underlies circadian expression of PepT1/Slc15a1.
Significance: Bile acids cause circadian change in the intestinal absorption of peptides.
Keywords: Bile Acid, Circadian Rhythm, Intestinal Epithelium, Peptide Transport, Peroxisome Proliferator-activated Receptor (PPAR)
Abstract
Digested proteins are mainly absorbed as small peptides composed of two or three amino acids. The intestinal absorption of small peptides is mediated via only one transport system: the proton-coupled peptide transporter-1 (PepT1) encoded from the soluble carrier protein Slc15a1. In mammals, intestinal expression of PepT1/Slc15a1 oscillates during the daily feeding cycle. Although the oscillation in the intestinal expression of PepT1/Slc15a1 is suggested to be controlled by molecular components of circadian clock, we demonstrated here that bile acids regulated the oscillation of PepT1/Slc15a1 expression through modulating the activity of peroxisome proliferator-activated receptor α (PPARα). Nocturnally active mice mainly consumed their food during the dark phase. PPARα activated the intestinal expression of Slc15a1 mRNA during the light period, and protein levels of PepT1 peaked before the start of the dark phase. After food intake, bile acids accumulated in intestinal epithelial cells. Intestinal accumulated bile acids interfered with recruitment of co-transcriptional activator CREB-binding protein/p300 on the promoter region of Slc15a1 gene, thereby suppressing PPARα-mediated transactivation of Slc15a1. The time-dependent suppression of PPARα-mediated transactivation by bile acids caused an oscillation in the intestinal expression of PepT1/Slc15a1 during the daily feeding cycle that led to circadian changes in the intestinal absorption of small peptides. These findings suggest a molecular clock-independent mechanism by which bile acid-regulated PPARα activity governs the circadian expression of intestinal peptide transporter.
Introduction
Dietary protein is digested into short chain peptides and single amino acids, which are then absorbed by various transport mechanisms in the gut mucosal cells. Approximately 70% of digested proteins are absorbed as small peptides composed of two (dipeptides) or three (tripeptides) amino acids, whereas the remaining 30% are absorbed as a single amino acid (1, 2). Although single amino acids are taken up into enterocytes through numerous amino acid transporters, the intestinal absorption of di- and tripeptides are mediated via only one transport system consisting of PepT12 (3, 4). PepT1 is encoded by soluble carrier protein family 15 member 1 (Slc15a1) and mainly expressed in the apical plasma membrane in intestinal epithelial cells (5, 6). Because PepT1 is capable of transporting most small peptides as well as several peptidomimetic drugs such as the β-lactam antibiotics, angiotensin-converting enzyme inhibitors, antivirals, and anticancer agents (7), much effort has been directed toward the regulation of intestinal PepT1 expression. For example, it has been reported that intestinal expression of PepT1 is regulated by insulin, thyroid hormone, epidermal growth factor, and some pharmacological agents (8–11). In laboratory rodents, the levels of Slc15a1 mRNA and PepT1 protein fluctuate in a time-dependent manner (12), suggesting that the expression of Slc15a1/PepT1 is under the control of a circadian clock.
Circadian rhythms are ∼24-h cycles that allow an adaptation of physiological and behavioral activities to environmental cues. The rhythmic changes in the physiological functions would help organisms to anticipate daily changes in environmental conditions and their feeding time. In mammals, circadian clock machinery consists of a hierarchical assembly of multiple endogenous oscillators (13). Among these oscillators, a major pacemaker is located in the suprachiasmatic nucleus of the hypothalamus. The suprachiasmatic nucleus circadian oscillators are entrained to a 24-h period by daily light input from the visual neural system and produce output signals for coordinating the phase of independent oscillators in peripheral tissues. The peripheral oscillators generate daily rhythms in output physiology through the periodic activation/repression of clock-controlled output genes (14–16).
Although the light/dark cycle is the most powerful “Zeitgeber” for the suprachiasmatic nucleus oscillators in mammals (17, 18), feeding regimen is also important for the generation and synchronization of peripheral circadian rhythms (19). The intestinal expressions of Slc15a1/PepT1 are induced by situations of fasting or starvation (20). Fasting-induced expression of Slc15a1 mRNA is suggested to be mediated via peroxisome proliferator-activated receptor-α (PPARα). A number of endogenous substrates such as polyunsaturated fatty acids serve as ligand activators of this nuclear receptor (21, 22). Because plasma levels of free fatty acids are elevated during the fasting state (23), the elevation is thought to trigger fasting-induced activation of PPARα. Conversely, several bile salts also have the ability to modulate the activity of PPARα and the expression of its target genes (24). The secretion of bile acids and intestinal absorption of fatty acids likely change during the daily feeding cycle. However, it remains to be clarified whether interaction between these compounds and PPARα is involved in the circadian regulation of the intestinal expression of Slc15a1/PepT1.
In this study, we found that the suppressing action of bile acids on PPARα activity caused oscillation in the intestinal expression of Slc15a1/PepT1 during the daily feeding cycle. In mammals, PepT1 is responsible for the absorption of small peptides arising from digestion of dietary proteins. Therefore, we investigated the physiological significance of the modulatory action of bile acids on PPARα activity for the function of PepT1 to absorb small peptides.
EXPERIMENTAL PROCEDURES
Animals and Feeding Schedule
PPARα-null mice (129S4-Svjae-PPARαmuGonzN12) with a JcI:ICR background, Period2 gene mutant (Per2m/m), and wild-type mice of the same strain were fed ad libitum before the experiments. Animals were treated in accordance with the guidelines stipulated by the Animal Care and Use Committee of Kyushu University. To investigate the influence of fasting on intestinal gene expression, mice were also forced to fast for 24-h starting at zeitgeber time (ZT) 0 and then fed ad libitum. During the manipulation of the feeding schedule, water was freely available.
Cell Culture and Treatment
The human colon carcinoma cell line Caco-2 was obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1× nonessential amino acids. Cells were transfected with siRNA (20 nm) against PPARα (Santa Cruz Biotechnology, Santa Cruz, CA) and used for experiments at 48 h after transfection. Down-regulation of PPARα expression was confirmed by Western blotting. The PPARα-down-regulated cells were treated with the PPAR ligand WY-14643, cholic acid (Wako Chemicals, Osaka, Japan), or 0.05% DMSO/0.25% ethanol as a control for the indicated times.
