Abstract
Purpose
Retinal degenerations are a heterogeneous group of diseases in which there is slow but progressive loss of photoreceptors (PR). There are currently no approved therapies for treating retinal degenerations. In an effort to identify novel small molecules that are (1) neuroprotective and (2) promote PR differentiation, we have developed microscale (1,536 well) cell culture assays using primary retinal neurons.
Methods
Primary murine retinal cells are isolated, seeded, treated with a 1,280 compound chemical library in a 7 point titration and then cultured under conditions developed to assay protection against an introduced stress or enhance PR differentiation. In the protection assays a chemical insult is introduced and viability assessed after 72 h using CellTiterGlo, a single-step chemiluminescent reagent. In the differentiation assay, cells are isolated from the rhodopsin-GFP knock-in mouse and PR differentiation is assessed by fixing cells after 21 days in culture and imaging with the Acumen plate-based laser cytometer (TTP Labtech) to determine number and intensity of GFP-expressing cells. Positive wells are re-imaged at higher resolution with an INCell2000 automated microscope (GE). Concentration-response curves are generated to pharmacologically profile each compound and hits identified by xx.
Results
We have developed PR differentiation and neuroprotection assays with a signal to background (S/B) ratios of 11 and 3, and a coefficient of variation (CV) of 20 and 9 %, suitable for chemical screening. Staurosporine has been shown in our differentiation assay to simultaneously increase the number of rhodopsin positive objects while decreasing the mean rhodopsin intensity and punctate rhodopsin fluorescent objects.
Conclusions
Using primary murine retinal cells, we developed high throughput assays to identify small molecules that influence PR development and survival. By screening multiple compound concentrations, dose-response curves can be generated, and the false negative rate minimized. It is hoped that this work will identify both potential preclinical candidates as well as molecular probes that will be useful for analysis of the molecular mechanisms that promote PR differentiation and survival.
Keywords: Neuroprotection, Photoreceptor, HTS, Screening, High content analysis
97.1 Introduction
The retinal degenerations, the prototype of which is retinitis pigmentosa (RP), are a group of genetically heterogeneous orphan diseases in which there is slow but progressive loss of Photoreceptor (PR) cells, resulting in concomitant loss of vision. The past several decades have witnessed tremendous strides to define many of the genes that when mutated can cause retinal degeneration. To date, more than 150 retinal degeneration related genes have been identified [1]. These findings have led to mechanistic insights and have opened up the possibility of new treatment strategies, such as gene therapy and related treatment approaches [2–6]. Although these avenues are exciting and have great potential, treatment strategies based on a particular gene or mutation have the limitation that even if they are effective, they generally are appropriate for only a small fraction of RP patients. An alternative and complementary approach is to develop so called “neuroprotective” treatments aimed at protecting PRs from degeneration, often independent of the nature of the initiating cellular insult. Neuroprotection may therefore be useful for a larger group of patients, since they may preserve vision in multiple different genetic forms of RP. Theoretically, such approaches could also be effective for the treatment of the “dry” (atrophic) form of age-related macular degeneration.
With the goal of identifying lead compounds for the development of such novel neuroprotective therapies, we have been carrying out phenotypic screens with primary retinal neurons to identify small molecules that promote the survival and/or differentiation of PR cells. Although not the traditional route to contemporary drug discovery, phenotypic screens actually have a well-established track record. Out of the 50 first-in-class small molecules given FDA approval between 1998 and 2008, 28 were discovered using phenotypic approaches and of these, 8 were approved without a known molecular mechanism of action [7].
For our phenotypic screens, we have developed cell-based assays that are compatible with quantitative high throughput screening (qHTS), which by assessing the compound library over multiple concentrations develops a full pharmacokinetic profile for each molecule tested [8]. We are using primary retinal neurons in a high content assay that measures the numbers of cells expressing endogenous levels of a knocked-in human rhodopsin-EGFP fusion protein [9]. For retinal cell survival following oxidative damage, we employ a single-step addition cell viability reagent that is amenable for ultra HTS (greater than 100,000 wells per day).
