Background: Cells lacking c-Myc demonstrate metabolic abnormalities marked by reduced glycolysis, oxidative phosphorylation, and proliferation.
Results: These cells preferentially utilize fatty acids as energy-generating substrates and reprogram other pathways to maximize acetyl-CoA and ATP production.
Conclusion: Despite these compensatory changes, basal levels of acetyl-CoA and ATP remained low.
Significance: Therapies that limit acetyl-CoA availability might represent novel ways of inhibiting tumor cell growth.
Keywords: AMP-activated Kinase (AMPK), Electron Transfer Complex, Mitochondria, Pyruvate Dehydrogenase Complex (PDC), Tricarboxylic Acid Cycle (TCA Cycle) (Krebs cycle), Warburg Effect
Abstract
myc−/− rat fibroblasts (KO cells) differ from myc+/+ (WT) cells and KO cells with enforced Myc re-expression (KO-Myc cells) with respect to mitochondrial structure and function, utilization of glucose and glutamine as energy-generating substrates, and ATP levels. Specifically, KO cells demonstrate low levels of glycolysis and oxidative phosphorylation, dysfunctional mitochondria and electron transport chain complexes, and depleted ATP stores. We examined here how these cells adapt to their energy-deficient state and how they differ in their uptake and utilization of long- and medium-chain fatty acids such as palmitate and octanoate, respectively. Metabolic tracing of these molecules showed that KO cells preferentially utilize them as β-oxidation substrates and that, rather than directing them into phospholipids, preferentially store them as neutral lipids. KO cell transcriptional profiling and functional assays revealed a generalized up-regulation of pathways involved in fatty acid transport and catabolism as well as evidence that these cells attempt to direct acetyl-CoA into the tricarboxylic acid (TCA) cycle for ATP production rather than utilizing it for anabolic purposes. Additional evidence to support this idea included the finding that AMP-dependent protein kinase was constitutively activated in KO cells. The complex control of pyruvate dehydrogenase, which links glycolysis to the TCA cycle, was also maximized to ensure the conversion of pyruvate to acetyl-CoA. Despite these efforts to maximize acetyl-CoA for energy-generating purposes, its levels remained chronically low in KO cells. This suggests that tumor cells with Myc deregulation might be susceptible to novel therapies that limit acetyl-CoA availability.
Introduction
Cells dividing rapidly in response to normal or oncogenic signals have metabolic profiles distinct from those of their quiescent counterparts. Because they must coordinate mass accretion and division, they devote considerable resources to generating macromolecular precursors (1, 2). To support these processes, they must also be equipped to generate large amounts of ATP, usually by increasing glucose and glutamine utilization by the TCA3 cycle (3). Given this increased demand for energy and the fact that many of the macromolecular precursors originate from glycolytic and TCA cycle intermediates (1, 2), dividing cells undergo a process of metabolic reprogramming whereby the shunting of these intermediates into anabolic pathways assumes a more prominent role than during quiescence. An example of this occurs with the Warburg effect whereby glycolysis, normally utilized by resting cells to generate ATP anaerobically, continues to function aerobically to supply certain essential amino acids, nucleotides, and pentose sugars for macromolecular biosynthesis (2–4).
The dependence of dividing cells on the Warburg effect has occasionally been misconstrued as indicating that they minimize energy production via oxidative phosphorylation (Oxphos). In fact, provided sufficient oxygen, both glycolysis and Oxphos are often concurrently increased in tumor cells (5, 6). A particularly instructive example of this occurs with Myc oncoprotein deregulation, which, in addition to stimulating glycolysis, also increases mitochondrial mass, Oxphos, and electron transport chain function (7, 8). Although basal ATP levels do not change, its half-life is shortened (8) and likely reflects its increased utilization for anabolic processes. In contrast, Myc-deficient cells, such as myc−/− rat fibroblasts (KO cells) (7), have dramatically lower ATP levels and turnover that correlate with reduced glycolysis, Oxphos, replication, and cell mass relative to their myc+/+ WT counterparts or to KO cells whose Myc expression is restored (KO-Myc cells) (8). The mitochondrial changes documented in KO cells include an overall paucity of these organelles, atrophy of those that remain, and structural and functional electron transport chain defects (7, 8).
The enhanced utilization of glucose and glutamine that accompanies Myc overexpression correlates with increased uptake of these substrates and their consumption in the glycolytic pathway and TCA cycle, respectively. Myc positively regulates a majority of glycolytic enzymes and increases the conversion of glutamine to glutamate and α-ketoglutarate by both transcriptional and post-transcriptional mechanisms (3, 4, 9).
Another highly efficient energy source derives from mitochondrial fatty acid β-oxidation (FAO), which, like glycolysis, yields acetyl-CoA, the entry level TCA cycle substrate. During proliferation, the immediate downstream product of acetyl-CoA, citrate, can also be converted back to acetyl-CoA in the cytoplasm where, in ATP-consuming processes, it can be used for de novo lipid or steroid biosynthesis (10, 11). Although considerable effort has been devoted to delineating the means by which glucose and glutamine metabolism are regulated by Myc (3, 4, 9), our understanding of how Myc supervises the transport, directionality, and metabolism of fatty acids and their catabolites remains incomplete. In the current work, we have studied how WT, KO, and KO-Myc rat fibroblasts differ in this regard. Our studies indicate that, in an apparent effort to compensate for their ATP deficit and poor utilization of glucose and glutamine as energy-generating substrates, KO cells preferentially transport and oxidize long-chain fatty acids (LCFAs) such as palmitate. The channeling of LCFAs into the TCA cycle is facilitated not only by the up-regulation of enzymes involved in their transport and β-oxidation but also by a concurrent down-regulation of acetyl-CoA consumption for anabolic purposes. Because KO cells oxidize LCFAs more rapidly, their rate of incorporation into neutral lipids is lower than that of WT or KO-Myc cells. These latter cells utilize their neutral lipid stores for anabolic purposes to a greater extent, whereas KO cells eventually accumulate a higher stored neutral lipid content. Similar studies, which traced the fate of the freely diffusible medium-chain fatty acid (MCFA) octanoate and the two-carbon molecule acetate, indicated that their metabolism was also altered to maximize their conversion to acetyl-CoA. The importance of acetyl-CoA as a critical metabolic intermediate that links these opposing functions was further underscored by demonstrating that its supply is also regulated by additional Myc-dependent enzymes including pyruvate dehydrogenase (PDH), which converts pyruvate to acetyl-CoA; acetyl-CoA acetyltransferase (Acat1/2), which participates in FAO and directs the catabolism of certain amino acids into acetyl-CoA; and acetyl-CoA synthase 2 (AceCS2) and cytoplasmic acetyl-CoA hydrolase (cACH), which regulate the balance between acetate and acetyl-CoA. Despite these compensatory changes, KO cells remained profoundly depleted of acetyl-CoA. Collectively, these studies identify adaptive pathways through which exogenous fatty acid substrates, ranging from LCFAs to simple two-carbon units, can be converted to acetyl-CoA, which in KO cells is then preferentially directed toward replenishing ATP. KO cells resort to multiple strategies to correct their acetyl-CoA and ATP deficits. These include generating acetyl-CoA from multiple sources, redirecting it into an otherwise compromised TCA cycle, and minimizing its use for purposes other than ATP generation.
EXPERIMENTAL PROCEDURES
Cell Culture
All cell lines were routinely maintained as described previously (8). KO-Myc cells were generated through the use of stable transduction with a lentiviral vector encoding a full-length human Myc cDNA (8). A549-shMyc cells were generated by infecting A549 human alveolar lung cancer cells with a pTRIPZ lentiviral vector encoding red fluorescent protein and an shRNA directed against human Myc, both of which were tetracycline-inducible (Thermo Fisher). All lentiviral packaging and infections were performed as described previously (12) under BSL2+ conditions and were approved by the University of Pittsburgh Biosafety Committee. Stable transfectants were selected and maintained in puromycin-containing medium (1 μg/ml) as described above.
