Abstract
Gene expression in tree shrew choroid was examined during the development of minus-lens induced myopia (LIM, a GO condition), after completion of minus-lens compensation (a STAY condition), and early in recovery (REC) from induced myopia (a STOP condition). Five groups of tree shrews (n = 7 per group) were used. Starting 24 days after normal eye-opening (days of visual experience [DVE]), one minus-lens group wore a monocular −5 D lens for 2 days (LIM-2), another minus-lens group achieved stable lens compensation while wearing a monocular −5 D lens for 11 days (LIM-11); a recovery group also wore a −5D lens for 11 days and then received 2 days of recovery starting at 35 DVE (REC-2). Two age-matched normal groups were examined at 26 DVE and 37 DVE. Quantitative PCR was used to measure the relative differences in mRNA levels in the choroid for 77 candidate genes that were selected based on previous studies or because a whole-transcriptome analysis suggested their expression would change during myopia development or recovery. Small myopic changes were observed in the treated eyes of the LIM-2 group (−1.0 ± 0.2 D; mean ± SEM) indicating eyes were early in the process of developing LIM. The LIM-11 group exhibited complete refractive compensation (−5.1 ± 0.2 D) that was stable for five days. The REC-2 group recovered by 1.3 ± 0.3 D from full refractive compensation. Sixty genes showed significant mRNA expression differences during normal development, LIM, or REC conditions. In LIM-2 choroid (GO), 18 genes were significantly down-regulated in the treated eyes relative to the fellow control eyes and 10 genes were significantly up-regulated. In LIM-11 choroid (STAY), 10 genes were significantly down-regulated and 12 genes were significantly up-regulated. Expression patterns in GO and STAY were similar, but not identical. All genes that showed differential expression in GO and STAY were regulated in the same direction in both conditions. In REC-2 choroid (STOP), 4 genes were significantly down-regulated and 18 genes were significantly up-regulated. Thirteen genes showed bi-directional regulation in GO vs. STOP. The pattern of differential gene expression in STOP was very different from that in GO or in STAY. Significant regulation was observed in genes involved in signaling as well as extracellular matrix turnover. These data support an active role for the choroid in the signaling cascade from retina to sclera. Distinctly different treated eye vs. control eye mRNA signatures are present in the choroid in the GO, STAY, and STOP conditions. The STAY signature, present after full compensation has occurred and the GO visual stimulus is no longer present, may participate in maintaining an elongated globe. The 13 genes with bi-directional expression differences in GO and STOP responded in a sign of defocus-dependent manner. Taken together, these data further suggest that a network of choroidal gene expression changes generate the signal that alters scleral fibroblast gene expression and axial elongation rate.
Keywords: myopia, animal models, refractive error, emmetropization, axial elongation, gene expression, choroid
1. INTRODUCTION
Studies of postnatal refractive development in both children and in animal models have found that there is a visually-guided emmetropization mechanism that uses refractive error to guide the growth of the eye so that the axial length eventually matches the location of the focal plane, producing visual images that are focused on the photoreceptors (emmetropia) (Mutti et al., 2005; Norton, 1999; Wallman & Winawer, 2004). In animal models, the emmetropization mechanism can be manipulated with lenses, held in front of one (or both) eyes in a goggle frame. Minus-power (negative) lenses shift the focal plane away from the cornea, making the eye hyperopic. This produces retinal GO signals that cause an increase in the axial (vitreous chamber) elongation rate, moving the retina to the shifted focal plane and restoring emmetropia while the lens is in place (Irving et al., 1991; Irving et al., 1995; Norton et al., 2010). In tree shrews, the eye remains elongated as long as the lens is left in place, evidently because some form of STAY signal reaches the sclera (Norton et al., 2010). When the lens is removed, the increased axial length causes the eye to experience lens-induced myopia (LIM). The retina then generates STOP signals that, in juvenile animals where the eyes are still growing, rapidly slow the axial elongation rate to below normal, producing recovery (REC) from the induced myopia (Norton et al., 2010).
Although the emmetropization mechanism performs more effectively in a fully intact animal, eyes can still respond to myopiagenic stimuli if the optic nerve is cut (Troilo, 1990; Wildsoet & McFadden, 2010) or output is functionally blocked with tetrodotoxin (Norton et al., 1994). In addition, covering only half of the visual field with a minus lens produces elongation and myopia only in the affected visual field (Diether & Schaeffel, 1997; Norton & Siegwart, 1991; Smith, III et al., 2010). Thus, there is a direct, spatially-localized signaling cascade from the retina to the sclera that must pass through the retinal pigment epithelium (RPE) and choroid. The choroid, in addition to being the vascular supply to the photoreceptors, RPE, and sclera (Birol et al., 2007; Lutty et al., 2010; Oyster, 1999), plays an important role in the emmetropization mechanism (Nickla & Wallman, 2010; Summers, 2013; Wallman & Winawer, 2004) as a way-station in the signaling cascade that conveys the GO, STAY, and STOP signals to the sclera. The extent to which some signaling molecules generated by the retina or RPE may simply pass through the choroid on the way to the sclera and other, new, signaling molecules are generated by the choroid is not known. Examining gene expression changes in the choroid alone, relative to changes in the retina and RPE, may help clarify the situation.
In species such as chick, the choroid is thick because there are no blood vessels on the vitreal side of the retina and the choroid is the sole retinal vascular supply. Chick choroid displays rapid changes within a few hours after the onset of LIM and REC, thinning during myopia development and thickening during recovery (Wallman et al., 1995). Previous studies of chick choroid have reported changes in gene expression during the development of LIM, form deprivation-induced myopia (FDM), and recovery from FDM, suggesting the choroid plays an active role producing new signaling molecules (Mertz & Wallman, 2000; Nickla et al., 2009; Nickla & Wildsoet, 2004; Nickla et al., 2006; Rada et al., 2012; Rada et al., 2001; Rada & Wiechmann, 2009; Rada et al., 2010; Simon et al., 2004). In mammals (guinea pig, tree shrew, marmoset, and macaque), the choroid is not as thick, proportionally, as in chicks and seems to undergo smaller changes in thickness during myopia development and recovery (Gentle & McBrien, 1999; Howlett & McFadden, 2009; Hung et al., 2000; Siegwart, Jr. & Norton, 1998; Troilo et al., 2000).
A previous study in the marmoset examined gene expression in the combined RPE and choroid (Shelton et al., 2008). While a number of changes were found, it is not possible to know if the changes occurred in the RPE or choroid. In tree shrews (mammals closely related to primates; Luckett, 1980) and guinea pigs, studies have examined changes of a few genes and proteins in the choroid in form-deprived animals (Cui et al., 2010; Jobling et al., 2009; Liu et al., 2007; McBrien et al., 2009). In the present study, we examined, in the choroid, changes in gene expression of a large sample of genes in animals treated with a minus lens.
During the development of LIM in tree shrews, the GO signals from the choroid produce remodeling of the scleral extracellular matrix that increases the viscoelasticity of the sclera (measured as increased creep rate), allowing normal intraocular pressure to expand the globe (Phillips et al., 2000; Siegwart, Jr. & Norton, 1999). With continued lens wear, STAY signals from the choroid must be present because tree shrew eyes remain in an elongated state until minus lens-wear is discontinued (Norton et al., 2010). During REC, the STOP signals from the choroid cause a rapid reduction the creep rate that slows the axial elongation rate (Siegwart, Jr. & Norton, 1999).
The time-course of remodeling in the tree shrew sclera is rapid, but not instantaneous. After one day of LIM, there is little alteration of mRNA levels (Gao et al., 2011). After 2 days of LIM, scleral gene expression is altered; a scleral GO signature is found (Guo et al., 2013) and creep rate is elevated (Siegwart, Jr. & Norton, 1999). Very similar, somewhat stronger gene expression changes are found after four days of LIM (Frost & Norton, 2012; Guo et al., 2013) and scleral creep rate reaches a peak at this time (Siegwart, Jr. & Norton, 1999). After 11 days of LIM, the eyes have fully compensated for the minus lens but scleral viscoelasticity remains slightly elevated (Siegwart, Jr. & Norton, 1999) and some gene expression differences remain in the sclera (Guo, personal communication, 2013) suggesting the presence of STAY signals in the choroid at this time point. During recovery, the scleral gene expression after one day is little changed (Guo et al., 2012); after two days of recovery, a scleral STOP remodeling response has developed (Guo, personal communication, 2013) and scleral creep rate has dropped to, or below, normal (Siegwart, Jr. & Norton, 1999). Based on this time-course, it is expected that GO and STOP signals should be detectable in the choroid after two days of LIM and after two days of REC respectively.
The goal of the present study was to examine alterations in gene expression in the choroid, measured as alterations in mRNA levels, after two days of LIM (GO), 11 days of LIM (STAY), and after two days of REC (STOP). Although changes in levels of proteins or other molecules presumably are key to actually transmitting signals from choroid to sclera, it has been found that changes in mRNA can help to identify the responses of the cells in tissues and are useful in identifying pathways of interest (Gao et al., 2011; Guo et al., 2013; He et al., 2011; Schippert et al., 2006; Shelton et al., 2008; Siegwart, Jr. & Norton, 2005; Stone et al., 2011; Zhang et al., 2012). Based on these previous studies, our hypothesis was not only that many of the genes examined would show changes in mRNA expression but also that the pattern of differential gene expression would differ in the GO, STAY, and STOP conditions.
2. MATERIALS AND METHODS
2.1. Experimental Groups
The juvenile tree shrews (Tupaia glis belangeri) used in this study were produced in our breeding colony and raised by their mothers on a 14 hr light/10 hr dark cycle. Tree shrew pups open their eyes about three weeks after birth. The day both eyes are open is the first day of visual experience (DVE). All procedures complied with the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research and were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. Experimental groups were balanced to include both males and females, and avoided pups from the same parents wherever possible.
Five groups of animals (n = 7 per group) were used in this study (Fig. 1). Starting at 24 ± 1 DVE, a minus-lens wear group (LIM-2) wore a monocular −5 D (spherical power) lens for 2 days; the animals in this group also provided scleral mRNA for another study (Guo et al., 2013). A second minus-lens group (LIM-11) wore a monocular −5 D lens for 11 days and fully compensated for the lens. A recovery group (REC-2) recovered for 2 days starting at 35 ± 1 DVE, after 11 days of minus-lens wear that produced full compensation to a −5 D lens. In the LIM and REC groups, the untreated fellow eye served as a control. Two age-matched (26 DVE and 37 DVE) normal groups were also studied.
Figure 1.
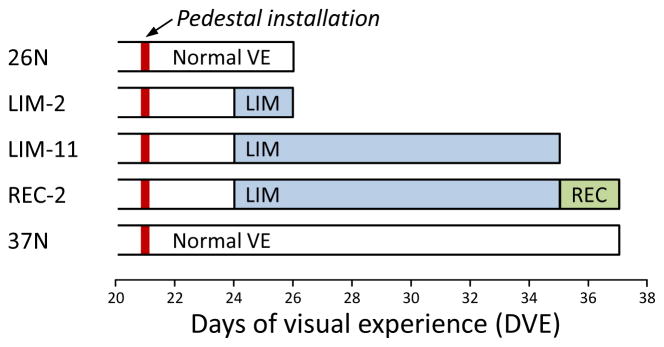
Experimental groups and duration of treatments. The red vertical bar indicates the point when a dental acrylic pedestal was installed under anesthesia. Filled regions indicate the type and duration of visual treatment. The right end of each bar indicates the time point when mRNA levels were measured.
2.2. Lens Treatment
Animals in all groups were anesthetized (17.5 mg ketamine, 1.2 mg xylazine; supplemented with 0.5 – 2.0% isoflurane as needed) and received a dental acrylic pedestal (Siegwart & Norton, 1994) at 21 ± 1 DVE (Fig. 1). After pedestal installation, all animals were placed in individual cages with standard colony fluorescent lighting, 100 – 300 lux on the floor of the cage. Three days later, in the LIM and REC groups, a goggle frame holding a −5 D lens (12 mm diameter PMMA contact lens; Conforma Contact Lenses, Norfolk, VA) was clipped to the pedestal, firmly holding the lens in front of the randomly selected treated eye. The untreated fellow control eye had unrestricted vision through an open goggle frame. Twice daily (approximately 9:30 AM and 4:30 PM), the goggles were briefly (< 3 min) removed to clean the lens under dim illumination. During goggle cleaning, animals were kept in a darkened nest box to minimize exposure to visual stimuli. For the REC group, after 11 days of minus lens compensation, the goggle was removed, and the treated eye was allowed to recover for 2 days. The normal groups received a pedestal but did not wear a goggle.
2.3. Refractive and Axial Measures
Non-cycloplegic refractive measures were made daily, in awake animals, throughout the treatment period with a Nidek ARK-700A infrared autorefractor (Marco Ophthalmic, Jacksonville, FL) (Norton et al., 2003). Normal animals were measured just before euthanasia. Cycloplegic refractive measures were omitted to prevent any interference by atropine on retino-scleral signaling (McKanna & Casagrande, 1981). However, previous studies have shown that non-cycloplegic measures provide a valid estimate of the refractive state and of induced myopia in tree shrews (Norton et al., 2006b; Norton et al., 2003). All refractive values were corrected for the small eye artifact (Glickstein & Millodot, 1970), previously shown to be approximately +4 D in tree shrews (Norton et al., 2003).