In addition to their potential as therapeutic lead compounds for further development, it is envisioned that small molecules discovered in this study may also be of use as molecular probes which will be useful for discovering and dissecting the mechanisms of PR development and survival.
97.2 Materials and Methods
97.2.1 Primary Cell Dissociation
All animal procedures were performed according to the guidelines of the ARVO statement for the “Use of Animals in Ophthalmic and Vision Research” and were approved by the Institutional Animal Care and Use Committees at the Johns Hopkins University School of Medicine. Retinas from postnatal day 0 (GFP differentiation assay) or 4 (cell survival) are isolated from homozygous rhodopsin-GFP knock-in mice [9] or C57BL/6 mice, respectively. Animals are sacrificed by hypothermia followed by decapitation, eyes are enucleated, and retinas are dissected. The retinas are dissociated into single cell suspensions by incubation with activated papain in Hibernate-E without Ca2+ (BrainBits) for 15 min at 37 °C. The solution is neutralized by adding Hibernate-E with Ca2+ plus B27 (Invitrogen), L-Glutamine, and Pen/Strep. The cells are then transported on ice to the screening facility.
97.2.2 1536 Well Cell Plating and Compound Library Treatment
Prior to cell plating, 1,536 well plates are filled with 4 µL of neuronal culture medium consisting of Neurobasal-E, B-27, L-Glutamine, pen/strep (all Life Technologies) using a Multidrop Combi peristaltic bulk dispenser (ThermoFisher). Each plate is then “pinned” with 23 nL of a compound at a set stock concentration in DMSO that is delivered from a library plate using a robotic pintool transfer tool (Wako). Cells are resuspended at a concentration of 2.5 × 105 cells/mL, filtered through a 10 µm nylon filter (Amazon.com: Small Parts), dispensed at a final well culture volume of 8 µL (1,000 cells/well), and incubated for up to 21 days. Since we are using such low volumes for extended cultures, the plates are covered with a Breathe-Easy gas-permeable membrane (Diversified Biotech) and a Microclime vapor barrier lid (Labcyte).
97.2.3 Fixation and Imaging
For the differentiation assay, cells are treated with 2 µL of 20 % (4 % final) fresh paraformaldehyde in PBS containing Hoechst 33342 (1:5000 final) for 15 min at room temperature. The plates are then aspirated and rinsed twice with PBS using an EL406 washer dispenser (BioTek Instruments) equipped with an aspirator and syringe dispenser for 1,536 well plates, and imaged with an Acumen Explorer (TTP Labtech) plate cytometer equipped with 405, 488, and 633 nm lasers [10]. PRs are defined as Hoecsht positive objects with size and GFP fluorescence intensity above defined threshold values. “On the fly” analysis allows for virtual ‘cherry-picking’ wells that display fraction of GFP positive cells or GFP intensity values greater or lesser than 3*SD relative to DMSO controls. These wells are then imaged with a microscope-based INCell2000 HCA platform (GE), with triple-channel images for nuclei, GFP, and brightfield acquired for each designated well.
97.2.4 CellTiterGlo Viability Assay
For the neuroprotection assay, following treatment with 0.2 mM paraquat (Sigma), cell cultures are treated with compounds and incubated for 72 h. Plates are then equilibrated at room temperature for 10 min treated with 4 µL CellTiterGlo (Promega), treated for 10 min at room temperature, and then imaged using a Viewlux uHTS CCD-based imager (Perkin Elmer).
97.2.5 Analysis and Curve Fitting
Plate data from GFP positive cell count data, GFP intensity, or viability (raw relative luminescence units) are exported from each respective instrument into .CSV files. Assay statistics are generated by analyzing activity of control compounds, as well as DMSO-treated control plates. The raw data is then normalized using the following formula: where Y = percent activity, x = specific data value, N = median DMSO control value, and P = median positive control value [8]. Following normalization, the data is then fitted with either NIH CurveFit (http://tripod.nih.gov/curvefit), an open source curve fitting and classification software, or with Prism (Graphpad). The data is then ranked according to efficacy, potency, and curve class as defined by Inglese et al. [8].