[14C]Palmitate and [14C]Octanoate Uptake and β-Oxidation Studies
FAO was quantified as described previously (13). Briefly, 2 × 104 WT and KO-Myc cells and 4 × 104 KO cells (all >90% viable) were seeded into 24-well tissue culture plates and allowed to attach overnight. The following day, medium was removed, and the cells were incubated at 37 °C for 30 min. in PBS. 200 μl of fresh PBS containing 1 mm carnitine (Santa Cruz Biotechnology, Santa Cruz, CA) and 0.2 μCi of BSA-bound [1-14C]-palmitate (specific activity = 32 mCi/mmol) (PerkinElmer) or 0.1 μCi of [14C]octanoate (specific activity = 55 mCi/mmol) (American Radiolabeled Chemicals, St. Louis, MO) were then added. 14CO2 was collected onto filters soaked in 0.6 n KOH, which were placed in a collection apparatus made from a 0.4-ml Eppendorf tube and maintained under an air-tight seal at 37 °C for 2 h (14). The medium was then acidified by adding 20 μl of 6 m perchloric acid to release additional dissolved CO2. Filters were removed after 60 min, and released 14CO2 was quantified from quadruplicate samples on a Beckman LS6500 scintillation counter. Counts were adjusted so as to normalize for any differences in total protein content among the three groups of cells (generally <10%). p values were calculated using one-way analysis of variance followed by Bonferroni's post hoc comparisons test. To measure [14C]palmitate and [14C]octanoate uptake, cells were plated as described above the day prior to labeling. Monolayers were washed twice with PBS and then incubated in fresh PBS (200 μl) for 30 min. Fatty acid-free BSA-bound [14C]palmitate (0.2 μCi/well) with 1 mm carnitine was incubated for the appropriate time periods. After thorough washing, the cells were lysed in 160 μl of 5% SDS and subjected to scintillation counting as described above. Results from triplicate samples were normalized to protein content. All experiments were repeated two additional times with comparable results.
Incorporation of [3H]Palmitate, [14C]Octanoate, and [14C] Acetate into Lipids
Cells were plated as described for oxidation studies except that tritiated palmitate was utilized (13). The next day, they were starved for 1 h in PBS and then labeled for 1 h in 200 μl of fresh PBS containing 0.5 μCi of BSA-bound [9,10-3H]palmitate (specific activity = 32 mCi/mmol) (PerkinElmer), [14C]octanoate (0.1 μCi) containing 1 mm carnitine, or 0.2 μCi of [14C]sodium acetate (PerkinElmer, specific activity = 54.3 mCi/mmol). The cells were then washed twice in warm PBS, and fresh PBS with 1 mm carnitine was added for 2 h. Cells were then lysed with 25 μl of 6 n KOH, and an organic extraction was performed in 1 ml of methanol:chloroform (1:1), which was then removed and dried under nitrogen. After adding ice-cold acetone to the dried lipid film and incubating at −20 °C overnight, the phospholipid pellet was separated by centrifugation (14,000 × g, 30 min at 4 °C). Both neutral lipids and phospholipids were redissolved in 50 μl of chloroform for quantification. The labeled fractions were quantified by scintillation counting as described above. Each assay was performed in triplicate, and p values were calculated using two-way analysis of variance followed by Bonferroni's post hoc comparison.
Enzyme Assays
Measurements of very long- and medium-chain acyl-CoA dehydrogenase activities (ACADVL and ACADM, respectively) in cell-free extracts were performed using the anaerobic electron transfer flavoprotein (ETF) fluorescence reduction assay as described previously (15). Measurements were performed with an LS50B fluorescence spectrophotometer (PerkinElmer) with a heated cuvette block set to 32 °C. One unit of activity was defined as the amount of enzyme necessary to completely reduce 1 μmol of ETF in 1 min. Each assay was performed in duplicate, and one-way analysis of variance tests followed by Bonferroni's post hoc comparisons tests were performed in all statistical analyses. Assay results were adjusted to account for previously determined differences in mitochondrial mass based on staining with 10-n-nonyl-acridine orange (8).
PDH activity was quantified using an enzyme activity microplate assay kit (MitoSciences, Eugene, OR). Five nearly confluent 100-mm plates of cells were harvested by trypsinization, washed twice in PBS, flash-frozen in liquid nitrogen, and stored at −80 °C until ready to assay. Each assay utilized 1 mg of total cellular protein in 200 μl of the buffer provided by the supplier. The remainder of the assay was completed according to the supplier's protocol.
Visualization and Quantification of Neutral Lipids
Cells were plated in 12-well dishes containing sterile glass coverslips and cultured overnight. They were then washed once in PBS and stained with BODIPY-493/503 neutral lipid stain (Molecular Probes/Thermo Fisher: final concentration = 10 μg/ml) in complete DMEM for 30 min at 37 °C. After washing in PBS, the cells were fixed for 30 min at room temperature in PBS, 2% paraformaldehyde, and permeabilized in PBS, 0.1% Triton X-100 for 5 min. After three additional washes in PBS, the coverslips were blocked in 1% BSA for 30 min and co-stained with 0.165 mg/ml Texas Red phalloidin (Life Technologies) and 0.1 mg/ml DAPI nuclear stain (Sigma-Aldrich) final concentration and then mounted. Representative fields were imaged on an Olympus IX81 motorized inverted confocal microscope using Olympus FluoView software (Version 2.1c).
Neutral lipid content was quantified using a modification of a previously described protocol (16). Briefly, 106 cells were trypsinized, collected in PBS, and stained at 25 °C for 30 min in the dark using BODIPY-493/503 (final concentration = 10 μg/ml in complete DMEM). Flow cytometry was performed using a FACStar flow cytometer (BD Biosciences). Each experiment was performed in triplicate with similar outcomes. Oil Red O staining was performed using standard histologic techniques following growth of cells on coverslips, which were air-dried and stained with both Oil Red O and H&E.
Immunoblotting
Western blots were performed as described previously (8). Antibodies used included those for total AMP-dependent protein kinase (AMPK) (rabbit polyclonal IgG, 1:1,000, Cell Signaling Technology, Beverly, MA, catalog number 2532); phospho-AMPK (phospho-Thr172) (rabbit monoclonal IgG, 1:2,000 Cell Signaling, catalog number 4188); PDHE1 (mouse monoclonal IgG, 1:1,000, Santa Cruz Biotechnology, catalog number SC-377092); phospho-PDH (phospho-Ser293) (rabbit monoclonal IgG, 1:10,000, EMD Millipore, Billerica, MA, catalog number ap1062); pyruvate dehydrogenase kinase 1 (PDK1) (rabbit monoclonal IgG, 1:1,000, Cell Signaling, catalog number 3820); and pyruvate dehydrogenase phosphatase 2 (PDP2) (rabbit polyclonal IgG, 1:500, BioVision, Inc., Milpitas, CA catalog number 3944). Endogenous Myc protein was assessed with the mouse monoclonal antibody 9E10 (Santa Cruz Biotechnology) as described previously (8). Protein loading was confirmed using a β-actin mAb (1:2,000, Santa Cruz Biotechnology catalog number 3700). Blots were developed using a SuperSignalTM West Pico chemiluminescent substrate kit (Thermo Fisher).