At the time the pedestal was attached, ocular component dimensions were measured whilst under anesthesia with A-scan ultrasound (Norton & McBrien, 1992) to ensure that the two eyes did not differ significantly in axial length before treatment began. Post-treatment A-scan measures were not made to eliminate any possibility that the time spent under anesthesia, required for the A-scan procedure, might alter gene expression. In all groups, post-treatment axial component measures were made with a Lenstar LS-900 optical biometer (Haag-Streit USA, Mason, OH). This instrument allowed measures to be quickly made, in awake animals, before euthanasia. Comparison of A-scan and Lenstar measures of the vitreous chamber in 32 animals in this laboratory, with between −1 D and −12 D of induced myopia, showed that the axial differences measured with the Lenstar were very similar to those measured with A-scan ultrasound (data not shown).
2.4. Choroid Dissection
On completion of the final refractive measures, approximately 2 - 4 hours into the light phase, animals were terminally anesthetized (17.5 mg ketamine and 1.2 mg xylazine, followed by 50 mg xylazine); both eyes were enucleated and placed into RNAlater solution (Life Technologies, Carlsbad, CA). Extraocular muscles, conjunctiva, and orbital fat were trimmed from the exterior surface of the eye and the cornea dissected away just behind the corneoscleral junction. While viewing through a surgical microscope, the lens and vitreous humor were removed; the retina and RPE, which were tightly bound to each other (Malik et al., 2003; Wang et al., 2012), were then lifted from the eyecup. While still immersed in RNAlater, choroid was teased from the scleral inner surface by gentle separation using the rounded ends of forceps, collected, and then frozen in liquid nitrogen. Because the dissection was extremely gentle, it is possible that small pieces of the lamina fusca, the outermost layer of the choroid, may have adhered to the sclera in some cases and, thus, not been included in our choroidal sample. Because the retina/RPE separated cleanly from the choroid without dissection, and because the inner surface of the sclera was not disrupted by forceful scraping of the surface (and most scleral fibroblasts are within the layered matrix, not on the inner surface), there is no reason to expect significant contamination of the choroidal samples from either retina/RPE or sclera.
2.5. Gene Expression Analysis
Each frozen choroid was homogenized with a disposable pellet pestle (Fisher Scientific, Pittsburgh, PA) from which total RNA was isolated using a RiboPure kit (Life Technologies) according to the manufacturer’s instructions, with the addition of an on-filter DNase treatment. The purified RNA was quantified (NanoDrop Technologies, Wilmington, DE), with an average yield per choroid of 5.0 ± 1.3 μg (mean ± SD). RNA quality was confirmed by denaturing gel electrophoresis (RNA FlashGel; Lonza, Rockland, ME). cDNA was synthesized from 1 μg of total RNA in a final reaction volume of 20 μL using a Superscript III RT kit (Life Technologies) with minor modifications (2.5 μM anchored oligo (dT)20 primers and DTT omitted). The resultant cDNA was diluted 5-fold and stored at −20°C until use.
Tree shrew-specific quantitative PCR (qPCR) primers were designed for 77 genes of interest (Table 1) and the reference gene RNA polymerase II (POLR2A) using Beacon Designer 7 (Premier Biosoft International, Palo Alto, CA). None of the treatment conditions affected the expression of the reference gene. Fold differences (right eye vs. left eye or treated vs. control; mean ± SEM) for a typical qPCR run were: 26N, 1.11 ± 0.07; LIM-2, 1.07 ± 0.08; LIM-11, −1.01± 0.04; REC-2, 1.10 ±0.08; 37N, 1.03 ± 0.14. Primer sequences, amplicon size, and efficiencies are listed in Supplementary Table S1. The selected candidate genes included representatives of three major groupings: signaling, metallopeptidases & TIMPs, and extracellular matrix (ECM) proteins. They were selected from genes that were found to change in preliminary studies of tree shrew choroid during LIM-2 along with additional genes that were suggested by a whole-transcriptome analysis of three of the LIM-2 animals. All primers were designed to work under the same cycling conditions. All amplicons were located within the coding region and most spanned at least one intron; amplicon identity was verified by gel electrophoresis and sequencing.
Table 1.
Genes examined by functional category, with cellular location of the protein encoded by the gene and its UniProt accession ID
| Gene symbol |
Protein name |
Location |
UniProt ID |
|---|---|---|---|
| Signaling – Cell surface | |||
| ADORA2A | Adenosine receptor A2a | Cell surface | P29274 |
| AQP4 | Aquaporin 4 | Cell surface | P55087 |
| CHRNA7 | Cholinergic receptor, nicotinic α7 | Cell surface | P36544 |
| DRD2 | Dopamine receptor D2 | Cell surface | P14416 |
| EPHA1 | EPH receptor A1 | Cell surface | P21709 |
| FGFR1 | FGF receptor 1 | Cell surface | P11362 |
| GFRA1 | GDNF family receptor α1 | Cell surface | P56159 |
| GRM5 | Metabotropic glutamate receptor 5 | Cell surface | P41594 |
| IGF2R | Insulin-like growth factor 2 receptor | Cell surface | P11717 |
| INSR | Insulin receptor | Cell surface | P06213 |
| OPN1LW | Opsin 1, long-wave-sensitive | Cell surface | P04000 |
| P2RY1 | Purinergic receptor P2Y, G-protein coupled, 1 | Cell surface | P47900 |
| SCUBE3 | Signal peptide, CUB and EGF-like domain-containing protein 3 | Cell surface | Q8IX30 |
| TNMD | Tenomodulin | Cell surface | Q9H2S6 |
| VIPR1 | VIP receptor 1 | Cell surface | P32241 |
| VIPR2 | VIP receptor 2 | Cell surface | P41587 |
| Signaling – Intracellular | |||
| BCO2 | Beta-carotene oxygenase 2 | Intracellular | Q9BYV7 |
| CABP5 | Calcium binding protein 5 | Intracellular | Q9NP86 |
| CAMP | Cathelicidin antimicrobial peptide | Intracellular | P49913 |
| CDC42 | Cell division cycle 42 | Intracellular | P60953 |
| CHAT | Choline O-acetyltransferase | Intracellular | P28329 |
| CYP26B1 | Cytochrome P450 26B1 | Intracellular | Q9NR63 |
| NOS1 | Nitric oxide synthase 1 | Intracellular | P29475 |
| RASGRF1 | Ras-specific guanine nucleotide-releasing factor 1 | Intracellular | Q13972 |
| RLBP1 | Retinaldehyde binding protein 1 | Intracellular | P12271 |
| RPE65 | Retinoid isomerohydrolase | Intracellular | Q16518 |
| S100A12 | Protein S100-A12 | Intracellular | P80511 |
| ZNF185 | Zinc finger protein 185 | Intracellular | O15231 |
| Signaling – Transcription regulators | |||
| EGR1 | Early growth response protein 1 | Intracellular | P18146 |
| HIF1A | Hypoxia-inducible factor 1α | Intracellular | Q16665 |
| PER2 | Period circadian clock 2 | Intracellular | O15055 |
| RXRB | Retinoid X receptor β | Intracellular | P28702 |
| VDR | Vitamin D receptor | Intracellular | P11473 |
| Signaling – Secreted | |||
| ANGPTL7 | Angiopoietin-related protein 7 | Extracellular | O43827 |
| APOE | Apolipoprotein E | Extracellular | P02649 |
| BMP2 | Bone morphogenetic protein 2 | Extracellular | P12643 |
| BMP4 | Bone morphogenetic protein 4 | Extracellular | P12644 |
| CILP | Cartilage intermediate layer protein 1 | Extracellular | O75339 |
| EGF | Epidermal growth factor | Extracellular | P01133 |
| FAM180A | Family with sequence similarity 180, member A | Extracellular | Q6UWF9 |
| IGF2 | Insulin-like growth factor 2 | Extracellular | P01344 |
| IL1B | Interleukin 1β | Extracellular | P01584 |
| LTBP1 | Latent TGFβ binding protein 1 | Extracellular | Q14766 |
| LTF | Lactotransferrin | Extracellular | P02788 |
| MEST | Mesoderm specific transcript | Extracellular | Q5EB52 |
| NRG1 | Neuregulin 1 | Extracellular | Q02297 |
| NTS | Neurotensin | Extracellular | P30990 |
| PENK | Proenkephalin A | Extracellular | P01210 |
| PI15 | Peptidase inhibitor 15 | Extracellular | O43692 |
| PTX3 | Pentraxin 3 | Extracellular | P26022 |
| SOSTDC1 | Sclerostin domain-containing protein 1 | Extracellular | Q6X4U4 |
| SST | Somatostatin | Extracellular | P61278 |
| TAC1 | Protachykinin 1 | Extracellular | P20366 |
| TGFB2 | Transforming growth factor β2 | Extracellular | P61812 |
| TGFB3 | Transforming growth factor β3 | Extracellular | P10600 |
| TGFBI | TGFβ-induced protein | Extracellular | Q15582 |
| VIP | Vasoactive intestinal peptide | Extracellular | P01282 |
| Signaling – Matricellular | |||
| CYR61 | Protein CYR61 | Extracellular | O00622 |
| NOV | Nephroblastoma overexpressed gene | Extracellular | P48745 |
| THBS1 | Thrombospondin 1 | Extracellular | P07996 |
| THBS2 | Thrombospondin 2 | Extracellular | P35442 |
| TNC | Tenascin C | Extracellular | P24821 |
|
| |||
| MPs/TIMPs | |||
| ADAMTS4 | ADAM metallopeptidase with thrombospondin motif, 4 | Extracellular | O75173 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin motif, 5 | Extracellular | Q9UNA0 |
| ADAMTSL3 | ADAMTS-like 3 | Extracellular | P82987 |
| MMP14 | Matrix metallopeptidase 14 | Cell surface | P50281 |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | Extracellular | P16035 |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | Extracellular | P35625 |
|
| |||
| Extracellular matrix | |||
| COL12A1 | Collagen type XII, α1 | Extracellular | Q99715 |
| COL6A6 | Collagen type VI, α6 | Extracellular | A6NMZ7 |
| DCN | Decorin | Extracellular | P07585 |
| FMOD | Fibromodulin | Extracellular | Q06828 |
| MXRA5 | Matrix remodeling associated protein 5 | Extracellular | Q9NR99 |
| NYX | Nyctalopin | Extracellular | Q9GZU5 |
| OGN | Mimecan | Extracellular | P20774 |
| PRELP | Prolargin | Extracellular | P51888 |
| SERPINH1 | Serpin H1 | Intracellular | P50454 |
Relative gene expression was measured by qPCR on a StepOnePlus Real-Time PCR System using Power SYBR Green PCR Master Mix (both, Life Technologies). Reactions were performed in triplicate in a 15 μL volume containing 300 nM each primer and 0.4 μL cDNA template. Cycling parameters were the same for all assays: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 62°C for 60 sec. Single gene products were obtained for all reactions as assessed by melt curve analysis. Relative gene expression was calculated using the ΔΔCt method (Livak & Schmittgen, 2001) to first normalize the expression level of the target gene to that of the reference gene, and then to compare the relative expression of the target gene for treated vs. control eyes, treated vs. normal eyes, and control vs. normal eyes. The geometric group mean (for the 7 biological replicates) of these expression ratios was used to calculate the fold change in gene expression for each of the target genes.
2.6. Statistical Analysis
One-way analysis of variance (ANOVA; Statistica, Statsoft, Tulsa, OK) was used to compare control and normal eye refractive data across groups of animals; paired t-tests were used to determine if significant myopia (treated eye vs. control eye) or recovery had occurred. For gene expression data, paired t-tests were used to assess treated-eye vs. control-eye differences; unpaired t-tests were used to test for gene expression differences between all independent groups. In all cases, p<0.05 was considered significant and no adjustment was applied for possible false discovery rate. Linear regressions between expression differences were made in SigmaPlot (Systat Software, San Jose, CA).