97.3 Results
97.3.1 Rhodopsin-GFP Count
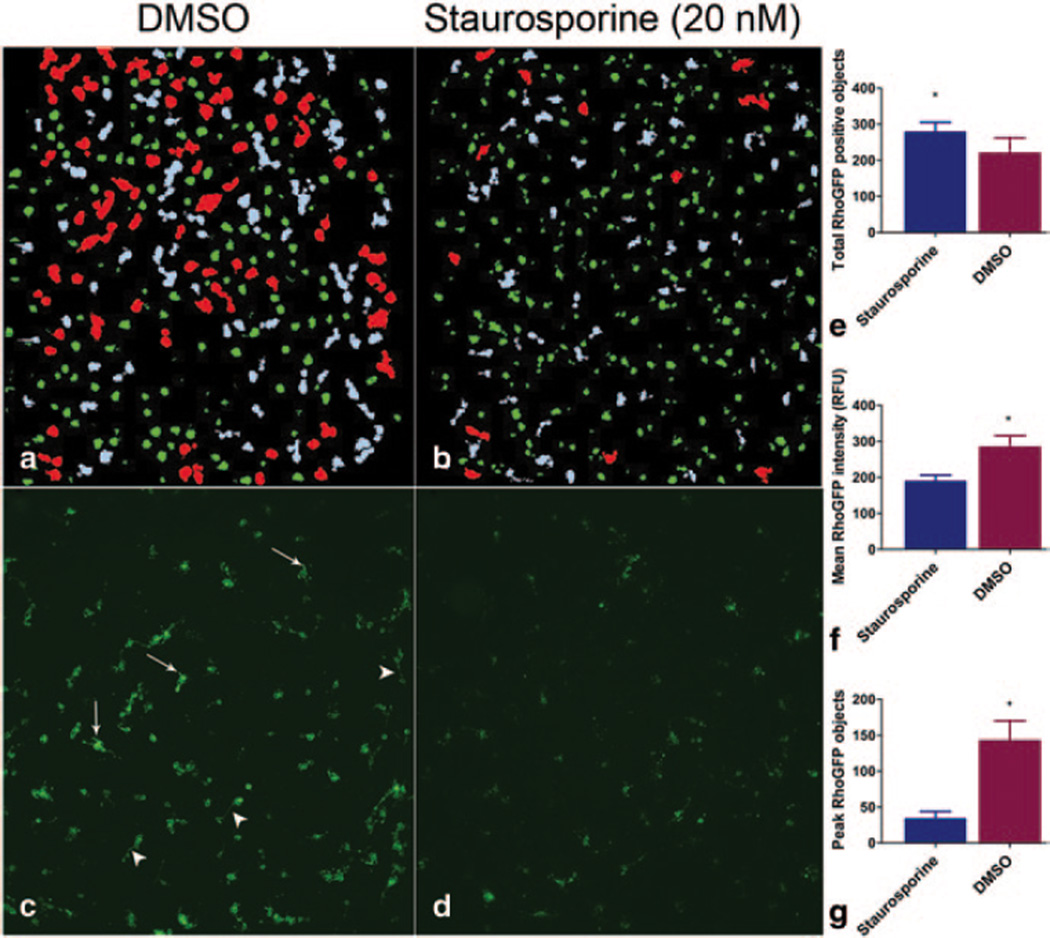
In an effort to observe the effects of small molecules on PR development, we chose to use a fluorescent reporter to follow PR differentiation. The hRhoGFP knock-in mouse was generated by replacing the mouse rhodopsin (rho) open reading frame with a human Rho-GFP fusion construct, thus creating a rhodopsin reporter controlled by native regulatory mechanisms [9]. Since the peak of PR cell genesis in the mouse is near the time of birth of the animal, for the assay we chose to harvest and culture cells from postnatal day 0 retinas. After 21 days of culture, PRs in the culture develop bright GFP fluorescence with bright punctate (‘peak rhodopsin’) objects, believed to be proto-outer segments, that have a signal to background of 11-fold over pre-differentiated RhoGFP cells. As a demonstration of the capability of this screen identifying modulators of PR differentation, we have found that culture in the presence of staurosporine, a broad spectrum kinase inhibitor implicated in rhodopsin expression modulation [11, 12], increases the number of rhodopsin positive cells (Fig 97.1a, c, e). However, the number of cells with peak rhodopsin objects (Fig 97.1f, arrows) is significantly decreased.