RNA Isolation and Real-time Quantitative RT-PCR
RNA was isolated from 80–90% confluent cell culture wells using a Qiagen RNeasy mini kit, according to the manufacturer's instructions (Qiagen, Inc. Chatsworth, CA). Residual DNA was removed using a TURBO DNA-freeTM DNase treatment and removal kit (Life Technologies). PCR reactions were conducted with a Power SYBR® Green RNA-to-CTTM 1-Step kit (Life Technologies), using a StepOnePlusTM real-time PCR system (Applied Biosystems, Inc. Carlsbad, CA). Quantitative RT-PCR primers were designed using Primer3web software and were synthesized by International DNA Technologies, Inc. (Coralville, IA). Reactions were optimized such that only single bands of the predicted size were visualized upon gel electrophoresis (primer sequences and conditions are available on request). Each quantitative RT-PCR reaction was performed in triplicate with variations among samples seldom exceeding 5%. Statistical analysis was performed using Student's t test.
Acetyl-CoA Assays
Acetyl-CoA was assayed using an acetyl-coenzyme A assay kit (Sigma-Aldrich catalog number MAK039) according to the manufacturer's instructions. Samples were isolated from fresh cell pellets and lysed, and protein was quantified using a PierceTM BCA protein assay kit (Thermo Fisher). Samples were deproteinized by adding 2 μl of 1 n perchloric acid/mg protein, and the resulting supernatant was neutralized by adding 3 m KHCO3. Standards were quantified on a 0–1 nmol scale, and 50 μl of sample was added to each well. The plate was read on a SpectraMax M2 fluorescence plate reader and normalized to protein content. Analysis was performed using a one-way analysis of variance and a post hoc Bonferroni's test of comparisons. Similar results were obtained when cells were simultaneously lysed and deproteinated or when lysates were incubated on ice for prolonged periods. Thus, results were not influenced by exogenous reactions that differentially depleted cells of acetyl-CoA.
RESULTS
Uptake and Oxidation of Fatty Acids by KO Cells
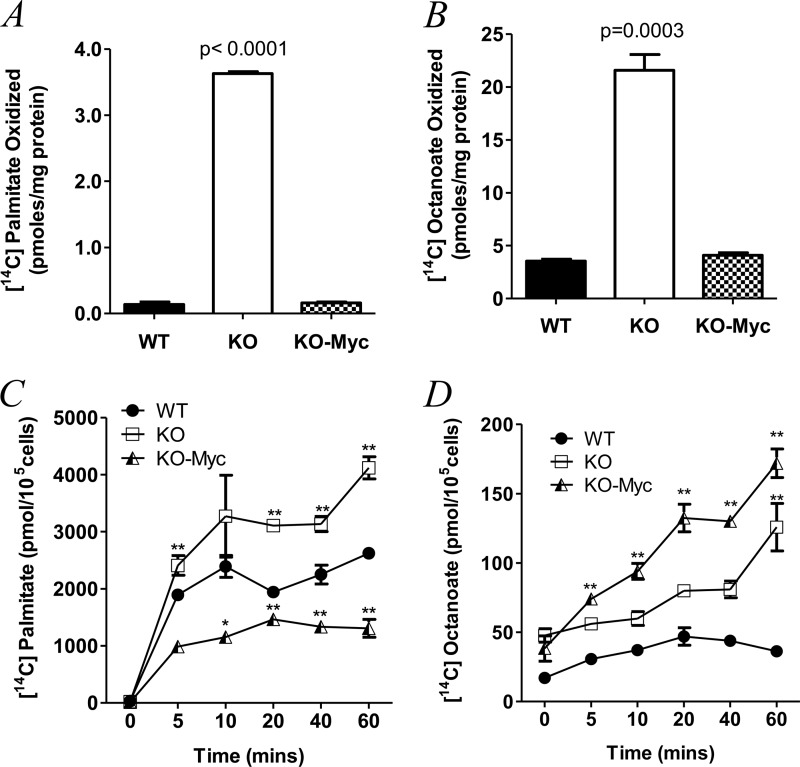
To quantify fatty acid utilization among WT, KO, and KO-Myc cells, we exposed them to 14C-radiolabeled palmitate or [14C]octanoate as representative LCFAs and MCFAs, respectively (13, 14). In each case, the 14C tag resided on the carboxylic acid moiety, which allowed us to test the integrity and interdependence of at least seven distinct enzymatic steps in the β-oxidation pathway. These include the placement of the trans-double bond between C2 and C3 by very long- or medium-chain acyl-CoA dehydrogenase, the production of l-B-hydroxyacyl-CoA by enoyl-CoA hydratase, the conversion of l-B-hydroxyacyl-CoA to B-ketoacyl-CoA by B-hydroxyacyl-CoA dehydrogenase, and thiolysis between C2 and C3 of B-ketoacyl-CoA to produce acetyl-CoA. Upon entry into the TCA cycle, 14C-tagged acetyl-CoA would need to be conjugated with oxaloacetate before eventually surrendering its tag as CO2 during the conversion of isocitrate to α-ketoglutarate. Importantly, LCFA oxidation is also dependent on the rate at which the substrate is actively transported across the plasma and mitochondrial membranes and into the mitochondrial matrix (17). These steps may not necessarily parallel CO2 production given that LCFAs can also be stored cytoplasmically as neutral lipids or utilized for anabolic rather than catabolic purposes. MCFAs can be utilized similarly although they enter mitochondria passively without contributing to neutral lipid pools (17, 18). Thus, differences in MCFA uptake should better reflect the rate of β-oxidation. As seen in Fig. 1A, the rate of [14C]palmitate oxidation was similar in WT and KO-Myc cells after adjusting for differences in mitochondrial mass (p = 0.58), but was nearly 25-fold higher in KO cells (p < 0.0001). Similar studies performed with [14C]octanoate (Fig. 1B) also showed a >5-fold higher rate of β-oxidation in KO cells. Thus, despite their markedly slower proliferation and their reduced mitochondrial function, KO cells actually utilize a larger amount of LCFAs and MCFAs for energy generation than do their Myc-replete counterparts.
FIGURE 1.
Differential utilization and uptake of LCFAs and MCFAs by WT, KO, and KO-Myc cells. A, β-oxidation of [14C]palmitate. B, β-oxidation of [14C]octanoate. C, uptake of [14C]palmitate. D, uptake of [14C]octanoate. Each point represents the mean of triplicate determinations ± 1 S.E. p values are expressed relative to WT cells (* = p < 0.05, ** = p < 0.001).
We next asked whether the observed differences in FAO among the above three cell lines were associated with differences in their fatty acid uptake rates. As seen in Fig. 1C, [14C]palmitate uptake was highest in KO cells, in keeping with their overall greater utilization of this substrate for FAO. A higher rate of [14C]octanoate uptake by KO-Myc cells was also consistent with their preferential utilization of this substrate for processes other than FAO (Fig. 1D). Interestingly, WT and KO-Myc cells showed distinct preferences for LCFAs and MCFAs, with the former cells demonstrating a greater uptake of palmitate than octanoate, whereas the reverse was true for KO-Myc cells. Thus, KO cells have a selective uptake for both LCFAs and MCFAs and utilize them more efficiently as FAO substrates. However, each cell line possesses a distinct pattern of LCFA and MCFA uptake that presumably reflects differential usage for processes other than FAO.