3. RESULTS
3.1. Refraction
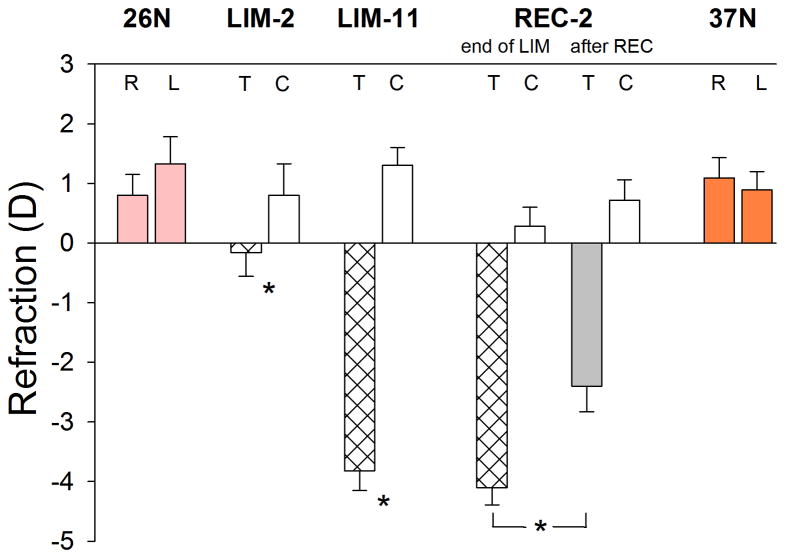
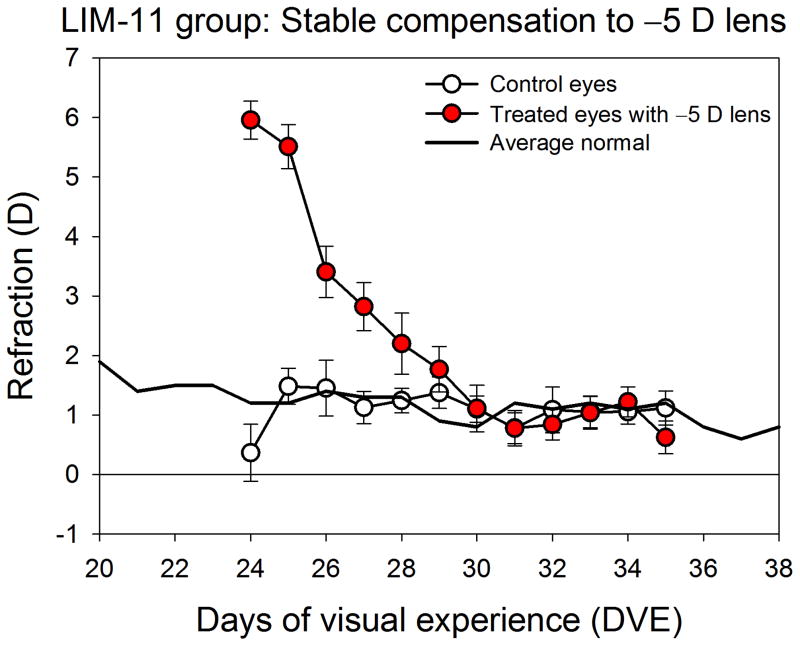
The final refractive values of the normal, minus-lens treated, control, and recovering eyes in the five groups are shown in Fig. 2. As expected in tree shrews at 26 DVE, both eyes of the normal group were slightly hyperopic (right eyes, 0.8 ± 0.4 D; left eyes, 1.3 ± 0.4 D; mean ± SEM). At 37 DVE both eyes of the normal group also were slightly hyperopic (right eyes, 1.1 ± 0.3 D; left eyes, 0.9 ± 0.3 D; mean ± SEM) as expected in juvenile animals of this age because the emmetropization process is nearly complete (Norton et al., 2006a). After two days of treatment, the LIM-2 treated eyes showed a small, statistically-significant myopic shift; the treated eyes were −1.0 ± 0.2 D myopic in comparison to the control eyes. After 11 days of treatment, the treated eyes in the LIM-11 group achieved complete compensation (−5.1 ± 0.2 D, treated eyes − control eyes). Fig. 3 shows the time-course of the compensation for the minus lens. As a group, refractions in the treated eyes, while wearing the lens, achieved and maintained a refractive match with their fellow control eyes for 5 days prior to the final measurements. The REC-2 group also had achieved complete refractive compensation (Fig. 2) at the end of lens wear. After 2 days of recovery from lens compensation, the treated eyes recovered by 1.3 ± 0.3 D. The control eyes in the LIM-2, LIM-11, and REC-2 groups did not differ significantly from the 26 DVE and 37 DVE normal eyes (one-way ANOVA, p = 0.56). Ocular component dimensions, measured with the Lenstar in the LIM-2 and LIM-11 groups confirmed that the vitreous chamber of the treated eyes had elongated, relative to the control eyes, by 0.016 ± 0.004 mm (LIM-2) and by 0.11 ± 0.01 mm (LIM-11). In the LIM-11 group the choroid thickness, measured with the Lenstar, was slightly, but significantly, thinner (0.058 ± 0.002 vs. 0.064 ± 0.004 mm) than in the control eyes.
Figure 2.
End-of-treatment refractive measures for the normal, minus lens (LIM-2 & LIM-11), and recovery (REC) groups. Values are the mean refraction ± SEM for the right (R) & left (L) eyes of the normal and for the treated (T) & control (C) eyes of the LIM and REC groups. Treated eyes in all groups were significantly different relative to control eyes. Treated eyes in the REC-2 group showed significant recovery from the refractions measured at the end of LIM (bracket).
Figure 3.
Daily refractive measurements of the treated and control eyes of the LIM-11 group showing that the lens-induced refractive hyperopia dissipated as the eyes compensated for the −5 D lens. Measurements of the treated eyes were taken while the −5 D lens was in place, providing a measure of the refractive state experienced by the treated eyes during treatment. The low control eye value at the beginning of treatment was caused by a low refraction in a single animal, perhaps from accommodation.
3.2. Gene Expression
3.2.1. Normal animals
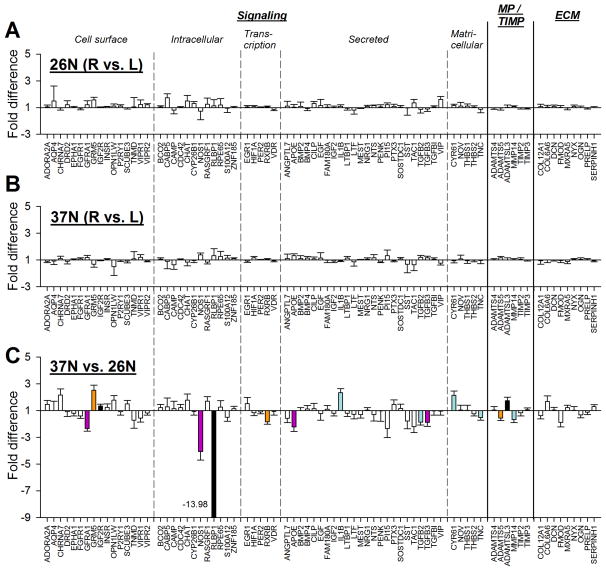
The mRNA levels in normal eyes, and the variability between normal left and right eyes, can provide a basis for comparison with the levels and variability found in treated and control eyes. Figs. 4A and 4B compare gene expression in the right and left eyes of the 26N and 37N groups. Group expression values for all 77 genes are provided in Table 2. Although considerable variability was occasionally seen for some genes between left- and right-eye mRNA levels in individual animals, for both the 26N and 37N groups, expression levels did not differ significantly between left and right eyes for any of the 77 genes.
Figure 4.
Gene expression fold differences. (A) Normal eyes (right eyes vs. left eyes) in the 26N group. (B) Normal eyes (right eyes vs. left eyes) in the 37N group. (C) Comparison of expression levels at 26N and 37N; down-regulation indicates lower expression in the 37N group. Headings separated by vertical dashed lines indicate functional grouping of the protein products of the genes. Filled bars represent statistically significant differences between the left and right eyes or between the two age groups (p < 0.05). Bar color is arbitrary and intended to help in comparing the same gene in the three different conditions. Error bars indicate SEM. In C, the off-scale fold difference for RLBP1 (−13.98) is indicated next to the bar.
Table 2.
Gene expression differences comparing treated vs. control, treated vs. normal, and control vs. normal eyes.
| 26 Normal |
37 Normal |
37N vs. 26N |
LIM-2 |
LIM-11 |
REC-2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RE vs. LE | RE vs. LE | T vs. C | T vs. N | C vs. N | T vs. C | T vs. N | C vs. N | T vs. C | T vs. N | C vs. N | ||
| Signaling – Cell surface | ||||||||||||
| ADORA2A | 1.06 | −1.10 | 1.46 | −1.24 | −1.88 | −1.52 | 1.03 | 1.39 | 1.36 | −1.03 | −2.00 | −1.94 |
| AQP4 | 1.50 | −1.08 | 1.00 | 1.46 | 1.65 | 1.12 | −1.48 | −4.20 | −2.84 | 1.93 | −1.11 | −2.14 |
| CHRNA7 | −1.03 | 1.09 | 2.15 | 1.15 | 1.42 | 1.23 | 1.19 | 1.26 | 1.06 | 1.28 | −1.57 | −2.02 |
| DRD2 | 1.21 | −1.11 | −1.09 | 1.68 | 1.70 | 1.01 | 1.19 | −1.03 | −1.22 | 1.95 | 2.17 | 1.11 |
| EPHA1 | 1.01 | 1.02 | −1.20 | −1.55 | −1.75 | −1.13 | −1.37 x | 1.07 | 1.46 | 1.77 | 2.08 | 1.18 |
| FGFR1 | −1.05 | 1.07 | −1.41 | −1.68 | −1.87 | −1.11 | −1.00 | 1.42 | 1.42 | 1.22 | 1.33 | 1.10 |
| GFRA1 | 1.22 | 1.18 | −2.34 | −1.12 | −1.18 | −1.05 | 1.15 | −1.03 | −1.18 | 1.48 | 2.39 | 1.62 |
| GRM5 | 1.57 | −1.32 | 2.51 | 1.34 | 1.50 | 1.12 | 1.29 | −2.13 | −2.74 | 2.67 | 1.02 | −2.61 |
| IGF2R | 1.01 | −1.02 | 1.33 | −1.16 | 1.01 | 1.17 | 1.05 | 1.02 | −1.03 | −1.06 | −1.63 | −1.53 |
| INSR | 1.07 | 1.05 | 1.23 | −1.49 | −1.76 | −1.18 | 1.04 | 1.16 | 1.11 | 1.01 | −1.93 | −1.96 |
| OPN1LW | 1.12 | −1.50 | 1.77 | 1.13 | 1.07 | −1.06 | 1.04 | −3.74 | −3.90 | 1.75 | 1.17 | −1.49 |
| P2RY1 | 1.12 | −1.02 | −1.08 | 1.29 | −1.24 | −1.60 | 1.22 | 1.70 | 1.39 | −1.16 | −1.53 | −1.32 |
| SCUBE3 | −1.00 | −1.09 | 1.49 | −1.62 | −1.28 | 1.27 | −1.31 x | 1.29 | 1.69 | 1.61 | 1.25 | −1.29 |
| TNMD | 1.06 | 1.07 | −1.72 | −1.27 | −2.23 | −1.76 | −1.42 | −1.06 | 1.34 | 1.37 | 1.13 | −1.21 |
| VIPR1 | 1.24 | 1.19 | −1.57 | 1.11 | 1.03 | −1.07 | −1.01 | −1.75 | −1.74 | 1.48 | 1.48 | 1.00 |
| VIPR2 | 1.22 | −1.08 | −1.18 | −1.55 | −1.63 | −1.05 | −1.08 | 1.17 | 1.25 | 1.13 | 1.22 | 1.08 |
| Signaling – Intracellular | ||||||||||||
| BCO2 | 1.06 | 1.06 | 1.21 | −1.48 | −1.33 | 1.11 | −1.26 | −1.05 | 1.20 | 1.53 | 1.70 | 1.11 |
| CABP5 | 1.74 | −1.10 | 1.28 | 2.14 | 2.14 | 1.00 | −1.01 | −1.71 | −1.70 | 3.05 | 2.62 | −1.16 |
| CAMP | −1.19 | −1.36 | 1.13 | 2.25 | 4.00 | 1.78 | 1.05 | −1.73 | −1.81 | −1.30 | 1.09 | 1.42 |
| CDC42 | 1.02 | 1.03 | 1.21 | −1.25 | −1.46 | −1.16 | 1.05 | 1.02 | −1.02 | −1.05 | −1.60 | −1.52 |
| CHAT | 1.49 | −1.17 | 1.77 | 1.22 | 1.70 | 1.39 | −1.31 | −1.61 | −1.23 | 3.37 | 1.78 | −1.89 |
| CYP26B1 | 1.31 | −1.04 | −1.06 | −4.05 | −3.28 | 1.24 | −1.60 | −1.06 | 1.51 | 1.43 | 1.11 | −1.29 |
| NOS1 | −1.27 | 1.37 | −4.07 | 1.53 | −1.62 | −2.46 | 1.03 | −1.89 | −1.96 | 3.34 | 4.35 | 1.30 |
| RASGRF1 | 1.26 | −1.17 | 1.69 | 1.06 | 1.43 | 1.34 | −1.03 | 1.08 | 1.12 | 2.20 | 1.28 | −1.72 |
| RLBP1 | 1.14 | 1.31 | −13.98 | 2.76 x | −1.22 | −3.37 | 3.79 | 1.83 | −2.07 | 3.58 | 23.43 | 6.55 |
| RPE65 | 1.20 | 1.26 | 1.26 | 2.28 | 1.59 | −1.43 | 6.36 x | 1.02 | −6.23 | 3.64 | 1.84 | −1.98 |
| S100A12 | −1.04 | 1.13 | −1.50 | 1.62 | 2.34 | 1.45 | 1.20 | 2.04 | 1.71 | −1.83 | 1.21 | 2.22 |
| ZNF185 | 1.01 | 1.09 | 1.13 | −1.35 | −1.62 | −1.20 | −1.12 | −1.03 | 1.08 | 1.37 | −1.07 | −1.47 |
| Signaling – Transcription | ||||||||||||
| EGR1 | 1.05 | −1.07 | 1.52 | −1.33 | −1.42 | −1.07 | −1.14 | −4.28 | −3.76 | −1.05 | −1.17 | −1.12 |
| HIF1A | 1.07 | 1.15 | −1.17 | 1.20 | 1.07 | −1.12 | 1.05 | 1.54 | 1.47 | 1.15 | −1.00 | −1.15 |
| PER2 | 1.02 | 1.01 | −1.10 | −1.35 | −1.36 | −1.01 | 1.06 | 1.14 | 1.07 | 1.04 | −1.06 | −1.10 |
| RXRB | 1.05 | 1.06 | −1.85 | −1.22 | −1.28 | −1.05 | 1.00 | 1.11 | 1.11 | −1.08 | 1.56 | 1.68 |
| VDR | −1.12 | −1.06 | −1.06 | −1.69 | −1.86 | −1.10 | −1.09 | 1.19 | 1.30 | −1.12 | −1.33 | −1.18 |
| Signaling – Secreted | ||||||||||||
| ANGPTL7 | 1.11 | 1.10 | −1.58 | 1.27 | 1.17 | −1.09 | 1.40 | 2.44 | 1.74 | 1.15 | 1.19 | 1.04 |
| APOE | 1.04 | 1.32 | −2.23 | 1.49 | 2.83 | 1.90 | 1.14 | 1.07 | −1.06 | 1.13 | 2.22 | 1.98 |