Determination of Lipid Contents in the Epithelial Cells of Mouse Small Intestine
Epithelial cells were collected from the small intestine of mice. For assessment of free fatty acids, 30 mg of cells were homogenized with 200 μl of chloroform/Triton X-100 (1% Triton X-100 in pure chloroform). After centrifugation at 15,000 rpm for 15 min, the organic phase (lower phase) was collected and dried at 50 °C to remove chloroform. The dried lipid samples were dissolved in assay buffer, and concentrations of free fatty acids were determined using the Free Fatty Acid Quantification kit (Abcam, Cambridge, UK). For assessment of bile acids, 30 mg of cells were homogenized with 1.0 ml of 70% ethanol and then incubated at 55 °C for 4 h. The ethanol extracts were evaporated to dryness and resuspended in 300 μl of 0.5 m phosphate buffer (pH 7.0). Concentrations of total bile acids were determined using the Total Bile Acid Assay kit (Diazyme Laboratories, CA).
Gas Chromatography/Mass Spectrometry (GC/MS) Analysis
The composition of free fatty acid and bile salt species in epithelial cells of the small intestine was analyzed by GC/MS. Fatty acids in epithelial cells were subjected to acid or alkaline methanolysis to produce fatty acid methyl esters. The methyl ester derivatives were extracted in an organic solvent and analyzed by GC/MS. The GC/MS analysis of bile acids was carried out by using selected ion monitoring of characteristic fragments of methyl ester dimethylethylsilyl ether methoxime bile acid derivatives. The lipid contents in epithelial cells were expressed as μmol/g of tissue.
Determination of mRNA Expression Levels
The epithelial cells were removed from the intestinal segments of mice and homogenized using RNAiso reagent (Takara Bio Inc.). Real time RT-PCR was performed using SYBR Premix Ex Taq II (Takara Bio Inc.) or THUNDERBIRD (Toyobo Co. Ltd.) using a LightCycler (Roche Diagnostics). The reaction conditions were 95 °C for 10 s followed by 45 cycles at 95 °C for 5 s, 57 °C for 10 s, and 72 °C for 10 s. Sequences for PCR primers are given in Table 1. The amount of target mRNA was corrected relative to that of β-actin.
TABLE 1.
Primer sets for RT-PCR analysis
| Gene | Primers |
|---|---|
| Mouse Bmal1 | |
| Forward | 5′-CCGATGACGAACTGAAACACCT-3′ |
| Reverse | 5′-TGCAGTGTCCGAGGAAGATAGC-3′ |
| Mouse Dbp | |
| Forward | 5′-GCGAGAAGTGCAAAATTGGC-3′ |
| Reverse | 5′-CGGGAGGCTCCTATAGTCTGG-3′ |
| Mouse Slc15a1 | |
| Forward | 5′-CGTGTGGTAAAAGATGGTCCTAA-3′ |
| Reverse | 5′-CGTTGTGACTTGTGAATTTTTCAT-3′ |
| Mouse β-actin | |
| Forward | 5′-CACACCTTCTACAATGAGCTGC-3′ |
| Reverse | 5′-CATGATCTGGGTCATCTTTTCA-3′ |
| Human SLC15A1 | |
| Forward | 5′-ATTGTGTCG CTCTCCATTGTCTAC-3′ |
| Reverse | 5′-ATGGTTGTGGTCTGTGAGGTCAT-3′ |
| Human β-actin | |
| Forward | 5′-AGAGCTACGAGCTGCCTGAC-3′ |
| Reverse | 5′-AGCACTGTGTTGGCGTACAG-3′ |
Luciferase Reporter Assay
The 5′-flanking region of the mouse Slc15a1 gene spanning from bp −1946 to +3 (the number is the distance in base pairs from the putative transcription start site, +1) was amplified by PCR, and the product was ligated into the pGL4.12 luciferase reporter vector (Slc15a1-luciferase). The peroxisome proliferator response element (PPRE) located between bp −1876 and −1864 with respect to the transcription start site of the mouse Slc15a1 gene was mutated from AGGTCAAAGAAGA to AGGTCTCGAGAGA using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The PPRE located between bp −1693 and −1681 with respect to the transcription start site was also changed from AGGTGATAAATGA to AGGTCTCGAGTGA using the same reagent. Expression vectors for mouse PPARα and retinoid X receptor-α (RXRα) were constructed using a cDNA generated from mouse liver RNA by RT-PCR. All coding regions were ligated into the pcDNA3.1 vector (Invitrogen). The NIH3T3 cells were seeded at a density of 1 × 105/24-well culture plate. The cells were transfected 18 h later with 100 ng/well reporter vectors and 1–2 μg/well (total) expression vectors. The pRL-TK vector (0.5 ng/well; Promega) was also co-transfected as an internal control reporter. The cells were then harvested, and the cell lysates were analyzed using a Dual-Luciferase reporter assay system (Promega). The ratio of firefly (expressed from the reporter construct) to Renilla (expressed from pRL-TK) luciferase activities in each sample served as a measure of normalized luciferase activity. The reporter vector-transfected cells were also treated with WY-14643, cholic acid, or 0.05% DMSO/0.25% ethanol as a control for the indicated times. Luciferase activities in each sample were assessed as described above.
Chromatin Immunoprecipitation (ChIP) Analysis
Cross-linked chromatin in epithelial cells from the small intestine was sonicated on ice, and nuclear fractions were obtained by centrifugation at 10,000 × g for 5 min. Supernatants were incubated with antibodies against PPARα (H-98), RXRα (D-20), CREB-binding protein (CBP)/p300 (N-15), or rabbit IgG (Santa Cruz Biotechnology). DNA was isolated using the GeneElute mammalian genomic DNA kit (Sigma) and amplified by PCR for the surrounding PPREs in the 5′-flanking region of the Slc15a1 gene. Primer sequences for amplification of the surrounding or outside PPREs are given in Table 2. The quantitative reliability of PCR was evaluated by kinetic analysis of the amplified products to ensure that signals were only derived from the exponential phase of amplification. ChIP in the absence of an antibody and in the presence of rabbit IgG served as negative controls. Ethidium bromide staining did not detect any PCR products in these samples.
TABLE 2.
Primer sets for ChIP analysis
| Gene | Primers |
|---|---|
| Set 1 | |
| Forward | 5′-GAGCTCTCAGAGGGAGCACA-3′ |
| Reverse | 5′-TGCACCGGACAAACGGAAAC-3′ |
| Set 2 | |
| Forward | 5′-TGTGACTCTCAGTGTAGAGG-3′ |
| Reverse | 5′-CATCTACCCCATTCATGCAC-3′ |
| Set 3 | |
| Forward | 5′-AACAACCAAAGGAAACCTCA-3′ |
| Reverse | 5′-GAGTCTTCCACATTCGCTTT-3′ |
Western Blotting
Cell membrane fractions were prepared from small intestine epithelial cells. Fractions containing 20 μg of total protein were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane that was incubated with antibodies against PepT1 (H-235, Santa Cruz Biotechnology) or Na+/K+-ATPase (Abcam). Specific antigen-antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibodies and Chemi-Lumi One (Nacalai Tesque Inc., Kyoto, Japan). Nuclear extracts were also prepared from Caco-2 cells. Protein levels of PPARα, RXRα, and β-actin were assessed by the same method. The antibodies against PPARα (H-98), RXRα (D-20), or β-actin (C4) were purchased from Santa Cruz Biotechnology.