Differential Utilization of Fatty Acids
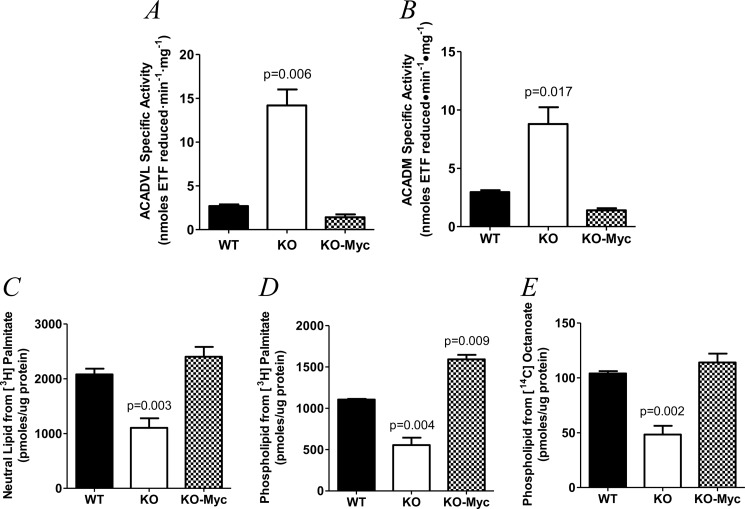
The initial stage of FAO involves the iterative insertion of a trans-double bond between the C2 and C3 carbon atoms of the acyl-CoA thioester substrate in a reaction that, for palmitate, is catalyzed by ACADVL and, for octanoate, by ACADM (19). To determine whether the preferential utilization of palmitate and octanoate for FAO by KO cells could be explained by differences in these enzymes, we measured their activities (15). As seen in Fig. 2, A and B, both ACADVL and ACADM activities were increased significantly in KO cells after adjusting to mitochondrial mass, thus providing an explanation for their more efficient utilization of these substrates.
FIGURE 2.
ETF assays for ACADVL and ACADM activities and incorporation of LCFAs and MCFAs into neutral and phospholipids in WT, KO, and KO-Myc cells. A, ACADVL enzymatic activity. Mean values are depicted ± 1 S.E. Results were normalized to account for differences in mitochondrial mass among the three cell types (8). B, ACADM activity. Results are presented as described for A. C, incorporation of [3H]palmitate into neutral lipids. D, incorporation [3H]palmitate of into phospholipids. E, incorporation of [14C]octanoate into phospholipids.
The foregoing studies were designed to evaluate the fate of fatty acids as energy-generating catabolic substrates but did not explain how the acetyl-CoA generated from their catabolism was utilized for anabolic purposes. To address this, we labeled the cell lines with [3H]palmitate and followed the incorporation of its tag into both phospholipid and neutral lipid pools (13). Because octanoate is not incorporated into neutral lipids, we measured the transfer of its 14C tag into phospholipids only. The rate of incorporation of the 3H tag of palmitate into both neutral lipids (Fig. 2C) and phospholipids (Fig. 2D) and the [14C]octanoate tag into phospholipids (Fig. 2E) was significantly lower in KO cells.
Neutral Lipid Accumulation in KO Cells
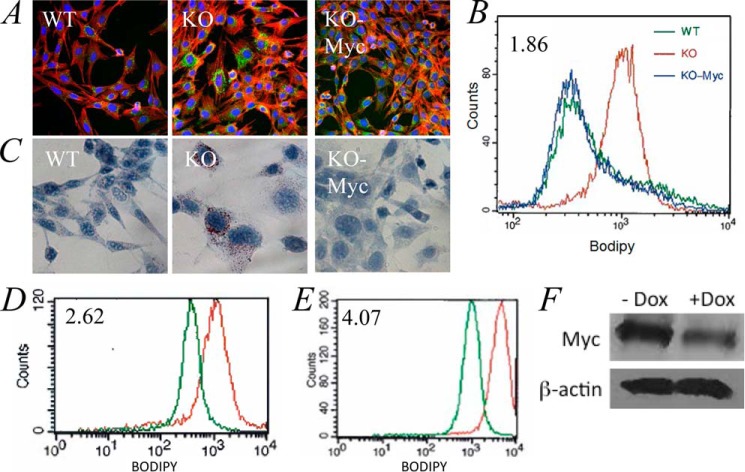
Previous studies have shown that N-Myc inhibition in neuroblastoma cells increases their neutral lipid content (20). We therefore next asked whether fatty acid uptake and utilization were balanced by assessing differences in basal neutral lipid content. Each cell line was stained with the neutral lipid-specific probe BODIPY-493/503 and visualized by fluorescence microscopy to assess its neutral lipid content. WT and KO-Myc cells demonstrated low level accumulation of BODIPY-493/503 in contrast to KO cells in which considerable amounts of the dye could be detected (Fig. 3, A and B). In other experiments, we confirmed the presence of excess neutral lipid staining in KO cells by Oil Red O staining (Fig. 3C). Using two different approaches, we confirmed that the accumulation of neutral lipids was a direct and rapid consequence of Myc inactivation. First, treatment of WT cells with the Myc inhibitor 10058-F4 (21) significantly increased BODIPY-493/503 uptake (Fig. 3D). Additionally, reduction of Myc protein levels in A549 human lung cancer cells using tetracycline-dependent conditional expression of a Myc shRNA produced a similar result (Fig. 3, E and F). Collectively, these findings support the idea that neutral lipid accumulation in KO cells is a direct consequence of Myc depletion and mitochondrial dysfunction.
FIGURE 3.
Neutral lipid accumulation in KO cells. A, staining of cells for neutral lipids. WT, KO, and KO-Myc cells were plated onto glass microscope slides and allowed to grow to subconfluency before being fixed and stained with BODIPY-493/503 and counterstained with Texas red-labeled phalloidin and DAPI. Representative fields are shown. B, quantification of neutral lipid staining. Each of the indicated cell types was stained with BODIPY-493/503 and assessed by flow cytometry. C, Oil Red O staining. Each of the indicated cell types was plated as in A and then stained with Oil Red O. D, 10058-F4-mediated inhibition of endogenous Myc leads to the accumulation of neutral lipids. WT cells in log-phase growth were exposed to 50 μm 10058-F4 (22) for 48 h before being stained with BIODIPY-493/503 and assessed by flow cytometry. The number in the upper left corner is the ratio of the mean intensity of staining of cells with (red) and without (green) 10058-F4 exposure. E, induction of shMyc in A549 cells leads to neutral lipid accumulation. A549 cells (∼10% confluency) were allowed to grow for an additional 3 days in the absence (green) or presence (red) of 2.5 μg/ml doxycycline before being stained with BODIPY-493/503 as described for B. F, immunoblots demonstrating a reduction in endogenous Myc protein levels following a 3-day exposure to doxycycline (Dox).
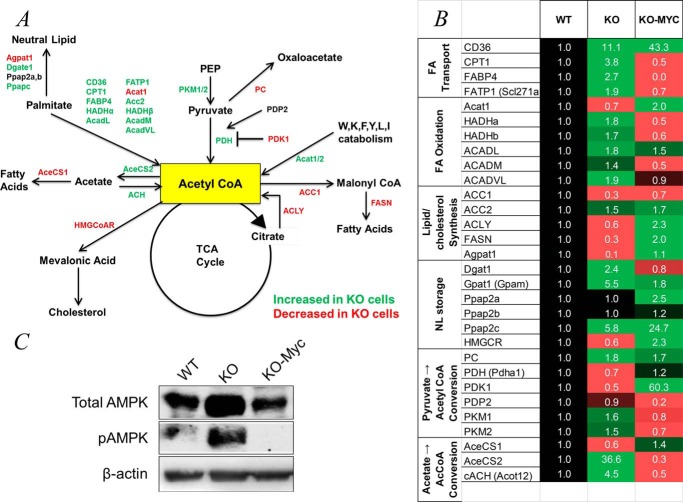
To better define the relationship between fatty acid transport and metabolism and the generation and utilization of acetyl-CoA, we utilized real-time quantitative RT-PCR to quantify transcripts encoding the enzymes described above plus select others to allow an overview of the activity of relevant pathways (Fig. 4A). Transcripts were grouped into six functional categories representing fatty acid transport and FAO, de novo lipid and steroid biosynthesis, neutral lipid storage, and the generation of acetyl-CoA from acetate and pyruvate. This last category included transcripts for the PDH E1 subunit and its regulators, PDK1 and PDP2, which are responsible for the phosphorylation-dependent inactivation and activation, respectively, of E1 (22). Also included were transcripts for pyruvate carboxylase, which catalyzes an anaplerotic reaction important for gluconeogenesis and lipid biosynthesis and irreversibly redirects pyruvate to oxaloacetate to limit the conversion of the former to acetyl-CoA (23). We also examined transcripts for the pyruvate kinase isoforms PKM1 and PKM2, which catalyze the irreversible conversion of phosphoenolpyruvate to pyruvate during glycolysis. PKM2 is typically more abundant in rapidly proliferating cells and has a significantly higher Km for phosphoenolpyruvate (24, 25). This may better allow for the accumulation of upstream glycolytic intermediates, thus facilitating their channeling into collateral, anabolic pathways in support of proliferation-associated mass accretion (25, 26). Consistent with this idea, the activity of PKM2 is subject to negative regulatory control by ATP and possibly by acetyl-CoA as well (1, 24, 27).