Fig. 97.1.
Extended culture of mature RhoGFP photoreceptors in 1536 qHTS format. Primary retinal neurons treated for 21 days with staurosporine (20 nM) show a small increase in total RhoGFP + cells (b: green objects, d, e). However total intensity and objects with a peak GFP area > 1200 RFU are decreased (b: red, f, g) a, b: Acumen GFP (green), ‘peak GFP’ objects (red), objects outside parameters (gray), c, d: corresponding INCell2000 images showing mature photoreceptors with ‘peak’ punctate GFP positive structures (arrows) and neurites (arrowheads). *p < 0.0001 (unpaired t-test)
97.3.2 Cell Titer Viability Screening
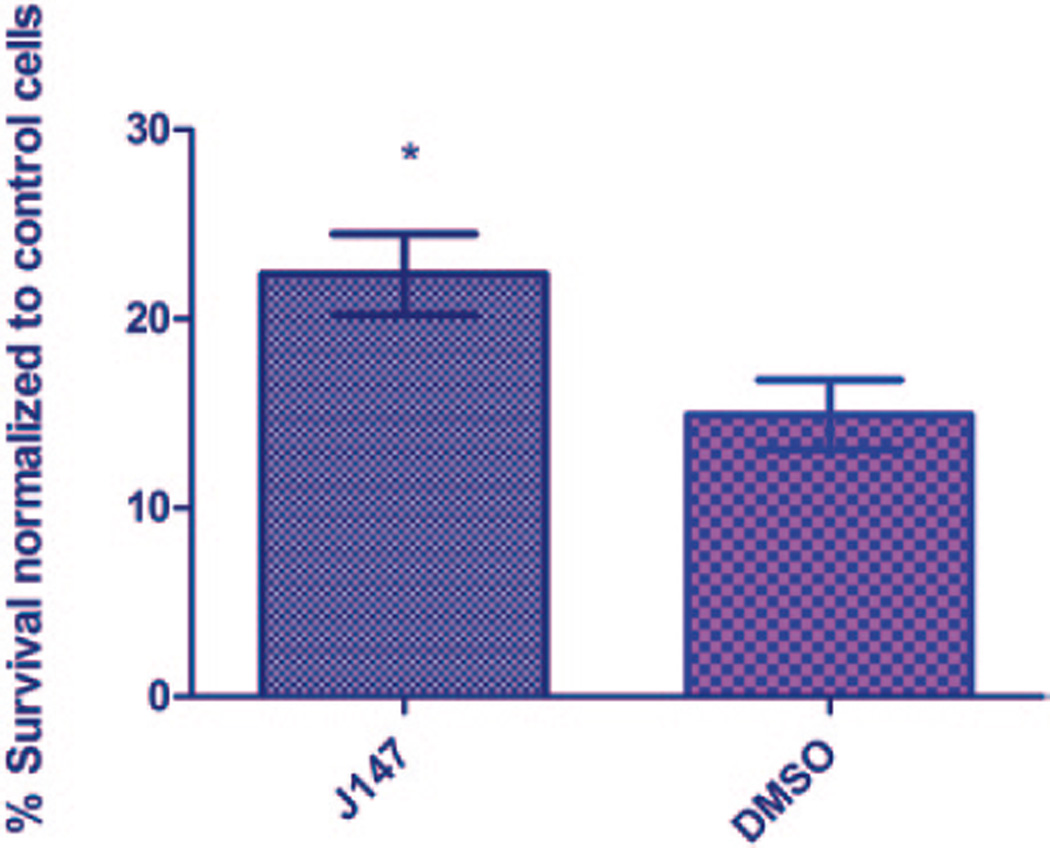
Oxidative and ER stress have been implicated in the pathogenesis of PR degeneration [13–18]. In an effort to find novel PR protective small molecules, we have developed a primary cell-based screen for compounds protective against this type of cell damage. For this assay, the herbicide paraquat, an inducer of both oxidative and ER stress that has been used to model retinal degeneration in vivo, is used to induce acute oxidative damage to retinal cell cultures [13, 19–23]. The cells are treated with 0.2 mM paraquat, a concentration found to induce 75 % cell death in 72 h. Cell survival is assayed at 72 h using CellTiterGlo (Promega), a single-step viability reagent. We have measured assay performance and have found a cell concentration correlation of 95 % and CV below 10 %. The difference between untreated and paraquat-treated cells as has a signal to background ratio of 3 and a Z’ of 0.61. We have found that J147 (Cellagen), a known neuroprotective compound [24], has maximal protective activity at 27 µM, and used as a positive control for this assay (Fig. 97.2).