Determination of Plasma Concentrations of Carnosine
Mice were orally administrated carnosine (1.75 mg/g), and blood samples were collected by cardiac puncture. Plasma was obtained by centrifugation at 10,000 rpm for 10 min. The supernatant was deproteinized with 0.36 m perchloric acid and centrifuged at 10,000 rpm for 5 min, and the supernatant was then used as a sample. The concentration of carnosine was determined using a high performance liquid chromatography (HPLC) system with a fluorescence detector. Carnosine was derivatized with o-phthalaldehyde (Sigma). The mobile phase consisting of 1.25% methanol and 0.75% sodium acetate (0.3 m, pH5.5) was eluted at 0.8 ml/min through a COSMOSIL 5C18-MS-II column (4.6 × 150 mm; Nacalai Tesque Inc.) maintained at 40 °C. The separated analyte was detected using an RF-550 spectrofluorometric detector (Hitachi) with excitation at 310 nm and emission at 375 nm.
Statistical Analysis
The significance of differences between groups was validated by the Bonferroni test for multiple comparisons and Student's t test for comparisons between two groups. The 5% level of probability was considered to be significant.
RESULTS
Circadian Oscillation in the Intestinal Expression of Slc15a1 mRNA Is Modulated by the Feeding State
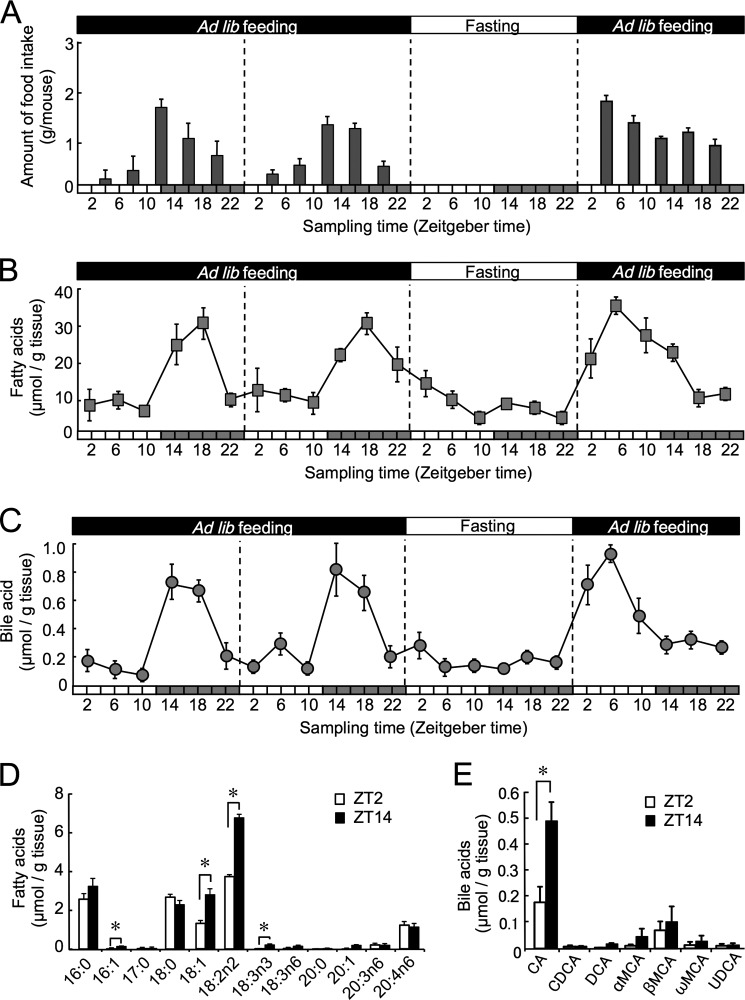
Bile salts are released from the gallbladder into the small intestine in response to food intake. When food is plentiful, nocturnally active mice ingest most of their food during the dark phase (25). We first investigated the relationship between temporal feeding patterns and the accumulation of fatty and bile acids in intestinal epithelial cells. As reported previously (25), mice consumed ∼80% of their food during the dark phase when they were fed ad libitum (Fig. 1A). Under this feeding condition, the amount of free fatty acids in the intestinal epithelial cells of mice exhibited an obvious circadian oscillation with higher levels being observed during the dark phase (Fig. 1B). This oscillation in the amount of free fatty acids was markedly dampened by 24 h of fasting; it remained at low levels throughout the fasting period. After mice resumed food intake, the amount of free fatty acids in the intestines increased during the light phase due to the resumption of feeding. A similar temporal variation was also observed in the accumulation of bile acids in the intestinal epithelial cells of mice during the manipulation of the feeding schedule (Fig. 1C). Because bile acids are secreted into the lumen of the small intestine by stimulation of food intake, the change in the temporal accumulation pattern of bile acids also appeared to be caused by the alteration in daily pattern of food intake.
FIGURE 1.
Time-dependent change in the accumulation of fatty and bile acids in the intestinal epithelial cells during the manipulation of feeding schedule. For A–C, mice were housed under a standardized light/dark cycle (ZT0, light on; ZT12, light off) with food and water ad libitum (Ad lib). They were deprived of food for 24-h and then refed ad libitum. A, changes in the temporal pattern of food intake by mice (n = 4). B, changes in the temporal pattern of fatty acid accumulation in the intestinal epithelial cells (n = 5). C, changes in the temporal pattern of bile acid accumulation in the intestinal epithelial cells (n = 5). D, temporal profiles for the composition of free fatty acids in the intestinal epithelial cells of mice under the ad libitum feeding condition (n = 3). Twelve types of fatty acids and their derivatives, palmitic acid (16:0), palmitoleic acid (16:1), heptadecanoic acid (17:0), stearic acid (18:0), oleic acid (18:1), linolenic acid (18:2n-2), α-linolenic Acid (18:3n-3), γ-linolenic acid (18:3n-6), arachidic acid (20:0), cis-11-eicosenoic acid (20:1), cis-8,11,14-eicosatrienoic acid (20:3n-6), and arachidonic (20:4n-6) acid, were detected in intestinal epithelial cells. E, temporal profiles for the composition of bile acids in the intestinal epithelial cells of mice under the ad libitum feeding condition (n = 3). Seven types of bile salts and their derivatives, CA, chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), α-muricholic acid (α-MCA), β-muricholic acid (β-MCA), ω-muricholic acid (ω-MCA), and ursodeoxycholic acid (UDCA), were detected in intestinal epithelial cells. For all panels, values shown are the mean ± S.E. (error bars). *, p < 0.05, compared between two groups.