| BMP2 | 1.09 | 1.11 | 1.00 | 1.54 | 1.18 | −1.30 | 1.93 x | 1.10 | −1.75 | 2.16 x | 1.12 | −1.92 |
| BMP4 | −1.08 | 1.14 | 1.11 | 1.48 | 1.48 | 1.00 | 1.58 | 1.90 | 1.20 | 1.18 | −1.21 | −1.43 |
| CILP | 1.31 | 1.07 | 1.14 | −3.00 | −2.52 | 1.19 | −1.57 | 1.19 | 1.86 | 1.17 | −1.10 | −1.29 |
| EGF | 1.16 | 1.13 | −1.23 | 2.09 | 1.38 | −1.51 | 6.83 x | 1.19 | −5.73 | 4.06 | 1.98 | −2.06 |
| FAM180A | 1.03 | −1.03 | 1.09 | −1.60 | −1.58 | 1.01 | −1.22 | 1.17 | 1.43 | 1.96 | 1.27 | −1.54 |
| IGF2 | 1.01 | −1.09 | −1.20 | 1.24 | 1.44 | 1.16 | 1.21 | 1.45 | 1.20 | −1.38 | −1.14 | 1.20 |
| IL1B | 1.10 | −1.06 | 2.33 | 1.05 | 1.16 | 1.11 | −1.03 | 1.12 | 1.16 | −1.28 | −3.26 | −2.54 |
| LTBP1 | −1.05 | 1.14 | −1.21 | 1.03 | −1.15 | −1.19 | 1.08 | 1.28 | 1.19 | 1.23 | −1.02 | −1.25 |
| LTF | −1.19 | −1.15 | −1.29 | 1.36 | 2.20 | 1.62 | 1.26 | 1.07 | −1.18 | −1.24 | 1.12 | 1.39 |
| MEST | −1.02 | 1.00 | −1.30 | −1.64 | −1.55 | 1.06 | −1.45 | 1.26 | 1.83 | 1.88 | 3.03 | 1.62 |
| NRG1 | 1.09 | 1.06 | 1.03 | 1.47 | 1.55 | 1.06 | 1.54 | 2.34 | 1.52 | −1.87 | −1.36 | 1.37 |
| NTS | 1.15 | 1.16 | −1.30 | 1.41 | 1.03 | −1.36 | 1.30 | 1.66 | 1.27 | 2.36 | 2.35 | −1.00 |
| PENK | 1.06 | −1.14 | −1.02 | −1.73 | −1.90 | −1.09 | −1.57 | 1.13 | 1.77 | −1.28 | 1.05 | 1.35 |
| PI15 | 1.22 | 1.32 | −2.35 | 3.41 | 2.87 | −1.19 | 4.18 | 10.83 | 2.59 | −5.73 | −6.52 | −1.14 |
| PTX3 | 1.10 | −1.06 | 1.45 | −3.48 | −2.25 | 1.55 | −2.21 | −1.65 | 1.34 | 1.53 | 1.39 | −1.10 |
| SOSTDC1 | 1.04 | 1.07 | 1.15 | −1.70 | −2.22 | −1.31 | −1.39 | −1.07 | 1.30 | 2.06 | 1.62 | −1.27 |
| SST | −1.02 | −1.33 | −1.79 | −1.45 | −1.17 | 1.25 | −1.16 | −1.67 | −1.44 | 2.92 | 3.67 | 1.26 |
| TAC1 | 1.35 | −1.36 | −2.20 | 1.22 | 1.28 | 1.05 | −1.25 | 1.48 | 1.85 | 1.59 | 2.76 | 1.73 |
| TGFB2 | −1.12 | 1.19 | −1.87 | −1.03 | −1.72 | −1.67 | −1.02 | −1.17 | −1.15 | 1.51 | 1.60 | 1.05 |
| TGFB3 | −1.10 | 1.16 | −1.89 | −1.07 | 1.03 | 1.10 | −1.11 | 1.39 | 1.54 | 1.61 | 2.60 | 1.61 |
| TGFBI | 1.06 | 1.06 | −1.03 | 1.46 | 1.48 | 1.02 | 1.32 | −1.11 | −1.46 | −1.21 | −1.44 | −1.19 |
| VIP | 1.62 | −1.24 | −1.05 | 1.03 | −1.10 | −1.13 | −1.02 | 1.00 | 1.02 | 1.27 | −1.14 | −1.45 |
| Signaling – Matricellular | ||||||||||||
| CYR61 | 1.15 | −1.06 | 2.14 | −1.63 | −1.20 | 1.35 | −1.09 | −2.06 | −1.89 | 1.43 | 1.29 | −1.10 |
| NOV | 1.11 | 1.07 | 1.03 | −2.14 | −2.75 | −1.28 | −1.55 | 1.18 | 1.84 | 1.37 | −1.06 | −1.45 |
| THBS1 | 1.11 | −1.06 | 1.01 | −2.97 | −3.08 | −1.04 | −1.59 | −1.10 | 1.45 | 1.11 | −1.05 | −1.17 |
| THBS2 | 1.04 | −1.03 | −1.24 | −1.98 | −2.25 | −1.14 | −1.03 | 1.08 | 1.11 | 1.00 | 1.03 | 1.03 |
| TNC | −1.12 | −1.04 | −1.52 | −1.24 | 1.21 | 1.49 | 1.17 | 1.46 | 1.24 | 1.44 | 1.78 | 1.24 |
|
| ||||||||||||
| MP/TIMP | ||||||||||||
| ADAMTS4 | −1.03 | 1.02 | 1.04 | −1.33 | 1.08 | 1.44 | 1.04 | 1.30 | 1.25 | −1.07 | 1.06 | 1.13 |
| ADAMTS5 | −1.03 | 1.16 | −1.56 | −1.06 | −1.20 | −1.13 | 1.26 | 1.29 | 1.02 | 1.15 | −1.01 | −1.16 |
| ADAMTSL3 | 1.02 | 1.00 | 1.73 | −1.99 | −1.20 | 1.65 | −1.35 | 1.17 | 1.57 | 1.60 | 1.44 | −1.11 |
| MMP14 | 1.07 | 1.01 | −1.68 | 1.14 | 1.05 | −1.09 | 1.07 | 1.73 | 1.62 | −1.06 | 1.27 | 1.35 |
| TIMP2 | −1.01 | 1.03 | −1.17 | −1.42 | −1.29 | 1.09 | 1.07 | 1.50 | 1.40 | −1.04 | −1.03 | 1.01 |
| TIMP3 | −1.05 | −1.00 | 1.06 | −1.28 | −1.53 | −1.20 | −1.09 | 1.11 | 1.21 | 1.04 | −1.18 | −1.23 |
|
| ||||||||||||
| Extracellular matrix | ||||||||||||
| COL12A1 | 1.08 | −1.06 | −1.38 | −1.90 | −1.97 | −1.04 | −1.25 | 1.63 | 2.03 | 1.22 | 1.18 | −1.03 |
| COL6A6 | 1.04 | −1.00 | 1.68 | −1.83 | −1.35 | 1.35 | −1.25 | 1.61 | 2.01 | 2.75 | 2.01 | −1.37 |
| DCN | 1.09 | −1.00 | 1.04 | 1.07 | 1.08 | 1.00 | 1.11 | 1.80 | 1.62 | −1.13 | −1.20 | −1.07 |
| FMOD | 1.01 | −1.05 | −1.89 | −1.19 | −1.82 | −1.52 | −1.03 | −1.00 | 1.03 | 1.04 | 1.57 | 1.52 |
| MXRA5 | −1.03 | −1.12 | 1.21 | −1.18 | −1.04 | 1.13 | 1.10 | 2.05 | 1.87 | 1.62 | 1.26 | −1.29 |
| NYX | 1.09 | 1.04 | 1.01 | −1.09 | −1.26 | −1.16 | −1.02 | 1.38 | 1.41 | −1.28 | −1.58 | −1.24 |
| OGN | 1.00 | 1.05 | −1.33 | −1.32 | −1.70 | −1.29 | −1.13 | 1.30 | 1.47 | 1.14 | 1.15 | 1.00 |
| PRELP | −1.02 | 1.01 | −1.13 | −1.08 | −1.29 | −1.20 | 1.08 | 1.31 | 1.22 | −1.13 | −1.04 | 1.09 |
| SERPINH1 | 1.00 | −1.06 | 1.29 | −1.12 | −1.09 | 1.03 | −1.01 | 1.19 | 1.20 | −1.01 | −1.52 | −1.51 |
Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant, “X” = differential expression likely due to control eye effect.
As expected in juvenile animals whose eyes are growing more slowly with increasing age, significant differences in the expression of some genes was found between the 26N and 37N groups. Expression levels were lower in the 37N group for 10 genes; five genes were up-regulated. One gene (RLBP1; retinaldehyde binding protein 1) showed a dramatic down-regulation (−13.98 fold). NOS1 (nitric oxide synthase 1) mRNA levels were −4.07 fold lower in the older normal group. All remaining differences were less than 2-fold.
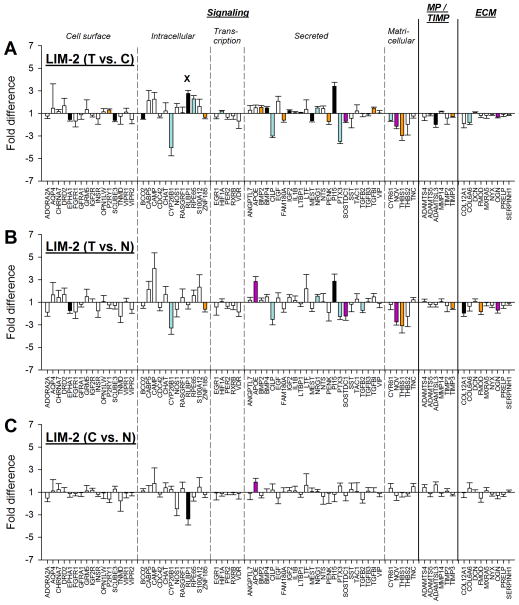
3.2.2. Two-day minus lens treatments (GO)
Two days of −5 D lens wear (LIM-2) produced significant differences in mRNA expression levels for 28 genes in the treated vs. control eyes (Fig. 5A); expression values are listed in Table 2. Eighteen of the 28 differentially expressed genes were down-regulated, while the other 10 were up-regulated. As may be seen by examining Figs. 5B and 5C, the significant up-regulation of one gene (RLBP1) occurred because the control eye mRNA levels were significantly reduced relative to age-matched normal eyes (26N; mean of right and left eye values).
Figure 5.
Gene expression fold differences. (A) Treated eyes vs. control eyes after 2 days of minus-lens wear. (B) Treated eyes vs. normal (26N) eyes. (C) Control eyes vs. normal (26N) eyes. Symbols as in Figure 4. In A, the “x” indicates that the significant treated-eye vs. control-eye up-regulation of RLBP1 was produced by decreased mRNA expression in the control eyes (shown in C).
Previous studies of mRNA and protein levels in tree shrew sclera have found alterations in expression levels not only in the treated eyes, but also in the untreated fellow control eyes when compared with age-matched normal eyes (Frost & Norton, 2012; Gao et al., 2011). Because (unpaired) comparisons of expression between groups of animals are less sensitive than (paired) comparisons between the two eyes within an animal, fewer significant differences are typically detected. In the treated vs. normal comparison (Fig. 5B), 16 genes differed significantly in their expression levels. When comparing control eyes with the 26N group (C vs. N), mRNA levels for 2 genes (RLBP1, down-regulated; APOE [apolipoprotein E], up-regulated) were significantly different from normal levels (Fig. 5C).
When mRNA levels in treated eyes were compared with normal eyes (Fig. 5B), significant differences were found that did not entirely match the differences found between treated and control eyes (Fig. 5A). In three instances (COL12A1 [collagen type XII, α1], FMOD [fibromodulin], and TGFB2 [transforming growth factor β2]), the treated eyes were significantly different from normal eyes but the treated vs. control eye differences were not significant. The reason is that the control eye mRNA levels also were different from the normal eyes in the same direction as the treated eyes. For a fourth gene, APOE, both the control eye and treated eye mRNA levels were significantly different from normal so that the treated vs. control eye difference was very small and was not statistically significant.
3.2.3. Eleven-day minus lens treatments (STAY)
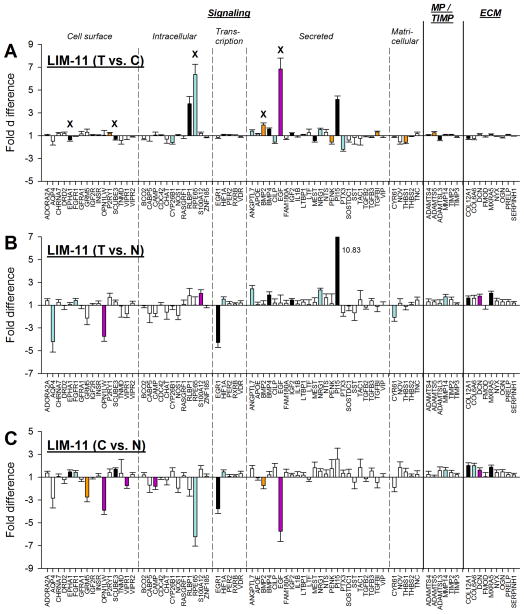
Although the treated eyes of each animal in the LIM-11 group had refractively compensated fully for the −5 D lens after 11 days of lens wear, significant differences in mRNA expression were found between treated vs. control eyes (Fig. 6A), and also between treated vs. normal (Fig. 6B) and control vs. normal eyes (Fig. 6C). Expression values are listed in Table 2. mRNA levels for 22 genes were significantly different between treated eyes and control eyes. Ten were down-regulated, and the other 12 were up-regulated. Five of these (BMP2 [bone morphogenetic protein 2], EGF [epidermal growth factor], EPHA1 [EPH receptor A1], RPE65 [retinoid isomerohydrolase], and SCUBE3 [signal peptide, CUB and EGF-like domain-containing protein 3]) were significant because the control eye mRNA levels were significantly affected (compared with normal) while the treated eye levels were not.