Fig. 97.2.
Neuroprotection of primary photoreceptors following paraquat-induced oxidative damage with J147, a reported neuroprotective molecule. J147 is a single point positive with activity at 27 µM. *p < 0.0001 (unpaired t-test)
97.4 Discussion
There is a clear need for drug therapies that target vision loss arising from retinal degeneration. Currently, the pharmaceutical and biotechnology sectors struggle with increasing R&D costs and dwindling development pipelines [25]. The so-called preclinical ‘valley of death’ particularly impacts rare disease therapeutic development due to increasing risks incurred in development with a reduced incentive (treatable patient populations). As such, there has been a trend towards increased involvement by academic and government laboratories in developing strategies for treating orphan diseases [26, 27]. One major benefit to this approach with respect to retinal degeneration will be the rapid establishment of new paradigms for discovery by pairing novel assay technologies (e.g. advanced high content imaging systems) with more appropriate models of retinal disease. In our work, rather than focusing on a single target in retinal degeneration, we are striving to find the most potent and efficacious molecules that are neuroprotective and modulate PR development.
High-throughput screening with retinal primary cells has many challenges to overcome. Issues such as day-to-day variability in cell preparation, variability in animal litters, and stochastic effects within each well can have tremendous effects on culture conditions. We have found that cells take at least 6 days to produce a measurable response with respect to RhoGFP numbers and rhodopsin intensity. In typical, 1,536 well assays in this time frame would be unacceptable due to variation arising from plate-edge and evaporation effects. However by taking precautions to mitigate edge and evaporation effects, we can consistently culture cells well over a period of three weeks under conditions in which the cells develop in a fashion that mirrors in vivo PR development.
Autofluorescence in the GFP emission spectrum is a major challenge for fluorescent protein-based reporter assay systems as many molecules in compound libraries are also fluorescent in the FITC spectrum. To distinguish false positives, we have employed a far-red conjugated primary antibody to stain positive wells after primary GFP object identification. However, high throughput immunostaining in a large screening campaign can be expensive and labor intensive. An alternative approach would be to develop a consensus biocircuit that employs nonhomologous reporters separated by a 2A polycistronic peptide [28].
In this study, we have used primary retinal neurons in high-throughput format for studying small molecule effects on PR differentiation and survival. In addition to their potential as therapeutic leads, we believe that bioactive molecules identified though our ongoing screens will have applicability as molecular probes that are involved in retinal cell biology will serve as useful reagents for future studies.
Acknowledgment
We thank John Wilson and Ted Wensel (Baylor College of Medicine) for generously providing Rho-GFP mice, and Patricia Dranchak, Sam Hasson, and Ryan MacArthur of the NIH NCATS DPI for their help and assistance. This work was funded by an FFB/Wynn-Gund Translational Research Acceleration Program Award.
List of Abbreviations
- DMSO
Dimethyl sulfoxide
- ER
Endoplasmic reticulum
- GFP
Green fluorescent protein
- HCA
High content analysis
- qHTS
Quantitative high throughput screening
- PBS
Phosphate buffered saline
- PR
Photoreceptor
- RP
Retinitis pigmentosa
Contributor Information
John A. Fuller, Email: jfulle19@jhmi.edu, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA.
Gillian C. Shaw, Email: gshaw6@jhmi.edu, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA.
Delphine Bonnet-Wersinger, Email: delphine.bonnet@inserm.fr, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA.
Baranda S. Hansen, Email: bhansen2@jhmi.edu, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA.