GC/MS analysis detected 12 types of fatty acids and their derivatives in the intestinal epithelial cells of wild-type mice (Fig. 1D). The main composition of these fatty acids was palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), linolenic acid (18:2n-2), and arachidonic acid (20:4n-6). Among them, the amount of oleic acid (18:1) and linoleic acid (18:2n-2) showed significant time-dependent variations in the epithelial cells of mice under the ad libitum feeding condition (p < 0.05, respectively). Composition analysis by GC/MS also detected seven types of bile acids and their derivatives in the intestinal epithelial cells (Fig. 1E). The main composition of these bile salts was cholic acid (CA), the amount of which exhibited significant time-dependent variations under the ad libitum feeding (p < 0.05). These results indicated that the intake of food also caused the daily accumulation of bile acids in intestinal epithelial cells. The temporal accumulation of bile acids was attributed to an increase in the amount of CA.
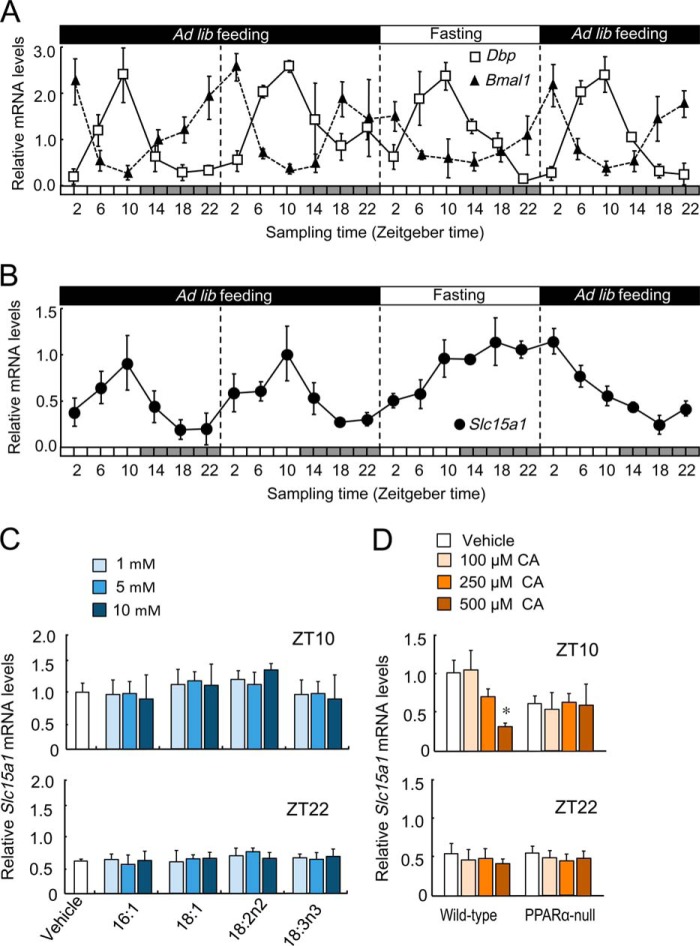
Under the ad libitum feeding condition, the mRNA levels of circadian clock genes Dbp and Bmal1 in the small intestine of mice exhibited obvious circadian oscillations (Fig. 2A). The rhythmic phase of Dbp mRNA was nearly antiphase to that of Bmal1 expression. The mRNA levels of Slc15a1 also showed obvious circadian oscillation under ad libitum feeding conditions (Fig. 2B); the mRNA levels gradually increased during the light phase and then decreased after the start of the dark phase. Although the mRNA levels of Dbp and Bmal1 still exhibited circadian oscillations during and after 24 h of fasting (Fig. 2A), the expression pattern of Slc15a1 mRNA markedly changed by the manipulation of the feeding schedule; Slc15a1 mRNA levels increased during 24 h of fasting, and expression levels decreased after the resumption of feeding (Fig. 2B). Increases and decreases in Slc15a1 mRNA levels during manipulation of the feeding schedule were in contrast to the accumulation pattern of fatty and bile acids in intestinal epithelial cells (Fig. 1, B and C).
FIGURE 2.
Comparison of the expression profiles of Slc15a1 and circadian clock genes in the intestinal cells of mice during manipulation of the feeding schedule. For A and B, the feeding schedule was manipulated as shown in Fig. 1, A–C. A, temporal mRNA expression profiles of the circadian clock genes Dbp and Bmal1 in the intestinal cells (n = 4–6). B, the temporal expression profile of Slc15a1 mRNA in the intestinal cells (n = 4–5). C, influence of fatty acids on the expression of Slc15a1 mRNA in the intestinal epithelial cells. Intestinal segments were prepared from wild-type mice at ZT10 and ZT22 and incubated with palmitoleic acid (16:1), oleic acid (18:1), linoleic acid (18:2n-2), α-linolenic acid (18:3n-3), or vehicle (0.05% DMSO and 0.25% ethanol) for 2 h. The levels of mRNA are expressed as a relative ratio to the vehicle group at ZT10 (set at 1.0). D, influence of CA on the expression of Slc15a1 mRNA in the intestinal epithelial cells of wild-type and PPARα-null mice. Intestinal segments were prepared at ZT10 and ZT22 and incubated with CA or vehicle (0.05% DMSO and 0.25% ethanol) for 2 h. The levels of mRNA are expressed as a relative ratio to the vehicle group at ZT10 (set at 1.0). Values shown are the mean ± S.E. (error bars) of four to six mice for all panels. *, p < 0.05, significantly different from the vehicle-treated group. Ad lib, ad libitum.
Bile Acids Act as a Negative Regulator of Slc15a1 Expression in the Intestinal Epithelial Cells
Next we investigated the possibility that fatty and bile acids could modulate the expression of Slc15a1 in the intestinal epithelial cells. To this end, intestinal segments were isolated from mice and then incubated for 2 h with fatty or bile acids whose intestinal concentrations exhibited significant time-dependent variation (Fig. 1, D and E). Treatment of wild-type mouse intestinal segments prepared at ZT10 and ZT22 with fatty acids had little effect on the mRNA levels of Slc15a1 (Fig. 2C). Furthermore, no significant change in the expression of Slc15a1 mRNA in the isolated intestinal segments was observed even when the segments were treated with 400 μm WY-14643 (data not shown). Conversely, treatment of wild-type mouse intestinal segments prepared at ZT10, but not ZT22, with CA dose-dependently suppressed the expression of Slc15a1 mRNA (Fig. 2D). The suppression effects of CA were significant at a concentration over 500 μm (p < 0.05).