Figure 6.
Gene expression fold differences. (A) Treated eyes vs. control eyes after 11 days of minus-lens wear. (B) Treated eyes vs. normal (37N) eyes. (C) Control eyes vs. normal (37N) eyes. Symbols as in Figure 4. In A, the “x” indicates genes in which the significant treated-eye vs. control-eye differences for five genes (EPHA1, SCUBE3, RPE65, BMP2, and EGF) were produced by altered mRNA expression in the control eyes. In B, the off-scale fold difference for PI15 (10.83) is indicated next to the bar.
As shown in Table 2 and in Figs. 6B and 6C, numerous significant differences in gene expression were found both between the treated eyes compared with 37N eyes and between control eyes compared with 37N eyes. mRNA levels of 16 genes differed between treated and normal eyes (Fig. 6B) and 17 genes were significantly different between control eyes and normal eyes (Fig. 6C). Potential reasons for the control eye gene expression to differ from normal will be presented in the Discussion. As in the LIM-2 group, some of the genes in which the treated eye mRNA levels were significantly different from normal were not found to be significantly different when treated eyes were compared with control eyes. This was because treated and control eyes both differed from normal in the same direction.
3.2.4. Comparison between GO and STAY patterns
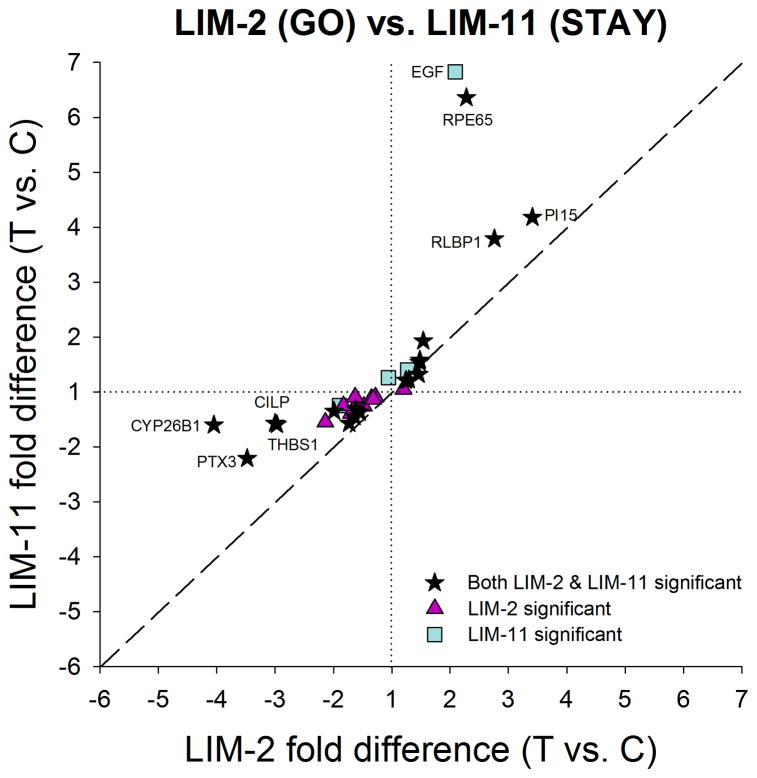
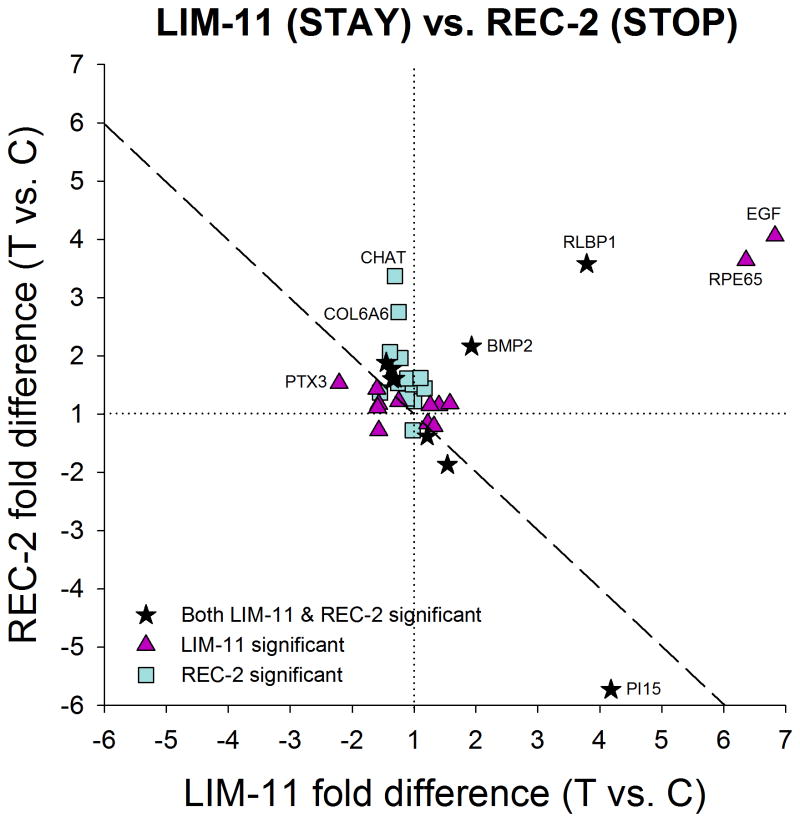
The similarities in the mRNA patterns between the LIM-2 group and LIM-11 group are of interest. After 2 days of minus-lens wear, retinally-derived signals must be passing through the choroid to produce the scleral remodeling that results in axial elongation. After 11 days of minus-lens wear, the treated eyes have completed their compensation and are maintaining an elongated state. mRNA differences that are unique to the LIM-2 group may be involved in actively promoting elongation (a GO signal) whereas mRNA differences that are unique to the LIM-11 group may be involved in maintaining the elongated state (a STAY signal). Shared mRNA differences may be involved in both GO and STAY. Fig. 7 compares the significant treated vs. control eye differences at LIM-2 with those at LIM-11. Eighteen genes had significantly different mRNA levels in the treated vs. control eyes at both LIM-2 and LIM-11. All were regulated in the same direction in both conditions; nine were down-regulated in both conditions and nine were up-regulated at both LIM-2 and LIM-11. The correlation was significant (p < 0.05) with a slope of 0.88 and r2 = 0.58. Although the direction of the shared differential expression was the same for these genes, the amplitude (fold differences) differed for several genes. Four genes labeled in Fig. 7 (CILP [cartilage intermediate layer protein 1], CYP26B1 [cytochrome P450 26B1], PTX3 [pentraxin 3], and THBS1 [thrombospondin 1]) were more strongly down-regulated in the LIM-2 group than in the LIM-11 group. Three additional genes (PI15 [peptidase inhibitor 15], RLBP1, and RPE65), also labeled in Fig. 7, were more strongly up-regulated in the LIM-11 group than in the LIM-2 group. In addition, there were 10 genes that were significantly affected in LIM-2 but not in LIM-11, and 4 genes that were significantly regulated only in the LIM-11 group.
Figure 7.
Comparison of the gene expression differences (treated eye vs. control eye) in Fig. 5A (LIM-2) with the differences in Fig. 6A (LIM-11) showing the similar differential expression patterns in GO and STAY. All genes that were up- or down-regulated in LIM-2 were regulated in the same direction at LIM-11. Highly-regulated genes are labeled. Values near the dashed line indicate genes that responded similarly in the two conditions. Stars = significant fold differences for both LIM-2 and LIM-11; triangles = significant fold differences only for LIM-2; squares = significant fold differences only for LIM-11.
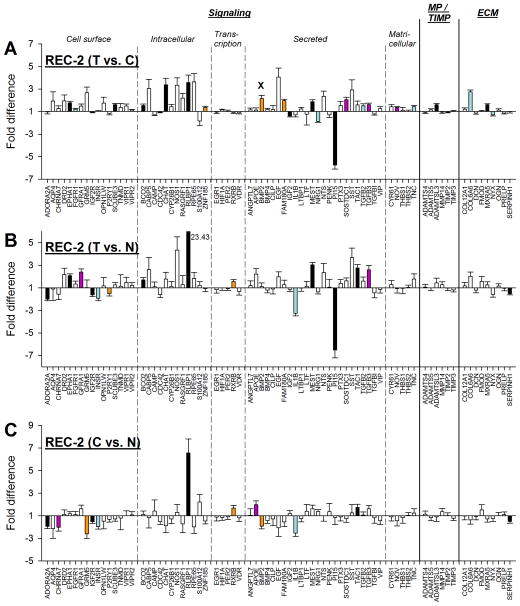
3.2.5. Two-day recovery (STOP)
As shown in Fig. 2, the treated eyes of the REC-2 group had begun to recover from the full compensation produced by 11 days of −5 D lens wear. The mRNA expression pattern at REC-2 presumably reflects STOP signals present in the choroid at this time point. Fig. 8A shows the fold differences in gene expression between the treated and control eyes in the REC-2 group. Expression values are also listed in Table 2. Twenty-two genes were significantly different in treated eyes compared with control eyes, 18 were up-regulated, and the other 4 were down-regulated. One of the up-regulated genes (BMP2) was significant because the control eye mRNA levels were significantly down-regulated (compared with normal) while the treated eye levels were not.
Figure 8.
Gene expression fold differences. (A) Treated eyes vs. control eyes after 2 days of recovery from full compensation to minus-lens wear. (B) Treated eyes vs. normal (37N) eyes. (C) Control eyes vs. normal (37N) eyes. Symbols as in Figure 4. In A, the “x” indicates that the significant treated-eye vs. control-eye up-regulation of one gene (BMP2) was produced by decreased mRNA expression in the control eyes. In B, the off-scale fold difference for RLBP1 (23.43) is indicated next to the bar.
When comparing treated eyes with age-matched normal eyes in the 37N group (T vs. N), there were 15 genes significantly different (Fig. 8B). Comparing control eyes with age-matched normal eyes in 37N group (C vs. N), there were 12 genes significantly different (Fig. 8C). As in the LIM-2 and LIM-11 groups, some of the genes in the REC-2 group in which the treated eye mRNA levels were significantly different from normal were not found to be significantly different when treated eyes were compared with control eyes. This was because treated and control eyes both differed from normal in the same direction.
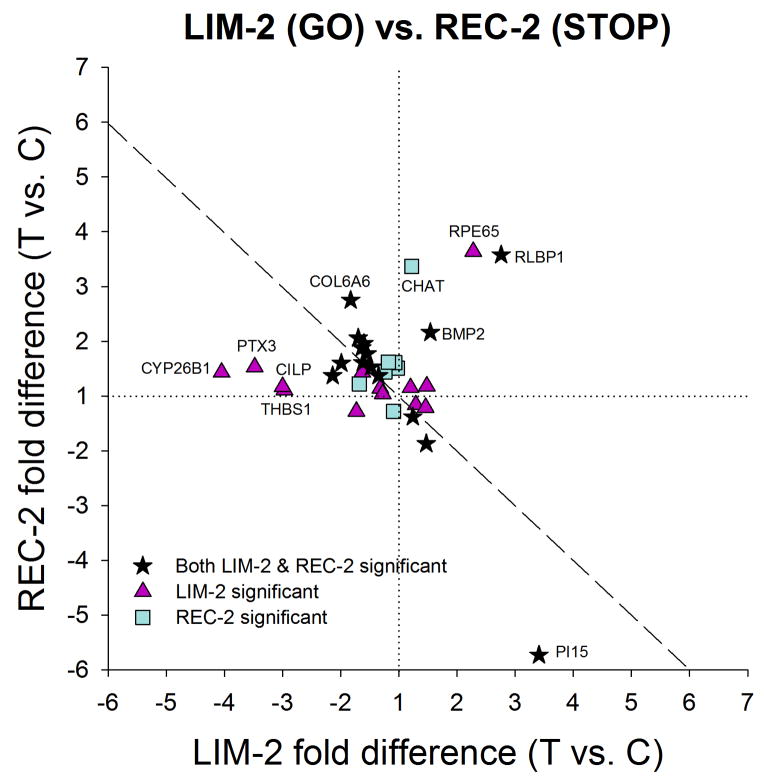
3.2.6. Comparison of GO and STOP patterns
The differences in the mRNA patterns between the LIM-2 group (GO) and REC-2 group (STOP) are of interest. As shown in Fig. 9, the pattern is very different from that in Fig. 7 in that the genes were generally regulated in opposite directions in STOP than they were in GO. Most of the 35 genes that showed significant regulation in one or both conditions either were down-regulated in LIM and up-regulated in REC (top left quadrant of Fig. 9) or the reverse (bottom right quadrant of Fig. 9). Thirteen genes showed bi-directional regulation; 20 additional genes showed significant differences in either LIM or REC, but not both; 2 genes (BMP2 and RLBP1) were up-regulated under both, but regulation of RLBP1 in LIM-2 and BMP2 in REC-2 are likely caused by a control-eye effect.
Figure 9.
Comparison of the gene expression differences (treated eye vs. control eye) in Fig. 5A (LIM-2) with the differences in Fig. 8A (REC-2), showing that the gene expression patterns in GO and STOP were very different. Highly-regulated genes are labeled. Values near the dashed line indicate genes that responded with opposite fold-differences in the two conditions. Stars = significant fold differences for both LIM-2 and REC-2; triangles = significant fold differences only for LIM-2; squares = significant fold differences only for REC-2.