Cynthia A. Berlinicke, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA
James Inglese, Email: jinglese@mail.nih.gov, National Center for Advancing Translational Sciences, NIH, Rockville, MD, USA; National Human Genome Institute, NIH, Bethesda, MD, USA.
Donald J. Zack, Email: dzack@jhmi.edu, Wilmer Eye Institute, Johns Hopkins University School of Medicine, 400 N Broadway, Baltimore, MD 21231, USA; Departments of Molecular Biology and Genetics, Neuroscience, and Institute of Genetic Medicine, The Johns Hopkins University School of Medicine, Baltimore, MD, USA; Institut de la Vision, Paris 75012, France.
References
- 1.RetNet: Retinal Information Network. https://sph.uth.edu/retnet/
- 2.Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, Pang JJ, Sumaroka A, Windsor EA, Wilson JM, Flotte TR, Fishman GA, Heon E, Stone EM, Byrne BJ, Jacobson SG, Hauswirth WW. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Nat Acad Sci USA. 2008;105(39):15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett PJ. Mathematical modeling of triamcinolone acetonide drug release from the I-vation intravitreal implant (a controlled release platform) Conference proceedings: annual international conference of the IEEE engineering in medicine and biology society IEEE engineering in medicine and biology society conference. 2009;2009:3087–3090. doi: 10.1109/IEMBS.2009.5332803. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson SG, Cideciyan AV, Ratnakaram R, Heon E, Schwartz SB, Roman AJ, Peden MC, Aleman TS, Boye SL, Sumaroka A, Conlon TJ, Calcedo R, Pang JJ, Erger KE, Olivares MB, Mullins CL, Swider M, Kaushal S, Feuer WJ, Iannaccone A, Fishman GA, Stone EM, Byrne BJ, Hauswirth WW. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch Ophthalmol. 2012;130(1):9–24. doi: 10.1001/archophthalmol.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell’Osso L, Hertle R, Ma JX, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N England J Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sieving PA, Caruso RC, Tao W, Coleman HR, Thompson DJ, Fullmer KR, Bush RA. Ciliary neurotrophic factor (CNTF) for human retinal degeneration: phase I trial of CNTF delivered by encapsulated cell intraocular implants. Proc Nat Acad Sci USA. 2006;103(10):3896–3901. doi: 10.1073/pnas.0600236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 8.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Nat Acad Sci USA. 2006;103(31):11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan F, Bradley A, Wensel TG, Wilson JH. Knock-in human rhodopsin-GFP fusions as mouse models for human disease and targets for gene therapy. Proc Nat Acad Sci USA. 2004;101(24):9109–9114. doi: 10.1073/pnas.0403149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen WP, Wylie PG. Application of laser-scanning fluorescence microplate cytometry in high content screening. Assay Drug Dev Technol. 2006;4(2):209–221. doi: 10.1089/adt.2006.4.209. [DOI] [PubMed] [Google Scholar]
- 11.Xie HQ, Adler R. Green cone opsin and rhodopsin regulation by CNTF and staurosporine in cultured chick photoreceptors. Invest Ophthalmol Vis Sci. 2000;41(13):4317–4323. [PubMed] [Google Scholar]
- 12.Bradford RL, Wang C, Zack DJ, Adler R. Roles of cell-intrinsic and microenvironmental factors in photoreceptor cell differentiation. Dev Biol. 2005;286(1):31–45. doi: 10.1016/j.ydbio.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani C, Rogers B, Lu L, Kachi S, Shen J, Campochiaro PA. Retinal degeneration from oxidative damage. Free Radical Biol Med. 2006;40(4):660–669. doi: 10.1016/j.freeradbiomed.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Hackam AS, Strom R, Liu D, Qian J, Wang C, Otteson D, Gunatilaka T, Farkas RH, Chowers I, Kageyama M, Leveillard T, Sahel JA, Campochiaro PA, Parmigiani G, Zack DJ. Identification of gene expression changes associated with the progression of retinal degeneration in the rd1 mouse. Invest Ophthalmol Vis Sci. 2004;45(9):2929–2942. doi: 10.1167/iovs.03-1184. [DOI] [PubMed] [Google Scholar]