Under the ad libitum feeding condition, the levels of Slc15a1 mRNA in the small intestine of mice increased during their rest period (light phase). Furthermore, the mRNA levels of Slc15a1 were also elevated by 24 h of fasting. Fasting-induced expression of Slc15a1 mRNA is suggested to be mediated via the nuclear receptor PPARα (20). Therefore, we also tested whether bile acids could suppress the Slc15a1 expression in PPARα-null mice. Although the mRNA levels of Slc15a1 in intestinal segments of PPARα-null mice were reduced at both ZT10 and ZT22, no significant suppression effects of CA on the expression of Slc15a1 were detected in PPARα-null mice (Fig. 2D). These results indicated that bile acids suppressed the intestinal expression of Slc15a1 by inhibiting PPARα-mediated transactivation.
Regulation of PPARα-mediated Expression of Slc15a1 by Bile Acids
To assess the potential involvement of PPARα in bile acid-repressed expression of Slc15a1 in intestinal cells, we also examined the influence of CA on the mRNA levels of SLC15A1 in PPARα-down-regulated Caco-2 cells, which are often used as an in vitro model of the intestinal system. The expression of endogenous PPARα proteins in Caco-2 cells was down-regulated by transfection with siRNA against PPARα (Fig. 3A), but PPARα protein levels were not obviously changed by transfection of cells with control siRNA. The mRNA levels of SLC15A1 in control siRNA-transfected Caco-2 cells were elevated significantly in response to treatment with the PPARα ligand WY-14643 (Fig. 3B). The PPARα ligand-induced expression of SLC15A1 was dose-dependently repressed by CA treatment (Fig. 3B). Conversely, treatment of PPARα-down-regulated cells with the same concentration of WY-14643 resulted in a moderate induction of SLC15A1 mRNA (Fig. 3B). Therefore, CA had little effect on the mRNA levels of SLC15A1 in PPARα-down-regulated cells.
FIGURE 3.
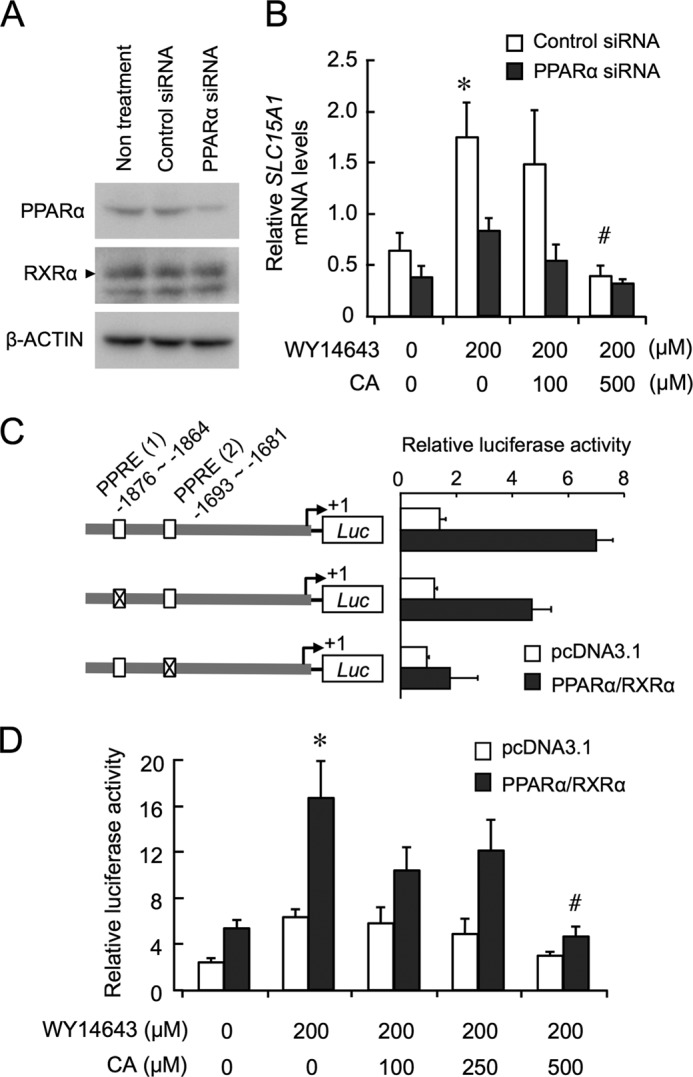
PPARα is involved in the bile acid-repressed expression of Slc15a1. A, expression of PPARα protein in siRNA-transfected Caco-2 cells. RXRα was used to confirm the specificity of PPARα-siRNA. β-Actin was used as a loading control. B, control or PPARα siRNA-transfected cells were treated with 200 μm WY-14643 for 6 h, and then CA or vehicle (0.05% DMSO and 0.25% ethanol) was added into the media at the indicated concentration. At 4 h after treatment, total RNA was extracted from cells, and Slc15a1 mRNA levels were assessed by RT-PCR. C, schematic representation of wild-type or PPRE-mutated Slc15a1 reporters is shown on the left, and their corresponding luciferase (Luc) activities are shown on the right. The numbers shown are the nucleotide residues in which PPREs were positioned relative to the transcription start site (+1). D, effects of WY-14643 and/or CA on the PPARα/RXRα-mediated promoter activity of Slc15a1 reporters containing wild-type PPREs. At 24 h after transfection, cells were treated with WY-14643 for 12 h in the presence or absence of CA. Values shown are the mean ± S.E. (error bars) of three to four experiments. *, p < 0.05, significantly different from the control group; #, p < 0.05, significantly different from the group that was treated with WY-14643 alone.
PPARα has been shown to heterodimerize with RXRα to activate the transcription of their target genes by binding to PPREs (26). Two nucleotide sequences showing homology with PPREs were detected within 2.0 kilobases (kb) of the 5′-flanking region of the mouse Slc15a1 gene, and similar PPRE sequences were also found in the upstream region of the human SLC15A gene. Thus, we also performed a transcription assay by transfecting NIH3T3 cells with the mouse Slc15a1-luciferase reporters. Although the mouse Slc15a1-luciferase reporter construct containing wild-type PPREs responded to PPARα/RXRα, mutation of PPREs attenuated the PPARα/RXRα-mediated transactivation of Slc15a1 (Fig. 3C).
The PPARα/RXRα-mediated transactivation of Slc15a1 was further enhanced by treatment with 200 μm WY-14643. However, the enhancement effect of WY-14643 on the transcriptional activity of Slc15a1 was dose-dependently suppressed by CA (Fig. 3D). Because such modulatory actions of WY-14643 and CA on the Slc15a1 transcription were undetectable in pcDNA3.1 (empty vector)-transfected cells, PPARα seemed to be involved in bile acid-repressed expression of Slc15a1 in intestinal cells.