FIGURE 4.
Alteration of metabolic pathways in KO cells. A, pathways depicting the generation and utilization of acetyl-CoA in KO cells. The major sources of mitochondrial acetyl-CoA include the glycolytic intermediate pyruvate; long- and medium-chain fatty acids such as palmitate and octanoate; acetate; and a subset of amino acids that includes tryptophan, lysine, phenylalanine, tyrosine, leucine, and isoleucine. Cytoplasmic acetyl-CoA can also be generated from the mitochondrial TCA substrate citrate in a reaction involving ATP citrate lyase (ACLY) and from acetate by AceCS2. The fate of acetyl-CoA in pathways other than the TCA cycle primarily includes its conversion to malonyl-CoA during fatty acid synthesis. In addition, pyruvate, the direct glycolytic precursor of acetyl-CoA, can be diverted from this pathway by an anaplerotic reaction involving its conversion to oxaloacetate that is catalyzed by pyruvate carboxylase (PC), and palmitate can be diverted into neutral lipids. The activity of PDH, which catalyzes the conversion of pyruvate to acetyl-CoA, is also negatively regulated by PDK1 and positively regulated by PDP2. Based on transcriptional profiling shown in B, enzymes whose transcripts are up-regulated in KO cells are depicted in green, and those that are down-regulated are depicted in red. PEP, phosphoenolpyruvate; FASN, fatty acid synthase; ACH, acyl-CoA hydrolase; HMG-CoAR, HMG-CoA receptor; Acat1/2, acetyl-CoA acetyltransferase 1 and 2, HADHα/HADHβ, mitochondrial trifunctional protein. B, transcript expression. For simplicity, transcripts and proteins are designated by common acronyms. Transcripts that were significantly up-regulated in KO cells are indicated in green, and those that are down-regulated are depicted in red. The values of transcripts in WT cells were arbitrarily set at 1 (black). Transcripts are arranged according to the functional categories of their representative enzymes. Each value represents the mean of triplicate determinations for each transcript. FA Transport, fatty acid transport; NL storage, neutral lipid storage. C, AMPK is up-regulated in KO cells. Immunoblots of total cell lysates from WT, KO, and KO-Myc cells were probed with antibodies for total AMPK or phospho-AMPK (pAMPK) (phospho-Thr172).
The results of transcriptional profiling (Fig. 4B) were largely consistent with our foregoing studies. First, they indicated that KO cells up-regulate transcripts encoding enzymes involved in the production of acetyl-CoA for energy generation while down-regulating those involved in anabolism such as de novo lipid and steroid biosynthesis (Fig. 4A). One example of the potential precision of this reprogramming in KO cells was seen in the case of the 5-fold change in the relative ratio of acetyl-CoA carboxylase 1 and 2 isoforms (ACC1 and ACC2), which function in fatty acid synthesis and β-oxidation, respectively (28). Also notably up-regulated in KO cells were several transcripts such as phosphatidic acid phosphatase types 2b and c (Ppap2b and Ppap2c) and diacylglycerol acyltransferase 1 (Dgat1), which encode key enzymes involved in the shunting of fatty acids into neutral lipid storage pools (29, 30).
AMPK Is Myc-responsive
Some of the above discussed enzymes are regulated post-translationally by AMPK, a serine/threonine kinase that is itself activated by phosphorylation in response to low ATP/(AMP+ADP) ratios (31). A role for AMPK in maintaining adenosine nucleotide homeostasis stems from its inhibition of ATP-consuming processes such as macromolecular synthesis and cell proliferation along with its stimulation of ATP-generating reactions such as glycolysis and Oxphos (32). Among the enzymes depicted in Fig. 4 whose activities are down-regulated by AMPK-mediated phosphorylation are ACC1, which catalyzes the conversion of acetyl-CoA to malonyl-CoA in the initial step of fatty acid synthesis; fatty acid synthase (FASN), which converts malonyl-CoA into palmitate; and HMG-CoA reductase (HMGCR), the rate-limiting step in the biosynthesis of cholesterol and other steroids (32). AMPK-mediated phosphorylation of the palmitate cell surface receptor CD36 has also been reported to increase its rate of cycling between the cell membrane and intracellular compartments, thereby affecting the normal balance between FAO and lipid accumulation in favor of the latter (33, 34). Finally, although not known to be a direct AMPK target, carnitine palmitoyltransferase I (CPT1) is suppressed by malonyl-CoA, such that ACC1 inhibition by AMPK would likely increase FAO (35).
To determine whether the altered metabolic pathways of KO cells might be susceptible to post-translational modulation by AMPK, we compared the levels of total and active (Thr172-phosphorylated) forms of AMPK in WT, KO, and KO-Myc cells. KO cells showed marked constitutive Thr172 phosphorylation as well as increased total AMPK levels (Fig. 4C). These findings are in keeping with the profound ATP deficit of KO cells (8) and suggest that, despite the constitutive activation of AMPK, it is unable to correct the energy deficit.
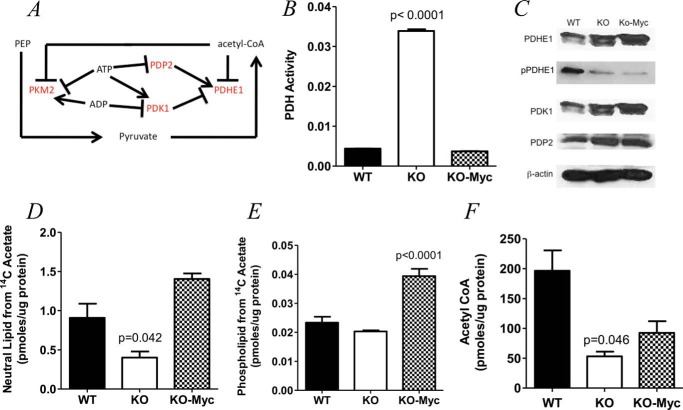
KO Cells Maximize Their Accumulation of Acetyl-CoA by Increasing Its Production and Decreasing Its Utilization for Purposes Other than TCA Cycle Utilization
The E1 subunit of the mitochondrial PDH complex catalyzes pyruvate decarboxylation, which is the first and rate-limiting step in its irreversible conversion to acetyl-CoA (Fig. 5A) (36). In addition to its regulation by PDK1 and PDP2 (22, 37, 38), PDHE1 is under additional negative feedback control by acetyl-CoA and positive control by ATP by virtue of the inhibitory effect of the latter on PDP2 (39). Furthermore, ATP and ADP exert positive and negative control, respectively, over PDK1 (Fig. 5A). Although the transcripts encoding these proteins were modestly down-regulated in KO cells (Fig. 4B), the complexity of PDHE1 post-translational regulation demanded that we actually measure its activity and thereby gauge the overall extent to which it was subject to control by these various and often opposing regulatory factors. As shown in Fig. 5B, KO cells contained approximately eight times as much PDH activity as WT and KO-Myc cells. Although immunoblotting showed modest differences in PDHE1 protein levels among the three cell lines (Fig. 5C), it demonstrated more dramatically, in both KO and KO-Myc cells, the relative under-phosphorylation of PDHE1 on Ser293, the site whose modification by PDK1 and PDP2 most affects its activity (31). Further consistent with the increased PDH activity in KO cells were their higher levels of PDP2 relative to WT cells. In contrast, no differences in the levels of PDK1 were observed between WT and KO cells. Although KO-Myc cells contained nearly 5-fold lower levels of PDP2 transcripts than WT cells and 60-fold higher levels of PDK1 transcripts (Fig. 4B), this was not reflected in PDH activity (Fig. 5B).