3.2.7. Comparison of STAY and STOP patterns
A final relevant comparison is between the treated-eye vs. control-eye differences at LIM-11 (STAY) and those present at REC-2 (STOP); this comparison is shown in Fig. 10. Not surprisingly, the pattern more closely resembles that seen comparing GO and STOP (Fig. 9) than that in Fig. 7, comparing GO vs. STAY. Seven genes showed significant bi-directional regulation, 13 genes were significantly regulated under LIM-11 but not REC-2, 13 other genes were significantly regulated under REC-2 but not LIM-11, and two genes (BMP2 and RLBP1) were significantly up-regulated under both LIM-11 and REC-2.
Figure 10.
Comparison of the gene expression differences (treated eye vs. control eye) in Fig. 6A (LIM-11) with the differences in Fig. 8A (REC-2) showing that the gene expression patterns in STAY and STOP were different. Highly-regulated genes are labeled. Values near the dashed line indicate genes that responded with opposite fold-differences in the two conditions. This correlation was significant (p = 0.02; slope = −0.55; r2 = 0.17). Stars = significant fold differences for both LIM-11 and REC-2; triangles = significant fold differences only for LIM-11; squares = significant fold differences only for REC-2.
4. DISCUSSION
4.1 Gene Expression Signatures
In addition to its important role as the vascular supply to the RPE and outer retina, the choroid participates in the emmetropization mechanism (Nickla & Wallman, 2010; Summers, 2013). It receives, presumably from RPE, unknown molecules that initiate responses in the choroid that, in turn, generate signals which pass to the sclera where they produce scleral remodeling and regulate the rate of axial elongation in juvenile eyes (Norton et al., 2008; Wallman & Winawer, 2004). We examined differential mRNA expression by choroidal cells 1) during normal development; 2) early in the development of LIM, a GO condition in which the eye is increasing its axial elongation; 3) after completing compensation to a minus lens, a STAY condition where the eye remains elongated until lens-wear is discontinued; and 4) early in the recovery process, a STOP condition in which the axial elongation rate slows to produce refractive recovery. We have found that distinct and unique constellations of differential mRNA expression occur amongst the sampled genes in each of these conditions as the cells in the choroid receive, integrate, and generate emmetropization-related signals that are transmitted to the sclera. Sixty of the 77 genes examined in the choroid showed significant mRNA expression differences during normal development, GO, STAY, or STOP conditions, including not only (treated eye vs. control eye) differential expression, but also expression differences between treated or control eyes relative to age-matched normal eyes, and as a function of age in normal development. Not only are the mRNA expression patterns different in each condition, only one of the genes examined (RLBP1) was differentially expressed in all of the conditions (normal growth, GO, STAY, and STOP). Thus, there is selective regulation with gene expression being altered uniquely in each condition. Table 3 lists the genes whose expression differs, exhibiting up- or down-regulation between the treated vs. control eyes in the GO, STAY, and STOP conditions. The patterns of gene expression are unique to each condition, constituting what may be considered gene expression “signatures”.
Table 3.
Genes that were significantly regulated in treated vs. control eyes under GO, STAY, or STOP conditions.
| Functional category | GO (LIM-2) | STAY (LIM-11) | STOP (REC-2) | |
|---|---|---|---|---|
| Signaling - Cell surface | B | EPHA1 | EPHA1 x | EPHA1 |
| B | SCUBE3 | SCUBE3 x | SCUBE3 | |
| P2RY1 | P2RY1 | |||
| FGFR1 | ||||
| Signaling - Intracellular | B | BCO2 | BCO2 | |
| B | ZNF185 | ZNF185 | ||
| CYP26B1 | CYP26B1 | |||
| RLBP1 x | RLBP1 | RLBP1 | ||
| RPE65 | RPE65 x | |||
| CHAT | ||||
| Signaling - Transcription | HIF1A | |||
| Signaling - Secreted | B | IGF2 | IGF2 | IGF2 |
| B | NRG1 | NRG1 | NRG1 | |
| B | PI15 | PI15 | PI15 | |
| B | FAM180A | FAM180A | ||
| B | MEST | MEST | MEST | |
| B | SOSTDC1 | SOSTDC1 | ||
| BMP2 | BMP2 x | BMP2 x | ||
| BMP4 | BMP4 | |||
| TGFBI | TGFBI | |||
| CILP | CILP | |||
| PENK | PENK | |||
| PTX3 | PTX3 | |||
| ANGPTL7 | ||||
| EGF x | ||||
| TGFB2 | ||||
| TGFB3 | ||||
| Signaling - Matricellular | B | NOV | NOV | |
| THBS1 | THBS1 | |||
| CYR61 | ||||
| TNC | ||||
| MPs/TIMPs | B | ADAMTSL3 | ADAMTSL3 | ADAMTSL3 |
| TIMP3 | ||||
| ADAMTS5 | ||||
| Extracellular matrix | B | COL6A6 | COL6A6 | |
| COL12A1 | ||||
| OGN | ||||
| MXRA5 | ||||
| NYX | ||||
“B” = bi-directional regulation between GO and STOP, red gene symbols = down-regulation, blue = up-regulation, “X” = differential expression likely due to control eye effect.
Although the concept of GO and STOP signals was first raised many years ago (Rohrer & Stell, 1994; Schaeffel & Howland, 1991), it has not been clear if they are separate signals, or even if there are two signals. Potentially, STOP could be the absence of GO, or vice versa. The gene expression differences in choroid suggest that there may, indeed, be two different signals at the level of the choroid, one for GO and a different one for STOP; some genes may participate in both, showing bi-directional regulation that is opposite in STOP than it is in GO. Comparing LIM-2 (GO) and REC-2 (STOP), there were 13 bi-directionally regulated genes (Table 3). In addition to the bi-directionally regulated genes, there are others that were significantly regulated in GO but not STOP and vice versa, suggesting that there may be a “core” of genes that act in a push-pull manner and others that participate only in GO or STOP.
The present study also suggests the presence of a STAY signature in the choroid that resembles, but is distinct from, the GO signature (Fig. 7). As shown in Fig. 3, the refractions of the treated eyes in the LIM-11 group, measured with the −5 D lens in place, were no longer hyperopic and, as a group, had been within 1 D of their fellow control eyes for five days. The refractive hyperopia that generated retinal GO signals and stimulated the emmetropization mechanism to produce the elongation had dissipated. The treated eyes, however, were elongated compared to their untreated fellow control eyes. In other tree shrews that wore a −5 D lens for 30 days, the eyes remained elongated until lens-wear was discontinued, whereupon refractive recovery commenced (Norton et al., 2010). It seems unlikely that the retina would still generate a GO signal because the eyes had achieved a refractive match to the control eyes, yet some signal evidently existed that kept the treated eye elongated. This may be a representation, at the mRNA level, of a “shape factor” or “eye-size factor” that has been hypothesized previously (Nickla et al., 2005; Schaeffel & Howland, 1991; Siegwart, Jr. & Norton, 2013; Troilo & Wallman, 1991). Perhaps this STAY signal is more evident in the choroid than in the sclera because the scleral remodeling returns toward normal but does not develop a recovery (STOP) pattern because of continued signals from the choroid. As shown in Table 3 and Fig. 7, this STAY signature includes many of the genes that are differentially expressed in GO (LIM-2) and are similarly expressed in both, but several genes that are significantly differentially expressed in GO are no longer significantly affected in STAY, along with a few additional genes expressed in STAY but not GO. Four genes, strongly up-regulated in STAY, RPE65 and EGF along with RLBP1 and PI15 (both also up-regulated in GO), may be of interest. Although four genes were up-regulated in both STAY and STOP, from Table 3 and Fig. 10, it also is clear that the STOP signature is very different from the STAY signature. Assuming that the REC-2 group also had a STAY signature at the end of lens-wear, the transition between STAY and STOP must be very rapid, occurring over a two-day period.
The presence of mRNA for RPE65 in the choroidal samples raises the question of whether or not there might be RPE tissue intermixed with the choroid. Several lines of evidence suggest that this was not the case. Although RPE65 is highly abundant in the RPE, it is also found in many locations, typically in association with vitamin A metabolism and/or with melanocytes. For instance, RPE65 is also found to be expressed, at mRNA and/or protein levels, in human cones (Tang et al., 2011), bovine ciliary epithelial tissue (Salvador-Silva et al., 2005), mouse hypothalamus (Helfer et al., 2012), and in keratinocytes (Hinterhuber et al., 2004) and melanocytes (Amann et al., 2012) in human skin. In tree shrews, preliminary results comparing differential mRNA expression in RPE with that in choroid have found that mRNA for RPE65 is not differentially regulated after one day of −5 D lens wear, whereas it is significantly up-regulated in the choroid. Thus, our finding mRNA for RPE65 in the choroid samples is not indicative of contamination from RPE cells. Although, in fresh tissue, retina is easily separated from RPE, which adheres to the choroid (Bruch’s membrane), treatment with RNAlater causes the RPE to adhere tightly to the retina (Malik et al., 2003; Wang et al., 2012). These factors, along with our observation during tissue dissection that retina/RPE together cleanly separated from the choroid lead us to conclude that there was little, if any, RPE contamination of the choroidal samples.
Examination of Table 2 shows that the GO, STAY, and STOP signatures included only 7 of the 15 genes whose expression changed in normal animals between 26 DVE and 37 DVE. If these normal changes are related to slowed axial elongation, it might be expected that they would show a similar direction of altered expression during recovery, when the elongation rate also is slowed. However, only one gene did so, ADAMTSL3 [ADAMTS-like 3], up-regulated in both. RLBP1, which was strongly down-regulated (−13.98 fold) at 37 DVE, was significantly up-regulated during STOP. Similarly, it might be expected that the normal change in age-related expression differences would be in the opposite direction to the expression changes found during GO or STAY. This was the case for four genes (ADAMTS5 [ADAM metallopeptidase with thrombospondin motif, 5], ADAMTSL3, CYR61 [protein CYR61], and RLBP1). Thus, it appears that a subset of the genes whose expression is related to emmetropization signaling are also involved in the normal slowing of eye growth with age.
The differential mRNA expression in the GO, STAY, and STOP conditions contrast sharply with the very similar mRNA levels found in the left and right eyes of the two normal groups. As seen in Fig. 4, mRNA levels are nearly identical in the two eyes of these normal groups. Of the 77 genes examined, only 2 (at 26 DVE) displayed right-eye vs. left-eye differences that exceeded 1.5 fold. The absence of any significant mRNA level differences in the two eyes of normal groups lends confidence to our decision to not use a correction for false discovery.
4.2 Components of the Choroidal Signatures
The group of 77 genes examined in this study is a subset of a much larger group of genes that probably show differential expression in normal development and/or in GO, STAY, and STOP. A preliminary whole-transcriptome analysis using mRNA from three of the LIM-2 and three of the REC-2 animals suggests that over 300 distinct genes (of the over 18,000 genes annotated in the low-coverage tree shrew genome) may be up- or down-regulated by at least 1.20 fold, with changes occurring in each direction (Frost, personal communication, 2013). The subset of genes we examined with qPCR is large enough to determine that the GO, STAY, and STOP signatures are distinct from one another, but there undoubtedly are other genes that also show differential expression in these conditions. While the expression of other genes that we did not measure may be as, or more, important in the receipt and re-transmission of emmetropization-related signals in the choroid, expression changes in this sample of genes provide useful information about the role of the choroid in emmetropization.
It is clear from examination of Table 2, Table 3, and Figs. 5, 6, & 8, that genes belonging to several functional categories are included in the GO, STAY, and/or STOP signatures. Table 2 may be particularly helpful in illustrating that differential expression was selective, involving some, but not all, of the candidate genes whose protein products include cell-surface receptors, intracellular signaling, transcription factors, and numerous secreted signaling proteins. If the protein products of these genes are altered in accordance with the mRNA levels, the defocus-dependent behavior of such genes suggests that their levels may be involved in regulating axial elongation in a bi-directional, push-pull manner. This is consistent with the hypothesis of Nickla and Wallman (2010) that “the choroid contains secretory cells which function in the visual regulation of ocular growth, by influencing the biosynthetic activity of the sclera.”
Among the genes for cell-surface signaling, EPHA1 (actin cytoskeleton-related signaling) and SCUBE3 (which activates TGFβ signaling via binding with TGFBR2 [TGFβ receptor 2]) were bi-directionally regulated, down in GO and STAY and up in STOP. The purinergic receptor gene P2RY1 was up-regulated in GO and STAY; however, ADORA2A (adenosine receptor A2a) was not significantly affected, a similar result to that found at the protein level in form-deprived guinea pig choroid (Cui et al., 2010).
Genes for several intracellular signaling proteins were differentially expressed. ZNF185 (zinc finger protein 185), an actin cytoskeleton-related protein enriched at focal adhesions, and BCO2 (beta-carotene oxygenase 2), which may play a role in retinoic acid synthesis, were down-regulated in GO and up-regulated in STOP. Other genes related to retinoid and retinoic acid metabolism and signaling (CYP26B1, RLBP1, and RPE65) also showed differential expression, consistent with the possible involvement of the retinoic acid signaling pathway in the choroidal component of the emmetropization mechanism. However, RXRB (retinoid X receptor β) was not affected in GO, STAY, or STOP. All-trans-retinoic acid (atRA), an important growth regulator, has been reported to be involved in the choroid in the regulation of axial elongation in chick, guinea pig, and primates. The synthesis of atRA was down-regulated in chick choroid during myopia development and may be up-regulated in mammalian choroid during myopia development (McFadden et al., 2004; Mertz & Wallman, 2000; Troilo et al., 2006). The mRNA expression for RALDH2 (retinal dehydrogenase 2), an enzyme involved in atRA synthesis that we did not examine, was up-regulated in chick choroid during recovery from induced myopia and during plus-lens wear (Rada et al., 2012; Simon et al., 2004). Of the sampled genes involved in transcriptional regulation, only HIF1A (hypoxia inducible factor 1α) was affected, being up-regulated in GO.