ChIP analysis of intestinal epithelial cells of wild-type mice also revealed that PPARα and RXRα bound consistently to PPREs in the Slc15a1 gene during both the light and dark phases under the ad libitum feeding condition (Fig. 4, A and B). Conversely, a time-dependent oscillation was observed in the recruitment of the transcriptional coactivator CBP/p300 to PPREs in the Slc15a1 gene (Fig. 4B). This oscillation in the recruitment of CBP/p300 was similar to the Slc15a1 mRNA rhythm (Fig. 2B), which was nearly opposite to the accumulation rhythm observed in bile acids (Fig. 1C). Furthermore, treating intestinal segments of wild-type mice with CA dose-dependently interfered with the recruitment of CBP/p300 to PPREs but not with the binding of PPARα and RXRα (Fig. 4C).
FIGURE 4.
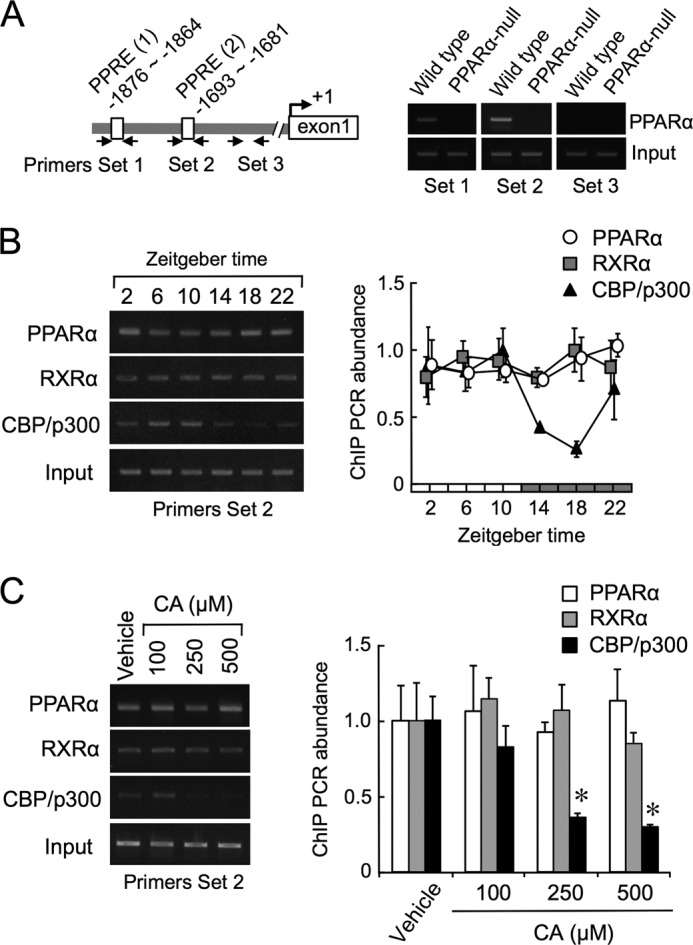
Bile acids interfere with the recruitment of CBP/p300 on Slc15a1 gene. A, schematic structures of the sequence of the 5′-flanking region of the Slc15a1 gene. Solid arrows indicate the primer sets for the amplification area by PCR. B, cross-linked chromatin was prepared from wild-type mice fed ad libitum and subjected to PCR using primers surrounding the PPRE (2) in the 5′-flanking region of the mouse Slc15a1 gene. The peak amounts of binding were set at 1.0. Values shown are the mean ± S.E. (error bars) of four to five experiments. There is a significant time-dependent variation in the binding of CBP/p300. C, suppression effects of CA on the recruitment of CBP/p300 on the PPRE (2) in the Slc15a1 gene. Intestinal segments were prepared from mice at ZT10 and then incubated in the presence of 100, 250, or 500 μm CA or vehicle for 2 h. Cross-linked chromatin was subjected to PCR using primers surrounding the PPRE (2) of the mouse Slc15a1 gene. Values in vehicle (0.05% DMSO and 0.25% ethanol)-treated groups were set at 1.0. Values shown are the mean ± S.E. (error bars) of four to six experiments. *, p < 0.05, significantly different from the vehicle-treated group.
Circadian Oscillation in the Intestinal Expression of Slc15a1 Is Independent from the Molecular Clock Machinery
The oscillations in the intestinal expressions of Dbp and Bmal1 were severely dampened in Per2m/m mice (Fig. 5A), suggested that the animals had a dysfunctional molecular clock in the intestinal epithelial cells. To explore whether a molecular circadian clock participates in the regulation of intestinal expression of Slc15a1, we investigated the temporal expression profiles of Slc15a1 mRNA in Per2m/m mice. Under the ad libitum feeding condition, the food intake pattern of Per2m/m mice was similar to that of wild-type mice (Fig. 5B). No significant difference was observed in the intestinal accumulation profile of bile acids between Per2m/m and wild-type mice (Fig. 5C), suggesting that Per2m/m mice have normal secretory function of bile acids in response to food intake. Although Per2m/m mice exhibited a dampened rhythm of circadian genes in the intestinal epithelial cells (Fig. 5A), the deletion of Per2 had a negligible effect on the oscillation in the expression of Slc15a1 (Fig. 5D). These results suggest that the oscillation in the intestinal expression of Slc15a1 is independent from the molecular clock machinery. The modulatory action of bile acids on PPARα activity seemed to underlie circadian expression of Slc15a1 in the small intestine of mice.
FIGURE 5.
Circadian oscillation in the intestinal expression of Slc15a1 is independent from the molecular clock machinery. Mice were housed under a standardized light/dark cycle (ZT0, light on; ZT12, light off) with food and water ad libitum. A, temporal mRNA expression profiles of the circadian clock genes Dbp and Bmal1 in the intestinal cells of wild-type and Per2m/m mice. B, temporal pattern of food intake by wild-type and Per2m/m mice. C, temporal accumulation profiles of bile acids in the intestinal cells of wild-type and Per2m/m mice. D, temporal mRNA expression profile of Slc15a1 in the small intestine of wild-type and Per2m/m mice. Values shown are the mean ± S.E. (error bars) of four to six mice for all panels.
Time-dependent Change in the PepT1 Function for Intestinal Absorption of Small Peptides
Under the ad libitum feeding condition, the expression of the PepT1 protein in the small intestine of wild-type mice also exhibited a circadian oscillation with peak levels being reached before the start of the dark phase (Fig. 6A). Conversely, the expression of PepT1 in PPARα-null mice remained at a low level throughout the day. In the final set of experiments, we used PPARα-null mice to investigate how the oscillation of PepT1 expression in the intestine affected the absorption of small peptide. The plasma concentration of carnosine, a typical substrate of PepT1 (27), peaked 30 min after its oral administration (Fig. 6B). The peak plasma concentration of carnosine was also significantly higher after its oral administration at ZT12 than after administration at ZT0 (p < 0.05). The intestinal absorption of carnosine appeared to be enhanced by administering it at times of the day when PepT1 expression was abundant. Such an administration time-dependent difference in the plasma concentration of carnosine was not observed in PPARα-null mice (Fig. 6C). After the administration of carnosine at ZT12, its plasma concentration in PPARα-null mice was markedly lower than that in wild-type mice. Taken together, these results suggest that bile acid-regulated PPARα activity underlies the time-dependent changes in PepT1 function for the intestinal absorption of small peptides.