FIGURE 5.
Myc-regulated control of acetyl-CoA generation from pyruvate. A, outline of the reaction and its regulatory network. The E1 subunit of the mitochondrial PDH complex is subject to negative feedback inhibition by acetyl-CoA and to enzymatic regulation by the inhibitory kinase PDK1 and the stimulatory phosphatase PDP2 via Ser293 phosphorylation on the E1 subunit of PDH. PDK1 is subject to additional positive control by ATP and to negative control by ADP, whereas PDP2 is subject to negative control by ATP. In addition, the PKM2 isoform, which is less efficient at converting phosphoenolpyruvate to pyruvate, is under positive regulatory control by ADP and negative regulatory control by ATP and acetyl-CoA. B, PDH activity in WT, KO, and KO-Myc cells after adjusting for differences in mitochondrial mass. C, immunoblots for PDHE1, phospho-Ser293-PDHE1 (pPDHE1), PDK1, and PDP2. A β-actin blot was included as a protein loading control. D and E, acetate incorporation in neutral lipids and phospholipids, respectively. F, total acetyl-CoA levels in WT, KO, and KO-Myc cells. Each point represents the mean of quadruplicate determinations ± 1 S.E.
Another source of acetyl-CoA is acetate, which, in the whole animal, is typically supplied by bacterial fermentation in the colon, by the metabolic breakdown of acetaldehyde, and by the action of enzymes such as sirtuins and histone deacetylases (40). Like octanoate, acetate is both freely diffusible and readily available for metabolism in both the cytosol and the mitochondria. Consistent with the notion that KO cells attempt unsuccessfully to normalize acetyl-CoA levels, we note that AceCS2 transcript levels were elevated by nearly 40-fold in KO cells, whereas those for AceCS1 were modestly decreased (Fig. 4B). AceCS2, a mitochondrial enzyme, converts acetate to acetyl-CoA for utilization by the TCA cycle whereas AceCS1, which is cytoplasmic, is more important for fatty acid synthesis (41). Thus, the >60-fold changes in the AceCS1:AceCS2 ratio described above would be expected to greatly favor acetate conversion into acetyl-CoA in the mitochondria. Further consistent with, and perhaps contributing to, the reduced utilization of acetate for fatty acid synthetic pathways was the finding that transcripts for cytoplasmic acetyl-CoA hydrolase, which converts acetyl-CoA back to acetate in the cytoplasm (42), were increased 4.5-fold in KO cells. Indeed, when cells were incubated with [14C]acetate, KO cells incorporated the least amount of 14C tag, particularly into phospholipids (Fig. 5, D and E).
Finally, we measured steady-state levels of acetyl-CoA. As seen in Fig. 5F, both KO and KO-Myc cells contained reduced levels of acetyl-CoA relative to WT cells, although the latter did not reach statistical significance. Together with the previous results (Fig. 4B), these findings are most compatible with the idea that KO cell metabolism is directed primarily at maximizing the generation of acetyl-CoA for use by the TCA cycle and minimizing its utilization for anabolic reactions. Despite maximal efforts to produce more acetyl-CoA, KO cells remain unable to maintain normal levels of this substrate.
DISCUSSION
Numerous studies support the idea that the importance of Myc in promoting cell proliferation derives in part from its ability to ensure the provision of adequate supplies of anabolic substrates and ATP to support macromolecular syntheses (2–4, 9, 43). The silencing of Myc is associated with numerous metabolic and proliferative consequences that ultimately can be traced to defects in glycolysis and mitochondrial structure and function (8, 12, 44). That these factors are rate-limiting for proliferation is supported by findings shown here and elsewhere that, even when provided with adequate energy-generating substrates such as glucose, glutamine, and fatty acids, KO cells remain chronically ATP-depleted and respond by activating AMPK in a futile attempt to remedy this energy deficit (Fig. 4C). However, because two of the major responses to AMPK activation include the up-regulation of glycolysis and Oxphos, both of which are Myc-dependent (3, 4, 8, 31), the AMPK response is ultimately abortive despite its chronicity.
KO cells respond to their ATP deficit by up-regulating FAO and many of the transcripts associated with LCFA uptake, transport, and metabolism (Fig. 4, A and B). Palmitate oxidation, which begins with ACADVL, is further enabled by the high activity of this enzyme in KO cell mitochondria. Thus, despite their reduced overall mass, atrophic structure, and relatively poor utilization of substrates such as glucose and glutamine (8, 44), KO cell mitochondria disproportionately oxidize LCFAs. They also contain higher ACADM activity and preferentially oxidize octanoate whose transport into cells and mitochondria, unlike that of palmitate, is passive.
Importantly, the transcriptional profiles depicted in Fig. 4B represent steady-state levels in otherwise isogenic cells that have adapted to long term differences in Myc expression and have distinct metabolic behaviors (7). Although these differences may not necessarily reflect direct Myc targets (45, 46), they are nonetheless useful in that they reveal long term strategies employed by Myc-compromised cells to compensate for their inherent metabolic disadvantages. In this regard AMPK, whose level of activation is clearly inversely related to Myc levels (Fig. 4C), is a well known regulator of many of the same process controlled by Myc such as FAO, fatty acid synthesis, glycolysis, and Oxphos (31). However, because the ultimate metabolic function of Myc is to increase ATP synthesis in support of anabolism and proliferation (8), whereas the function of AMPK is to conserve energy until the ATP:ADP/AMP balance is restored, the integrated effects we observe based on steady-state transcripts may reflect a metabolic compromise between these opposing actions (47).
In a seemingly paradoxical finding, KO cells were found to possess the highest stores of neutral lipids (Fig. 3, A–C) despite incorporating the least amount of [3H]palmitate into this compartment (Fig. 2C). This latter finding is likely attributable to the fact that a large proportion of palmitate entering KO cells is immediately utilized for FAO (Fig. 1A), whereas in Myc-replete cells, larger amounts of the fatty acid are directed into neutral lipid pools (Fig. 2C). The extremely slow rate of KO cell proliferation (7) minimizes the need for these neutral lipid pools to serve as sources for phospholipid synthesis. Reduced demand for neutral lipid mobilization thus permits KO cells to accumulate higher neutral lipid stores despite their lower rates of accumulation. In contrast, WT and KO-Myc cells synthesize phospholipids at higher rates (Fig. 2C) and thus mobilize neutral lipids much more rapidly for this purpose (Fig. 2D), thus preventing their accumulation. The highly dynamic nature of neutral lipid stores is further evidenced by the rapidity with which they accumulate following Myc inhibition (Fig. 3, D and E) (20).