- 15.Komeima K, Rogers BS, Campochiaro PA. Antioxidants slow photoreceptor cell death in mouse models of retinitis pigmentosa. J Cell Physiol. 2007;213(3):809–815. doi: 10.1002/jcp.21152. [DOI] [PubMed] [Google Scholar]
- 16.Punzo C, Xiong W, Cepko CL. Loss of daylight vision in retinal degeneration: are oxidative stress and metabolic dysregulation to blame? J Biol Chem. 2012;287(3):1642–1648. doi: 10.1074/jbc.R111.304428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui S, Oveson BC, Lee SY, Jo YJ, Yoshida T, Miki A, Miki K, Iwase T, Lu L, Campochiaro PA. NADPH oxidase plays a central role in cone cell death in retinitis pigmentosa. J Neurochem. 2009;110(3):1028–1037. doi: 10.1111/j.1471-4159.2009.06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Usui S, Oveson BC, Iwase T, Lu L, Lee SY, Jo YJ, Wu Z, Choi EY, Samulski RJ, Campochiaro PA. Overexpression of SOD in retina: need for increase in H2O2-detoxifying enzyme in same cellular compartment. Free Radical Biol Med. 2011;51(7):1347–1354. doi: 10.1016/j.freeradbiomed.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinta SJ, Rane A, Poksay KS, Bredesen DE, Andersen JK, Rao RV. Coupling endoplasmic reticulum stress to the cell death program in dopaminergic cells: effect of paraquat. Neuromolecular Med. 2008;10(4):333–342. doi: 10.1007/s12017-008-8047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CL, Lee YC, Yang YC, Kuo TY, Huang NK. Minocycline prevents paraquat-induced cell death through attenuating endoplasmic reticulum stress and mitochondrial dysfunction. Toxicol Lett. 2012;209(3):203–210. doi: 10.1016/j.toxlet.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Singh BK, Ahmad I, Shukla S, Patel DK, Srivastava G, Kumar V, Pandey HP, Singh C. Involvement of NADPH oxidase and glutathione in zinc-induced dopaminergic neurodegeneration in rats: similarity with paraquat neurotoxicity. Brain Res. 2012;1438:48–64. doi: 10.1016/j.brainres.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 22.Somayajulu-Nitu M, Sandhu JK, Cohen J, Sikorska M, Sridhar TS, Matei A, Borowy-Borowski H, Pandey S. Paraquat induces oxidative stress, neuronal loss in substantia nigra region and parkinsonism in adult rats: neuroprotection and amelioration of symptoms by water-soluble formulation of coenzyme Q10. BMC Neurosci. 2009;10:88. doi: 10.1186/1471-2202-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Tiffany-Castiglioni E, Koh HC, Son IH. Paraquat activates the IRE1/ASK1/JNK cascade associated with apoptosis in human neuroblastoma SH-SY5Y cells. Toxicology Lett. 2009;191(2–3):203–210. doi: 10.1016/j.toxlet.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, Prior M, Dargusch R, Roberts A, Riek R, Eichmann C, Chiruta C, Akaishi T, Abe K, Maher PA. Novel neurotrophic drug for cognitive enhancement and Alzheimer’s disease. PloS one. 2011;6(12):e27865. doi: 10.1371/journal.pone.0027865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silber BM. Driving drug discovery: the fundamental role of academic labs. Sci Transl Med. 2010;2(30):30cm16. doi: 10.1126/scitranslmed.3000169. [DOI] [PubMed] [Google Scholar]
- 26.Reed JC, White EL, Aubé J, Lindsley C, Li M, Sklar L, Schreiber S. The NIH’s role in accelerating translational sciences. Nat Biotechnol. 2012;30(1):16–19. doi: 10.1038/nbt.2087. [DOI] [PubMed] [Google Scholar]
- 27.Frearson J, Wyatt P. Drug discovery in academia: the third way? Expert Opin Drug Discov. 2010;5(10):909–919. doi: 10.1517/17460441.2010.506508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng KCC, Inglese J. A coincidence reporter-gene system for high-throughput screening. Nat Methods. 2012;9(10):937. doi: 10.1038/nmeth.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]