FIGURE 6.

Time-dependent changes in PepT1 function in mice. A, temporal expression profiles of PepT1 in the intestinal epithelial cells of wild-type and PPARα-null mice fed ad libitum. B, time course of plasma carnosine concentrations in wild-type mice after its oral administration (1.75 mg/g) at ZT0 or ZT12. *, p < 0.05, significantly different from that at the corresponding times. C, plasma carnosine concentrations in wild-type and PPARα-null mice after its oral administration (1.75 mg/g) at ZT0 or ZT12. Values shown in B and C are the mean ± S.E. (error bars) of four to six mice. *, p < 0.05, significantly different between groups.
DISCUSSION
After food intake, bile acids are secreted into the lumen of the small intestine in which they act not only as a digestive detergent of lipids but also as a modulator of gene expression in epithelial cells. Previous studies using PPARα-null mice demonstrated that Slc15a1 is typical of PPARα target genes whose expression oscillates in a circadian time-dependent as well as feeding state-dependent manner (28). Although a molecular component of circadian clock has been shown to govern the daily expression of Slc15a1 in laboratory rats (12), the present study demonstrated that the suppressing action of bile acids on PPARα activity is involved in the circadian regulation of the expression of Slc15a1 in mouse intestinal epithelial cells.
When food was plentiful, nocturnally active mice ingested most of their food during the dark phase. The levels of Slc15a1 mRNA in the epithelial cells of wild-type mice were elevated during the light phase. In addition, the intestinal expression of Slc15a1 mRNA was also increased by 24 h of fasting. These results confirm previous observations that situations of fasting or starvation enhance the PPARα-mediated transactivation of Slc15a1 (20). Because a number of polyunsaturated fatty acids serve as ligands of PPARα, the elevation of plasma levels of free fatty acids is thought to trigger fasting-induced activation of PPARα (23). However, under the ad libitum feeding condition, the amounts of free fatty acids in the intestinal epithelial cells of nocturnally active mice remained low in the light phase during which the mRNA levels of Slc15a1 were gradually elevated. Although the intestinal content of free fatty acids fluctuated depending on the daily feeding cycle, the basal level may be maintained in the range sufficient to activate the PPARα-mediated expression of Slc15a1. This notion was also supported by the results of the ChIP experiment. Both PPARα and RXRα were constantly bound to PPREs in the promoter region of the Slc15a1 gene in the intestinal epithelial cells of wild-type mice throughout the daily feeding cycle. Because PPARα/RXRα heterodimers can bind to PPREs after being stimulated by their ligands (26), PPARα binding to PPREs on the Slc15a1 gene is unlikely affected by the fluctuation in the intestinal amount of free fatty acids. Conversely, treatment of intestinal segments with 500 μm CA caused a significant decrease in the mRNA levels of Slc15a1. The same concentration of CA also suppressed the WY-14643-induced expression of SLC15A1 in Caco-2 cells. The concentrations of CA are equivalent to its intestinal contents during the dark phase (500 μmol/kg of tissue), and this seems to be sufficient to suppress the expression of Slc15a1 mRNA (Fig. 1D).
In mammals, PPARα is highly expressed in the liver, heart, kidney, brown adipose tissue, muscle, and small intestine (29, 30). This nuclear receptor has critical roles in energy metabolism, hepatic steatosis, inflammation, cardiac pathophysiology, cell cycle regulation, and oncogenesis (31, 32). In hepatic cells, the modulatory action of bile acids on PPARα-mediated transactivation has been shown to affect the expression of genes, including those involved in lipid homeostasis (24, 33). Therefore, the modulatory action of bile acids on PPARα-mediated transactivation in intestinal epithelial cells appears to have a different physiological significance in hepatic cells (34).
In an intact animal, peripheral circadian oscillators in many tissues can allow adaptation of their physiological and behavioral rhythms to maximize the opportunity to find food sources. However, food entrainment of circadian gene expression is required for several days (35, 36). In natural environments, a few days of starvation is a common threat for small mammals that may result in death. Thus, circadian rhythms in the intestinal expression of nutrient transporters should change rapidly for efficient absorption of food sources. In fact, the rhythmic phase of intestinal expression of Slc15a1 was rapidly changed by manipulation of the feeding schedule. The molecular clock-independent mechanism regulating temporal expression of PepT1/Slc15a1 seemed to be beneficial for animals to adapt the nutrient absorption function depending on feeding states.
Because PepT1 is capable of transporting not only small peptides but also several peptidomimetic drugs (7), the oscillation observed in PepT1 function appears to underlie the dosing time-dependent changes in the oral availability of peptidomimetic drugs. The therapeutic effects of many drugs are dependent on their pharmacokinetics. Therefore, the oscillation in PepT1 function may also be beneficial for the achievement of rational pharmacotherapy for the treatment of disease.
Acknowledgments
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University for technical support and Steven Sabotta for proofreading the paper.
This work was supported by Grant-in-aid for Scientific Research on Innovative Areas 25136716 (to S. O.), Grant-in-aid for Scientific Research (A) 25253038 (to S. O.), Grant-in-aid for Challenging Exploratory Research 25670079 (to S. O.), and 26670317 (to S. K.) from the Japan Society for the Promotion of Science.
- PepT1
- proton-coupled peptide transporter-1
- PPARα
- peroxisome proliferator-activated receptor-α
- Slc15a1
- soluble carrier protein family 15 member 1
- CBP
- cAMP-response element-binding protein (CREB)-binding protein
- ZT
- zeitgeber time
- PPRE
- peroxisome proliferator response element
- RXRα
- retinoid X receptor-α
- CA
- cholic acid
- Per2
- Period2.