A major finding of the current study is that KO cells, in addition to deriving a considerable fraction of their acetyl-CoA from FAO, also maximize its production from other sources and minimize its utilization for purposes other than energy production. For example, reduced levels of transcripts involved in fatty acid synthesis such as those for ATP citrate lyase (ACLY), ACC1, and fatty acid synthase support the notion that KO cells minimize their incorporation of acetyl-CoA into lipids. That this down-regulation occurs throughout the pathway and involves its most proximal enzyme (ATP citrate lyase) would seem to favor the retention of citrate within the TCA cycle to ensure its utilization for Oxphos. Further consistent with this was the finding that ACC2, proposed to be more important than ACC1 for FAO (28), was up-regulated in KO cells, whereas ACC1 was down-regulated. High levels of palmitate within KO cells might further inhibit fatty acid synthesis by virtue of the well known tendency of the substrate to suppress ACC1 (48). A similar attempt to direct acetyl-CoA away from synthetic pathways was observed with the down-regulation in KO cells of transcripts for HMG-CoA reductase, the rate-limiting enzyme of the mevalonate pathway (49). Both ACC1 and HMG-CoA reductase are also further suppressed by AMPK (31). The down-regulation of pyruvate carboxylase also serves indirectly to maximize the availability of pyruvate for conversion into acetyl-CoA by diverting it away from the anaplerotic pathway that furnishes oxaloacetate. Other pathways through which acetyl-CoA production is maximized are up-regulated in KO cells and include those involving its AceCS2-mediated synthesis directly from acetate and the catabolism of selected amino acids by Acat1 and Acat2.
The PDH-mediated conversion of pyruvate to acetyl-CoA provides yet another example of how KO cells attempt to selectively utilize acetyl-CoA for ATP generation (Fig. 5A). This reaction is particularly noteworthy as it illustrates the complex and interdependent regulation that Myc and adenine nucleotides may exert over acetyl-CoA levels as well as the negative feedback control that acetyl-CoA itself provides. PDH activity, which is increased in KO cells (Fig. 5B), is positively controlled by the PDP2 phosphatase and negatively controlled by the PDK1 kinase (22, 36–38). The net result is PDHE1 dephosphorylation and activation (Fig. 5C). Nonenzymatic control is exerted by the repressive action of acetyl-CoA; by ATP, which inhibits PDP2 and stimulates PDK1; and by ADP, which represses PDK1 (Fig. 5A) (22, 36–38). Given that the intracellular milieu of KO cells is one in which both acetyl-CoA and ATP levels are low, these small molecules likely exert significant additional influence on PDH activity. The relatively normal PDH activity in KO-Myc cells, despite its hypo-phosphorylation, suggests that factors other than those examined here may play additional roles in its regulation (22).
Another factor that might also influence acetyl-CoA levels in KO cells is PKM2, whose ability to catalyze the conversion of phosphoenolpyruvate to pyruvate is accelerated by ADP and inhibited by ATP and acetyl-CoA (Fig. 5A) (24, 25, 50). Because this reaction is one of only three in the entire glycolytic pathway that is irreversible, it provides a relatively stable source of the substrate. It is noteworthy that both PKM2 and PKM1 are equally up-regulated in KO cells in contrast to KO-Myc cells where they are coordinately down-regulated. In the latter case, where ATP generated by Oxphos is already abundant, this may permit phosphoenolpyruvate and its upstream precursors to accumulate and be diverted into anabolic pathways.
Although both KO and KO-Myc cells have lower levels of acetyl-CoA (Fig. 5F), the origins and consequences of these deficits are likely quite different. In KO cells, we believe that this arises primarily from reduced ability to produce acetyl-CoA within atrophic and dysfunctional mitochondrial that reduces the overall acetyl-CoA supply, despite an increase in PDH activity as discussed above. In contrast, the acetyl-CoA deficiency of KO-Myc cells likely reflects the proliferative strain imposed upon them as they attempt to keep pace with high levels of fatty acid synthesis and high rates of ATP turnover (8). Thus, the reduced level of acetyl-CoA in KO-Myc cells more likely represents the accelerated rate at which this substrate is utilized in contrast to KO cells in which acetyl-CoA production is compromised. This suggests that the supply of acetyl-CoA may represent a potential proliferative and metabolic bottleneck that might be exploited in a therapeutic setting, particularly in cancers that are Oxphos-dependent.
This work was supported, in whole or in part, by National Institutes of Health RO1 Grants DK78775 (to J. V.), DK090242 (to E. S. G.), DK065149 (to D. S.), and CA140624-05 (to E. V. P.).
- TCA
- tricarboxylic acid
- Oxphos
- oxidative phosphorylation
- FAO
- fatty acid β-oxidation
- LCFA
- long-chain fatty acid
- MCFA
- medium-chain fatty acid
- PDH
- pyruvate dehydrogenase
- ETF
- electron transfer flavoprotein
- AMPK
- AMP-dependent protein kinase
- AceCS2
- acetyl-CoA synthase 2
- ACADVL
- very long-chain acyl-CoA dehydrogenase
- ACADM
- medium-chain acyl-CoA dehydrogenase
- PDK1
- pyruvate dehydrogenase kinase 1
- PKM1
- pyruvate kinase isozyme M1
- PDP2
- pyruvate dehydrogenase phosphatase 2
- ACC1
- acetyl-CoA carboxylase 1.