Many genes for secreted signaling proteins were differentially expressed, including six that were bi-directionally regulated. The mRNA levels for IGF2 (insulin-like growth factor 2) were up-regulated in GO and STAY and down-regulated in STOP. So, too, was NRG1 (neuregulin 1), which can be involved in inducing acetylcholine receptor expression at neuromuscular junctions, and PI15, a cysteine-rich secretory protein of unknown function. Three other bi-directionally regulated genes (FAM180A [family with sequence similarity 180, member A], MEST [mesoderm specific transcript], and SOSTDC1 [sclerostin domain-containing protein 1]) showed the opposite pattern, down-regulation in GO and up-regulation in STOP. The down-regulation of SOSTDC1, an inhibitor of BMP activity, combined with the up-regulation of BMP2 and BMP4 (members of the TGFβ superfamily) may significantly increase BMP signaling in GO. In addition, TGFB2 and TGFB3 (also members of the TGFβ superfamily) were up-regulated in the treated eyes in STOP. The absence of significant regulation of these two genes during GO agrees with the results of Jobling et al. (2009) in choroid of form-deprived tree shrews. In the choroid of minus-lens-treated chicks (GO), Simon et al. (2004) found a down-regulation of TGFB2. The up-regulation we found during STOP may be consistent with that report. The combined effect of up-regulation of TGFB2, TGFB3, and SCUBE3 may increase TGFβ signaling in STOP. mRNA for TGFBI (TGFβ-induced protein) was up-regulated in GO and STAY, a finding that is consistent with a report in marmoset (Shelton et al., 2008), showing that TGFBI in RPE/choroid was significantly up-regulated in minus-lens treated eyes compared with plus-lens treated eyes. Other signaling-related genes (ovotransferrin and avian thymic hormone) that have been found to have altered expression in chick choroid (Rada et al., 2001; Rada & Wiechmann, 2009) were not examined in this study.
mRNA for several matricellular proteins (NOV [nephroblastoma overexpressed gene], THBS1, and CYR61) were down-regulated during GO; one (NOV) was up-regulated in STOP. Down-regulation of THBS1 has been also found in tree shrew sclera along with a reduction in its protein during GO (Frost & Norton, 2012; Gao et al., 2011). In the sample of genes coding for metallopeptidases or TIMPs, the metallopeptidase ADAMTSL3 was bi-directionally regulated, down in GO and STAY and up in STOP.
Of the extracellular matrix genes in our sample, the general pattern was for down-regulation in GO or STAY and up-regulation in STOP. An exception was NYX (nyctalopin), which was down-regulated only in STOP. In chicks, HAS2 (hyaluronan synthase 2) mRNA expression was reported to increase significantly in the choroid of eyes recovering from myopia (Rada et al., 2010). We were unable to design reliable qPCR primers for tree shrew HAS2; preliminary whole-transcriptome analysis data on three animals from the REC-2 group (Frost, personal communication, 2013) suggest that HAS2 may be up-regulated during STOP.
Overall, the signatures for GO, STAY, and STOP in tree shrew choroid are both complex and unique. A better understanding of the roles the affected genes may play will require an improved knowledge of how the genes interact as part of a complex network in vivo. However, our results are consistent, during GO, with an up-regulation of the BMP pathway and down-regulation of the TGFβ pathway. During STOP there may be a partial reversal of that pattern along with up-regulation of the TGFβ pathway. Finally, considering that the choroid is a highly vascularized tissue, the involvement of a group of 13 genes, spanning several functional groupings, with roles in vascular regulation and/or angiogenesis (BMP2, BMP4, CYR61, EGF, FGFR1, HIF1A, IGF2, NRG1, PTX3, TGFB2, THBS1, TIMP3, and TNC), may occur due either to blood-flow regulation and/or as part of emmetropization signaling.
4.3 Control Eye Effects
As noted in Table 2 and Figs. 5, 6, & 8, there were numerous instances in which the gene expression in the control eyes differed from age-matched normal eyes. Similar control-eye effects have been found in previous studies of mRNA and protein expression in tree shrew sclera in which the control and treated eyes both differ from normal in the same direction (Frost & Norton, 2012; Gao et al., 2011; Guo et al., 2013). Among the possible causes of these binocular changes, one that seems likely is binocular regulation of blood flow in the choroid, which could produce similar effects in both choroid and sclera in both eyes. Choroidal blood flow during the development of monocular induced myopia has not been measured in mammals; in chicks there is a reduction in choroidal blood flow in form-deprived eyes. However, several studies have found evidence for a binocular reduction in choroidal blood flow during monocular treatment (Jin & Stjernschantz, 2000; Shih et al., 1993). According to Jin and Stjernschantz (Jin & Stjernschantz, 2000), during form deprivation the choroidal blood flow was markedly reduced (significantly) in the form-deprived eye, and tended to be reduced (non-significantly) in the contralateral eye, compared with the normal group; while during recovery, choroidal blood flow in both recovering and contralateral eyes were significantly increased compared with the normal group. Indeed, some of the sampled genes whose expression in the control eyes differed from normal eyes included ones related to vascular regulation and/or angiogenesis. To the extent that regulation of choroidal blood flow in mammals is binocular, alterations in choroidal blood flow in the control eye choroid may help to explain the effects that we observed in the control eyes in GO, STAY, and STOP conditions. Another potential reason for binocular effects on choroidal mRNA expression in the control eye is that the open goggle frame around the control eye may affect the peripheral visual field (Amedo & Norton, 2012). Although this is not sufficient to produce refractive effects, measured on the pupillary axis, it may produce small changes in peripheral areas of the choroid that were included in our choroidal samples.
In the Results, we noted several genes that were significantly different in the treated eyes vs. control eyes because mRNA levels in the control eye choroid differed from normal but the levels in the treated eye choroids did not. Comparing treated eye vs. normal eye and control eye vs. normal eye mRNA differences (for example, Figs. 6B and 6C), many of the fold-differences (both significant and non-significant) moved in the same direction of up- or down-regulation. The cases where the treated eyes did not change but the control eyes did differ from normal were included as differentially expressed genes under the assumption that the control eye change reflected binocular effects in the choroid. That the mRNA levels for these genes in treated eye choroid did not change in concert with the control eye choroid implied active regulation that prevented a binocular shift. We examined treated vs. normal eye mRNA differences comparing LIM-2 (GO) vs. LIM-11 (STAY), LIM-2 vs. REC-2 (STOP), and LIM-11 vs. REC-2, analogous to the treated vs. control eye differences shown in Figures 7, 9, and 10. GO and STAY produced a similar treated vs. normal gene expression pattern, that was very weakly correlated. GO vs. STOP and STAY vs. STOP, as in Figs 9 and 10, showed expression patterns that differed, but are not opposite, so there was not a significant correlation between GO vs. STOP or STAY vs. STOP.
4.4 Summary
This study examined differential mRNA expression by cells in mammalian choroid during normal development; early in minus-lens wear, a GO condition; after full compensation for a minus lens, a STAY condition; and early in refractive recovery from lens-induced myopia, a STOP condition. We found that cells in the choroid respond with different mRNA expression signatures to the different emmetropization conditions. The STAY signature resembled, but differed from, the GO signature and both were very distinct from the STOP signature. Expression signatures were different from those seen in the RPE (Frost et al., 2013) and in the sclera (Guo et al., 2013). Thus, the choroid is a unique way-station that receives emmetropization-related signals from the RPE and generates signals that produce changes in the sclera. Because a limited number of genes were examined, these signatures are incomplete; however, the large number of genes that are altered suggest that the emmetropization-related signals in choroid are complex and unlikely to depend on the regulation of a single gene, or even a small number of genes.
Supplementary Material
Highlights (for review).
We examined emmetropization-related gene expression in mammalian choroid
Gene expression was examined in GO, STAY, and STOP conditions
GO and STAY conditions produced similar, but distinct, gene expression signatures
Both the GO and STAY signatures are very different from STOP
The choroid actively receives and transmits emmetropization-related signals
Acknowledgments
This study was supported by NIH grants EY005922 and EY003039 (P30). Li He was supported in part by a supplement to EY005922 and by funds from the Department of Vision Sciences. This work was performed in partial fulfillment of the requirements for the degree of Doctor of Philosophy at the University of Alabama at Birmingham (Li He). Preliminary results were presented in abstract form. We thank Lin Guo for technical contributions including demonstrating a way to dissect choroid away from both retina/RPE and from sclera.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amann PM, Luo C, Owen RW, Hofmann C, Freudenberger M, Schadendorf D, Eichmuller SB, Bazhin AV. Vitamin A metabolism in benign and malignant melanocytic skin cells: importance of lecithin/retinol acyltransferase and RPE65. J Cell Physiol. 2012;227:718–728. doi: 10.1002/jcp.22779. [DOI] [PubMed] [Google Scholar]
- Amedo AO, Norton TT. Visual guidance of recovery from lens-induced myopia in tree shrews (Tupaia glis belangeri) Ophthalmic Physiol Opt. 2012;32:89–99. doi: 10.1111/j.1475-1313.2011.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol G, Wang S, Budzynski E, Wangsa-Wirawan ND, Linsenmeier RA. Oxygen distribution and consumption in the macaque retina. Am J Physiol Heart Circ Physiol. 2007;293:H1696–H1704. doi: 10.1152/ajpheart.00221.2007. [DOI] [PubMed] [Google Scholar]
- Cui D, Trier K, Zeng J, Wu K, Yu M, Ge J. Adenosine receptor protein changes in guinea pigs with form deprivation myopia. Acta Ophthalmol. 2010;88:759–765. doi: 10.1111/j.1755-3768.2009.01559.x. [DOI] [PubMed] [Google Scholar]
- Diether S, Schaeffel F. Local changes in eye growth induced by imposed local refractive error despite active accommodation. Vision Res. 1997;37:659–668. doi: 10.1016/s0042-6989(96)00224-6. [DOI] [PubMed] [Google Scholar]
- Frost MR, He L, Norton TT. Differential gene expression in tree shrew retinal pigment epithelium (RPE) in response to six hours of minus-lens wear. Ophthalmic Physiol Opt. 2013;33:667. [Google Scholar]
- Frost MR, Norton TT. Alterations in protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Invest Ophthalmol Vis Sci. 2012;53:322–336. doi: 10.1167/iovs.11-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Frost MR, Siegwart JT, Jr, Norton TT. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis. 2011;17:903–919. [PMC free article] [PubMed] [Google Scholar]
- Gentle A, McBrien NA. Modulation of scleral DNA synthesis in development of and recovery from induced axial myopia in the tree shrew. Exp Eye Res. 1999;68:155–163. doi: 10.1006/exer.1998.0587. [DOI] [PubMed] [Google Scholar]
- Glickstein M, Millodot M. Retinoscopy and eye size. Science. 1970;168:605–606. doi: 10.1126/science.168.3931.605. [DOI] [PubMed] [Google Scholar]
- Guo L, Frost MR, He L, Siegwart JT, Jr, Norton TT. Gene expression signatures in tree shrew sclera in response to three myopiagenic conditions. Invest Ophthalmol Vis Sci. 2013;54:6806–6819. doi: 10.1167/iovs.13-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Frost MR, Siegwart JT, Jr, Norton TT. Gene expression signatures in tree shrew sclera in response to two STOP visual conditions: recovery from minus-lens wear, and plus-lens wear. Invest Ophthalmol Vis Sci. 2012;53:ARVO E-Abstract 3455. [Google Scholar]
- He L, Frost MR, Siegwart JT, Jr, Filios S, Norton TT. Retinal gene expression signatures in tree shrew in response to three myopiagenic visual conditions: minus lens, form deprivation, and darkness. Invest Ophthalmol Vis Sci. 2011;52:ARVO E-Abstract 6301. [Google Scholar]
- Helfer G, Ross AW, Russell L, Thomson LM, Shearer KD, Goodman TH, McCaffery PJ, Morgan PJ. Photoperiod regulates vitamin A and Wnt/beta-catenin signaling in F344 rats. Endocrinology. 2012;153:815–824. doi: 10.1210/en.2011-1792. [DOI] [PubMed] [Google Scholar]
- Hinterhuber G, Cauza K, Brugger K, Dingelmaier-Hovorka R, Horvat R, Wolff K, Foedinger D. RPE65 of retinal pigment epithelium, a putative receptor molecule for plasma retinol-binding protein, is expressed in human keratinocytes. J Invest Dermatol. 2004;122:406–413. doi: 10.1046/j.0022-202X.2004.22216.x. [DOI] [PubMed] [Google Scholar]