REFERENCES
- 1. Matthews D. M. (1975) Intestinal absorption of peptides. Physiol. Rev. 55, 537–608 [DOI] [PubMed] [Google Scholar]
- 2. Herrera-Ruiz D., Knipp G. T. (2003) Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharm. Sci. 92, 691–714 [DOI] [PubMed] [Google Scholar]
- 3. Adibi S. A. (1997) The oligopeptide transporter (Pept-1) in human intestine: biology and function. Gastroenterology 113, 332–340 [DOI] [PubMed] [Google Scholar]
- 4. Palacín M., Estévez R., Bertran J., Zorzano A. (1998) Molecular biology of mammalian plasma membrane amino acid transporters. Physiol. Rev. 78, 969–1054 [DOI] [PubMed] [Google Scholar]
- 5. Daniel H., Kottra G. (2004) The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch. 447, 610–618 [DOI] [PubMed] [Google Scholar]
- 6. Ogihara H., Saito H., Shin B. C., Terado T., Takenoshita S., Nagamachi Y., Inui K., Takata K. (1996) Immuno-localization of H+/peptide cotransporter in rat digestive tract. Biochem. Biophys. Res. Commun. 220, 848–852 [DOI] [PubMed] [Google Scholar]
- 7. Meredith D., Price R. A. (2006) Molecular modeling of PepT1—towards a structure. J. Membr. Biol. 213, 79–88 [DOI] [PubMed] [Google Scholar]
- 8. Berlioz F., Maoret J. J., Paris H., Laburthe M., Farinotti R., Rozé C. (2000) α2-Adrenergic receptors stimulate oligopeptide transport in a human intestinal cell line. J. Pharmacol. Exp. Ther. 294, 466–472 [PubMed] [Google Scholar]
- 9. Nielsen C. U., Amstrup J., Steffansen B., Frokjaer S., Brodin B. (2001) Epidermal growth factor inhibits glycylsarcosine transport and hPepT1 expression in a human intestinal cell line. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G191–G199 [DOI] [PubMed] [Google Scholar]
- 10. Ashida K., Katsura T., Motohashi H., Saito H., Inui K. (2002) Thyroid hormone regulates the activity and expression of the peptide transporter PEPT1 in Caco-2 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G617–G623 [DOI] [PubMed] [Google Scholar]
- 11. Gangopadhyay A., Thamotharan M., Adibi S. A. (2002) Regulation of oligopeptide transporter (Pept-1) in experimental diabetes. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G133–G138 [DOI] [PubMed] [Google Scholar]
- 12. Saito H., Terada T., Shimakura J., Katsura T., Inui K. (2008) Regulatory mechanism governing the diurnal rhythm of intestinal H+/peptide cotransporter 1 (PEPT1). Am. J. Physiol. Gastrointest. Liver Physiol. 295, G395–G402 [DOI] [PubMed] [Google Scholar]
- 13. Inouye S. T., Kawamura H. (1979) Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 76, 5962–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin X., Shearman L. P., Weaver D. R., Zylka M. J., de Vries G. J., Reppert S. M. (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68 [DOI] [PubMed] [Google Scholar]
- 15. Maemura K., de la Monte S. M., Chin M. T., Layne M. D., Hsieh C. M., Yet S. F., Perrella M. A., Lee M. E. (2000) CLIF, a novel cycle-like factor, regulates the circadian oscillation of plasminogen activator inhibitor-1 gene expression. J. Biol. Chem. 275, 36847–36851 [DOI] [PubMed] [Google Scholar]
- 16. Ripperger J. A., Shearman L. P., Reppert S. M., Schibler U. (2000) CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 [PMC free article] [PubMed] [Google Scholar]
- 17. Caldelas I., Poirel V. J., Sicard B., Pévet P., Challet E. (2003) Circadian profile and photic regulation of clock genes in the suprachiasmatic nucleus of a diurnal mammal Arvicanthis ansorgei. Neuroscience 116, 583–591 [DOI] [PubMed] [Google Scholar]
- 18. Dardente H., Cermakian N. (2007) Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol. Int. 24, 195–213 [DOI] [PubMed] [Google Scholar]
- 19. Feillet C. A., Albrecht U., Challet E. (2006) “Feeding time” for the brain: a matter of clocks. J. Physiol. Paris. 100, 252–260 [DOI] [PubMed] [Google Scholar]
- 20. Shimakura J., Terada T., Saito H., Katsura T., Inui K. (2006) Induction of intestinal peptide transporter 1 expression during fasting is mediated via peroxisome proliferator-activated receptor α. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G851–G856 [DOI] [PubMed] [Google Scholar]
- 21. Göttlicher M., Widmark E., Li Q., Gustafsson J. A. (1992) Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 89, 4653–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller H., Dreyer C., Medin J., Mahfoudi A., Ozato K., Wahli W. (1993) Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc. Natl. Acad. Sci. U.S.A. 90, 2160–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leone T. C., Weinheimer C. J., Kelly D. P. (1999) A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U.S.A. 96, 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinal C. J., Yoon M., Gonzalez F. J. (2001) Antagonism of the actions of peroxisome proliferator-activated receptor-α by bile acids. J. Biol. Chem. 276, 47154–47162 [DOI] [PubMed] [Google Scholar]
- 25. Ritskes-Hoitinga M. (2004) in The Laboratory Mouse (Hedrich H., ed) pp. 463–479, Elsevier Academic Press, New York [Google Scholar]
- 26. Forman B. M., Chen J., Evans R. M. (1997) Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. U.S.A. 94, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Son D. O., Satsu H., Kiso Y., Shimizu M. (2004) Characterization of carnosine uptake and its physiological function in human intestinal epithelial Caco-2 cells. Biofactors 21, 395–398 [DOI] [PubMed] [Google Scholar]
- 28. Pan X., Terada T., Okuda M., Inui K. (2003) Altered diurnal rhythm of intestinal peptide transporter by fasting and its effects on the pharmacokinetics of ceftibuten. J. Pharmacol. Exp. Ther. 307, 626–632 [DOI] [PubMed] [Google Scholar]
- 29. Bookout A. L., Jeong Y., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. (2006) Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Issemann I., Green S. (1990) Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347, 645–650 [DOI] [PubMed] [Google Scholar]
- 31. Lefebvre P., Chinetti G., Fruchart J. C., Staels B. (2006) Sorting out the roles of PPARα in energy metabolism and vascular homeostasis. J. Clin. Investig. 116, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee Y., Yu X., Gonzales F., Mangelsdorf D. J., Wang M. Y., Richardson C., Witters L. A., Unger R. H. (2002) PPARα is necessary for the lipopenic action of hyperleptinemia on white adipose and liver tissue. Proc. Natl. Acad. Sci. U.S.A. 99, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. (1999) Identification of a nuclear receptor for bile acids. Science 284, 1362–1365 [DOI] [PubMed] [Google Scholar]
- 34. Bünger M., van den Bosch H. M., van der Meijde J., Kersten S., Hooiveld G. J., Müller M. (2007) Genome-wide analysis of PPARα activation in murine small intestine. Physiol. Genomics 30, 192–204 [DOI] [PubMed] [Google Scholar]
- 35. Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le Minh N., Damiola F., Tronche F., Schütz G., Schibler U. (2001) Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 20, 7128–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]