REFERENCES
- 1. Vander Heiden M. G., Lunt S. Y., Dayton T. L., Fiske B. P., Israelsen W. J., Mattaini K. R., Vokes N. I., Stephanopoulos G., Cantley L. C., Metallo C. M., Locasale J. W. (2011) Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol. 76, 325–334 [DOI] [PubMed] [Google Scholar]
- 2. Ward P. S., Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dang C. V. (2009) MYC, microRNAs and glutamine addiction in cancers. Cell Cycle 8, 3243–3245 [DOI] [PubMed] [Google Scholar]
- 4. Dang C. V. (2010) Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 70, 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodríguez-Enríquez S., Gallardo-Pérez J. C., Marín-Hernández A., Aguilar-Ponce J. L., Mandujano-Tinoco E. A., Meneses A., Moreno-Sánchez R. (2011) Oxidative phosphorylation as a target to arrest malignant neoplasias. Curr. Med. Chem. 18, 3156–3167 [DOI] [PubMed] [Google Scholar]
- 6. Jose C., Bellance N., Rossignol R. (2011) Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim. Biophys. Acta 1807, 552–561 [DOI] [PubMed] [Google Scholar]
- 7. Mateyak M. K., Obaya A. J., Adachi S., Sedivy J. M. (1997) Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8, 1039–1048 [PubMed] [Google Scholar]
- 8. Graves J. A., Wang Y., Sims-Lucas S., Cherok E., Rothermund K., Branca M. F., Elster J., Beer-Stolz D., Van Houten B., Vockley J., Prochownik E. V. (2012) Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One 7, e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang R., Dillon C. P., Shi L. Z., Milasta S., Carter R., Finkelstein D., McCormick L. L., Fitzgerald P., Chi H., Munger J., Green D. R. (2011) The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hatzivassiliou G., Zhao F., Bauer D. E., Andreadis C., Shaw A. N., Dhanak D., Hingorani S. R., Tuveson D. A., Thompson C. B. (2005) ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8, 311–321 [DOI] [PubMed] [Google Scholar]
- 11. Sato R., Takano T. (1995) Regulation of intracellular cholesterol metabolism. Cell Struct. Funct. 20, 421–427 [DOI] [PubMed] [Google Scholar]
- 12. Wang H., Mannava S., Grachtchouk V., Zhuang D., Soengas M. S., Gudkov A. V., Prochownik E. V., Nikiforov M. A. (2008) c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene 27, 1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rardin M. J., He W., Nishida Y., Newman J. C., Carrico C., Danielson S. R., Guo A., Gut P., Sahu A. K., Li B., Uppala R., Fitch M., Riiff T., Zhu L., Zhou J., Mulhern D., Stevens R. D., Ilkayeva O. R., Newgard C. B., Jacobson M. P., Hellerstein M., Goetzman E. S., Gibson B. W., Verdin E. (2013) SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delmastro-Greenwood M. M., Votyakova T., Goetzman E., Marre M. L., Previte D. M., Tovmasyan A., Batinic-Haberle I., Trucco M. M., Piganelli J. D. (2013) Mn porphyrin regulation of aerobic glycolysis: implications on the activation of diabetogenic immune cells. Antioxid. Redox Signal. 19, 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vockley J., Mohsen al-W. A., Binzak B., Willard J., Fauq A. (2000) Mammalian branched-chain acyl-CoA dehydrogenases: molecular cloning and characterization of recombinant enzymes. Methods Enzymol. 324, 241–258 [DOI] [PubMed] [Google Scholar]
- 16. Gocze P. M., Freeman D. A. (1994) Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry 17, 151–158 [DOI] [PubMed] [Google Scholar]
- 17. Goetzman E. S. (2011) Modeling disorders of fatty acid metabolism in the mouse. Prog Mol. Biol. Transl. Sci. 100, 389–417 [DOI] [PubMed] [Google Scholar]
- 18. Papamandjaris A. A., MacDougall D. E., Jones P. J. (1998) Medium chain fatty acid metabolism and energy expenditure: obesity treatment implications. Life Sci. 62, 1203–1215 [DOI] [PubMed] [Google Scholar]
- 19. Gregersen N., Andresen B. S., Corydon M. J., Corydon T. J., Olsen R. K., Bolund L., Bross P. (2001) Mutation analysis in mitochondrial fatty acid oxidation defects: exemplified by acyl-CoA dehydrogenase deficiencies, with special focus on genotype-phenotype relationship. Hum. Mutat. 18, 169–189 [DOI] [PubMed] [Google Scholar]
- 20. Zirath H., Frenzel A., Oliynyk G., Segerström L., Westermark U. K., Larsson K., Munksgaard Persson M., Hultenby K., Lehtiö J., Einvik C., Påhlman S., Kogner P., Jakobsson P. J., Henriksson M. A. (2013) MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10258–10263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yin X., Giap C., Lazo J. S., Prochownik E. V. (2003) Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22, 6151–6159 [DOI] [PubMed] [Google Scholar]
- 22. Patel M. S., Korotchkina L. G. (2006) Regulation of the pyruvate dehydrogenase complex. Biochem. Soc. Trans. 34, 217–222 [DOI] [PubMed] [Google Scholar]
- 23. Wallace J. C., Jitrapakdee S., Chapman-Smith A. (1998) Pyruvate carboxylase. Int. J. Biochem. Cell Biol. 30, 1–5 [DOI] [PubMed] [Google Scholar]
- 24. Gupta V., Bamezai R. N. (2010) Human pyruvate kinase M2: a multifunctional protein. Protein Sci. 19, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tamada M., Suematsu M., Saya H. (2012) Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin. Cancer Res. 18, 5554–5561 [DOI] [PubMed] [Google Scholar]
- 26. Chaneton B., Gottlieb E. (2012) Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem. Sci. 37, 309–316 [DOI] [PubMed] [Google Scholar]
- 27. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barber M. C., Price N. T., Travers M. T. (2005) Structure and regulation of acetyl-CoA carboxylase genes of metazoa. Biochim. Biophys. Acta 1733, 1–28 [DOI] [PubMed] [Google Scholar]
- 29. Athenstaedt K., Daum G. (2006) The life cycle of neutral lipids: synthesis, storage and degradation. Cell Mol. Life Sci. 63, 1355–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujimoto T., Parton R. G. (2011) Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3, a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hardie D. G., Ross F. A., Hawley S. A. (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fullerton M. D., Galic S., Marcinko K., Sikkema S., Pulinilkunnil T., Chen Z. P., O'Neill H. M., Ford R. J., Palanivel R., O'Brien M., Hardie D. G., Macaulay S. L., Schertzer J. D., Dyck J. R., van Denderen B. J., Kemp B. E., Steinberg G. R. (2013) Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 19, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aguer C., Foretz M., Lantier L., Hebrard S., Viollet B., Mercier J., Kitzmann M. (2011) Increased FAT/CD36 cycling and lipid accumulation in myotubes derived from obese type 2 diabetic patients. PLoS One 6, e28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holloway G. P., Luiken J. J., Glatz J. F., Spriet L. L., Bonen A. (2008) Contribution of FAT/CD36 to the regulation of skeletal muscle fatty acid oxidation: an overview. Acta Physiol. (Oxf.) 194, 293–309 [DOI] [PubMed] [Google Scholar]
- 35. Cuthbert K. D., Dyck J. R. (2005) Malonyl-CoA decarboxylase is a major regulator of myocardial fatty acid oxidation. Curr. Hypertens. Rep. 7, 407–411 [DOI] [PubMed] [Google Scholar]
- 36. Sugden M. C., Holness M. J. (2006) Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Arch. Physiol. Biochem. 112, 139–149 [DOI] [PubMed] [Google Scholar]
- 37. Kolobova E., Tuganova A., Boulatnikov I., Popov K. M. (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 358, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaplon J., Zheng L., Meissl K., Chaneton B., Selivanov V. A., Mackay G., van der Burg S. H., Verdegaal E. M., Cascante M., Shlomi T., Gottlieb E., Peeper D. S. (2013) A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 [DOI] [PubMed] [Google Scholar]
- 39. Pettit F. H., Pelley J. W., Reed L. J. (1975) Regulation of pyruvate dehydrogenase kinase and phosphatase by acetyl-CoA/CoA and NADH/NAD ratios. Biochem. Biophys. Res. Commun. 65, 575–582 [DOI] [PubMed] [Google Scholar]
- 40. Shimazu T., Hirschey M. D., Huang J. Y., Ho L. T., Verdin E. (2010) Acetate metabolism and aging: An emerging connection. Mech Ageing Dev. 131, 511–516 [DOI] [PubMed] [Google Scholar]
- 41. Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. (2001) Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 276, 11420–11426 [DOI] [PubMed] [Google Scholar]
- 42. Ziegler D. M. (1985) Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu. Rev. Biochem. 54, 305–329 [DOI] [PubMed] [Google Scholar]
- 43. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li F., Wang Y., Zeller K. I., Potter J. J., Wonsey D. R., O'Donnell K. A., Kim J. W., Yustein J. T., Lee L. A., Dang C. V. (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25, 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. (2003) Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Connell B. C., Cheung A. F., Simkevich C. P., Tam W., Ren X., Mateyak M. K., Sedivy J. M. (2003) A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278, 12563–12573 [DOI] [PubMed] [Google Scholar]
- 47. Faubert B., Boily G., Izreig S., Griss T., Samborska B., Dong Z., Dupuy F., Chambers C., Fuerth B. J., Viollet B., Mamer O. A., Avizonis D., DeBerardinis R. J., Siegel P. M., Jones R. G. (2013) AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 17, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faergeman N. J., Knudsen J. (1997) Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 323, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jo Y., Debose-Boyd R. A. (2010) Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mazurek S., Grimm H., Boschek C. B., Vaupel P., Eigenbrodt E. (2002) Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br. J. Nutr. 87, Suppl. 1, S23–S29 [PubMed] [Google Scholar]