- Howlett MH, McFadden SA. Spectacle lens compensation in the pigmented guinea pig. Vision Res. 2009;49:219–227. doi: 10.1016/j.visres.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Hung LF, Wallman J, Smith EL., III Vision-dependent changes in the choroidal thickness of macaque monkeys. Invest Ophthalmol Vis Sci. 2000;41:1259–1269. [PubMed] [Google Scholar]
- Irving EL, Callender MG, Sivak JG. Inducing myopia, hyperopia, and astigmatism in chicks. Optom Vis Sci. 1991;68:364–368. doi: 10.1097/00006324-199105000-00007. [DOI] [PubMed] [Google Scholar]
- Irving EL, Callender MG, Sivak JG. Inducing ametropias in hatchling chicks by defocus--aperture effects and cylindrical lenses. Vision Res. 1995;35:1165–1174. doi: 10.1016/0042-6989(94)00235-e. [DOI] [PubMed] [Google Scholar]
- Jin N, Stjernschantz J. Regional blood flow in the myopic chick eye during and after form deprivation: a study with radioactively-labelled microspheres. Exp Eye Res. 2000;71:233–238. doi: 10.1006/exer.2000.0871. [DOI] [PubMed] [Google Scholar]
- Jobling AI, Wan R, Gentle A, Bui BV, McBrien NA. Retinal and choroidal TGFβ in the tree shrew model of myopia: isoform expression, activation and effects on function. Exp Eye Res. 2009;88:458–466. doi: 10.1016/j.exer.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wu J, Wang X, Zeng J. Changes in muscarinic acetylcholine receptor expression in form deprivation myopia in guinea pigs. Mol Vis. 2007;13:1234–1244. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luckett WP. Comparative Biology and Evolutionary Relationships of Tree Shrews. New York: Plenum Press; 1980. [Google Scholar]
- Lutty GA, Hasegawa T, Baba T, Grebe R, Bhutto I, McLeod DS. Development of the human choriocapillaris. Eye (Lond) 2010;24:408–415. doi: 10.1038/eye.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik KJ, Chen CD, Olsen TW. Stability of RNA from the retina and retinal pigment epithelium in a porcine model simulating human eye bank conditions. Invest Ophthalmol Vis Sci. 2003;44:2730–2735. doi: 10.1167/iovs.02-1120. [DOI] [PubMed] [Google Scholar]
- McBrien NA, Jobling AI, Truong HT, Cottriall CL, Gentle A. Expression of muscarinic receptor subtypes in tree shrew ocular tissues and their regulation during the development of myopia. Mol Vis. 2009;2009:464–475. [PMC free article] [PubMed] [Google Scholar]
- McFadden SA, Howlett MH, Mertz JR. Retinoic acid signals the direction of ocular elongation in the guinea pig eye. Vision Res. 2004;44:643–653. doi: 10.1016/j.visres.2003.11.002. [DOI] [PubMed] [Google Scholar]
- McKanna JA, Casagrande VA. Atropine affects lid-suture myopia development. Doc Ophthalmol Proc Ser. 1981;28:187–192. [Google Scholar]
- Mertz JR, Wallman J. Choroidal retinoic acid synthesis: a possible mediator between refractive error and compensatory eye growth. Exp Eye Res. 2000;70:519–527. doi: 10.1006/exer.1999.0813. [DOI] [PubMed] [Google Scholar]
- Mutti DO, Mitchell GL, Jones LA, Friedman NE, Frane SL, Lin WK, Moeschberger ML, Zadnik K. Axial growth and changes in lenticular and corneal power during emmetropization in infants. Invest Ophthalmol Vis Sci. 2005;46:3074–3080. doi: 10.1167/iovs.04-1040. [DOI] [PubMed] [Google Scholar]
- Nickla DL, Damyanova P, Lytle G. Inhibiting the neuronal isoform of nitric oxide synthase has similar effects on the compensatory choroidal and axial responses to myopic defocus in chicks as does the non-specific inhibitor L-NAME. Exp Eye Res. 2009;88:1092–1099. doi: 10.1016/j.exer.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickla DL, Sharda V, Troilo D. Temporal integration characteristics of the axial and choroidal responses to myopic defocus induced by prior form deprivation versus positive spectacle lens wear in chickens. Optom Vis Sci. 2005;82:318–327. doi: 10.1097/01.opx.0000159368.31481.de. [DOI] [PubMed] [Google Scholar]
- Nickla DL, Wallman J. The multifunctional choroid. Prog Retin Eye Res. 2010;29:144–168. doi: 10.1016/j.preteyeres.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickla DL, Wildsoet CF. The effect of the nonspecific nitric oxide synthase inhibitor NG-nitro-L-arginine methyl ester on the choroidal compensatory response to myopic defocus in chickens. Optom Vis Sci. 2004;81:111–118. doi: 10.1097/00006324-200402000-00009. [DOI] [PubMed] [Google Scholar]
- Nickla DL, Wilken E, Lytle G, Yom S, Mertz J. Inhibiting the transient choroidal thickening response using the nitric oxide synthase inhibitor l-NAME prevents the ameliorative effects of visual experience on ocular growth in two different visual paradigms. Exp Eye Res. 2006;83:456–464. doi: 10.1016/j.exer.2006.01.029. [DOI] [PubMed] [Google Scholar]
- Norton TT. Animal models of myopia: Learning how vision controls the size of the eye. ILAR J. 1999;40:59–77. doi: 10.1093/ilar.40.2.59. [DOI] [PubMed] [Google Scholar]
- Norton TT, Amedo AO, Siegwart JT., Jr Darkness causes myopia in visually experienced tree shrews. Invest Ophthalmol Vis Sci. 2006a;47:4700–4707. doi: 10.1167/iovs.05-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TT, Amedo AO, Siegwart JT., Jr The effect of age on compensation for a negative lens and recovery from lens-induced myopia in tree shrews (Tupaia glis belangeri) Vision Res. 2010;50:564–576. doi: 10.1016/j.visres.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TT, Essinger JA, McBrien NA. Lid-suture myopia in tree shrews with retinal ganglion cell blockade. Vis Neurosci. 1994;11:143–153. doi: 10.1017/s0952523800011184. [DOI] [PubMed] [Google Scholar]
- Norton TT, McBrien NA. Normal development of refractive state and ocular component dimensions in the tree shrew (Tupaia belangeri) Vision Res. 1992;32:833–842. doi: 10.1016/0042-6989(92)90026-f. [DOI] [PubMed] [Google Scholar]
- Norton TT, Metlapally R, Young TL. Myopia. In: Garner A, Klintworth GK, editors. The Pathobiology of Ocular Disease. New York: Taylor & Francis; 2008. [Google Scholar]
- Norton TT, Siegwart JT. Local myopia produced by partial visual-field deprivation in tree shrew. Soc Neurosci Abstr. 1991;17:558. [Google Scholar]
- Norton TT, Siegwart JT, Jr, Amedo AO. Effectiveness of hyperopic defocus, minimal defocus, or myopic defocus in competition with a myopiagenic stimulus in tree shrew eyes. Invest Ophthalmol Vis Sci. 2006b;47:4687–4699. doi: 10.1167/iovs.05-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TT, Wu WW, Siegwart JT., Jr Refractive state of tree shrew eyes measured with cortical visual evoked potentials. Optom Vis Sci. 2003;80:623–631. doi: 10.1097/00006324-200309000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster CW. The Human Eye. Sunderland, MA: Sinauer Associated, Inc; 1999. [Google Scholar]
- Phillips JR, Khalaj M, McBrien NA. Induced myopia associated with increased scleral creep in chick and tree shrew eyes. Invest Ophthalmol Vis Sci. 2000;41:2028–2034. [PubMed] [Google Scholar]
- Rada JA, Hollaway LR, Lam W, Li N, Napoli JL. Identification of RALDH2 as a visually regulated retinoic acid synthesizing enzyme in the chick choroid. Invest Ophthalmol Vis Sci. 2012;53:1649–1662. doi: 10.1167/iovs.11-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada JA, Huang Y, Rada KG. Identification of choroidal ovotransferrin as a potential ocular growth regulator. Curr Eye Res. 2001;22:121–132. doi: 10.1076/ceyr.22.2.121.5525. [DOI] [PubMed] [Google Scholar]
- Rada JA, Wiechmann AF. Ocular expression of avian thymic hormone: changes during the recovery from induced myopia. Mol Vis. 2009;15:778–792. [PMC free article] [PubMed] [Google Scholar]
- Rada JAS, Wiechmann AF, Hollaway LR, Baggenstoss BA, Weigel PH. Increased hyaluronan synthase-2 mRNA expression and hyaluronan accumulation with choroidal thickening: Response during recovery from induced myopia. Invest Ophthalmol Vis Sci. 2010;51:6172–6179. doi: 10.1167/iovs.10-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer B, Stell WK. Basic fibroblast growth factor (bFGF) and transforming growth factor beta (TGF-β) act as stop and go signals to modulate postnatal ocular growth in the chick. Exp Eye Res. 1994;58:553–561. doi: 10.1006/exer.1994.1049. [DOI] [PubMed] [Google Scholar]
- Salvador-Silva M, Ghosh S, Bertazolli-Filho R, Boatright JH, Nickerson JM, Garwin GG, Saari JC, Coca-Prados M. Retinoid processing proteins in the ocular ciliary epithelium. Mol Vis. 2005;11:356–365. [PubMed] [Google Scholar]
- Schaeffel F, Howland HC. Properties of the feedback loops controlling eye growth and refractive state in the chicken. Vision Res. 1991;31:717–734. doi: 10.1016/0042-6989(91)90011-s. [DOI] [PubMed] [Google Scholar]
- Schippert R, Brand C, Schaeffel F, Feldkaemper MP. Changes in scleral MMP-2, TIMP-2 and TGFβ-2 mRNA expression after imposed myopic and hyperopic defocus in chickens. Exp Eye Res. 2006;82:710–719. doi: 10.1016/j.exer.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Shelton L, Troilo D, Lerner MR, Gusev Y, Brackett DJ, Rada JS. Microarray analysis of choroid/RPE gene expression in marmoset eyes undergoing changes in ocular growth and refraction. Mol Vis. 2008;14:1465–1479. [PMC free article] [PubMed] [Google Scholar]
- Shih YF, Fitzgerald MEC, Norton TT, Gamlin PDR, Hodos W, Reiner A. Reduction in choroidal blood flow occurs in chicks wearing goggles that induce eye growth toward myopia. Curr Eye Res. 1993;12:219–227. doi: 10.3109/02713689308999467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart JT, Norton TT. Goggles for controlling the visual environment of small animals. Lab Animal Sci. 1994;44:292–294. [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. The susceptible period for deprivation-induced myopia in tree shrew. Vision Res. 1998;38:3505–3515. doi: 10.1016/s0042-6989(98)00053-4. [DOI] [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Regulation of the mechanical properties of tree shrew sclera by the visual environment. Vision Res. 1999;39:387–407. doi: 10.1016/s0042-6989(98)00150-3. [DOI] [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Selective regulation of MMP and TIMP mRNA levels in tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol Vis Sci. 2005;46:3484–3492. doi: 10.1167/iovs.05-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Response to interrupted hyperopia after restraint of axial elongation in tree shrews. Optom Vis Sci. 2013;90:131–139. doi: 10.1097/OPX.0b013e31827cda85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P, Feldkaemper M, Bitzer M, Ohngemach S, Schaeffel F. Early transcriptional changes of retinal and choroidal TGFβ-2, RALDH-2, and ZENK following imposed positive and negative defocus in chickens. Mol Vis. 2004;10:588–597. [PubMed] [Google Scholar]
- Smith EL, III, Hung LF, Huang J, Blasdel TL, Humbird TL, Bockhorst KH. Effects of optical defocus on refractive development in monkeys: evidence for local, regionally selective mechanisms. Invest Ophthalmol Vis Sci. 2010;51:3864–3873. doi: 10.1167/iovs.09-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RA, McGlinn AM, Baldwin DA, Tobias JW, Iuvone PM, Khurana TS. Image defocus and altered retinal gene expression in chick: Clues to the pathogenesis of ametropia. Invest Ophthalmol Vis Sci. 2011;52:5765–5777. doi: 10.1167/iovs.10-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers JA. The choroid as a sclera growth regulator. Exp Eye Res. 2013;114:120–127. doi: 10.1016/j.exer.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Buhusi MC, Ma JX, Crouch RK. RPE65 is present in human green/red cones and promotes photopigment regeneration in an in vitro cone cell model. J Neurosci. 2011;31:18618–18626. doi: 10.1523/JNEUROSCI.4265-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troilo D. Myopia and the Control of Eye Growth. New York: Wiley & Sons; 1990. Experimental studies of emmetropization in the chick. [DOI] [PubMed] [Google Scholar]
- Troilo D, Nickla DL, Mertz JR, Summers Rada JA. Change in the synthesis rates of ocular retinoic acid and scleral glycosaminoglycan during experimentally altered eye growth in marmosets. Invest Ophthalmol Vis Sci. 2006;47:1768–1777. doi: 10.1167/iovs.05-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troilo D, Nickla DL, Wildsoet CF. Choroidal thickness changes during altered eye growth and refractive state in a primate. Invest Ophthalmol Vis Sci. 2000;41:1249–1258. [PubMed] [Google Scholar]
- Troilo D, Wallman J. The regulation of eye growth and refractive state: an experimental study of emmetropization. Vision Res. 1991;31:1237–1250. doi: 10.1016/0042-6989(91)90048-a. [DOI] [PubMed] [Google Scholar]
- Wallman J, Wildsoet C, Xu A, Gottlieb MD, Nickla DL, Marran L, Krebs W, Christensen AM. Moving the retina: choroidal modulation of refractive state. Vision Res. 1995;35:37–50. doi: 10.1016/0042-6989(94)e0049-q. [DOI] [PubMed] [Google Scholar]
- Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43:447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Wang CXZ, Zhang K, Aredo B, Lu H, Ufret-Vincenty RL. Novel method for the rapid isolation of RPE cells specifically for RNA extraction and analysis. Exp Eye Res. 2012;102:1–9. doi: 10.1016/j.exer.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildsoet CF, McFadden SA. Optic nerve section does not prevent form deprivation-induced myopia or recovery from it in the mammalian eye. Invest Ophthalmol Vis Sci. 2010;51:ARVO E-Abstract 1737. [Google Scholar]
- Zhang Y, Liu Y, Wildsoet CF. Bidirectional, optical sign-dependent regulation of BMP2 gene expression in chick retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2012;53:6072–6080. doi: 10.1167/iovs.12-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.