Abstract
The smeT-smeDEF region and the smeT gene, which encodes the smeDEF repressor, are highly polymorphic. Few changes in smeT might be associated with smeDEF overexpression. The results obtained with cellular extracts suggest that mutant SmeT proteins cannot bind to the operator and that other transcription factors besides SmeT are involved in the regulation of smeDEF expression.
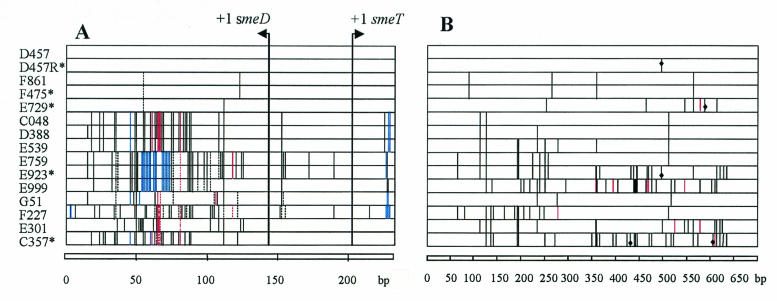
The expression of the Stenotrophomonas maltophilia multidrug resistance (MDR) pump SmeDEF is transcriptionally regulated by SmeT (12), a local repressor encoded by the smeT gene, which is located upstream of smeD and which is divergently transcribed. SmeT binds to the intergenic smeT-smeD region, where the promoters of smeT and smeD are located. It has previously been found that a mutation in smeT is responsible for smeDEF overproduction in MDR strain S. maltophilia D457R (1, 12). However, nothing is known about the molecular basis of smeDEF overproduction in clinical S. maltophilia isolates. For that goal, the intergenic smeT-smeD region, which contains both the smeT and smeD promoters, as well as the smeT gene, were cloned from a collection of clinical S. maltophilia strains, 33% of which were SmeDEF overproducers (2), by PCR and sequenced as described previously (12). The strains used in this work are listed in Table 1. The intergenic region and the smeT gene were highly polymorphic (Fig. 1). However, only some changes might account for smeDEF overproduction. Most nucleotide changes were located outside of the region between the transcriptional origins of smeT and smeD, which suggests that their operator sequences are located between the smeD-smeT transcriptional origins. Only one smeDEF-overproducing strain (strain E923) contained modifications within the smeT-smeD region compared with the sequence of wild-type strain D457. The nucleotide changes in E923 were exactly the same as those in antibiotic-susceptible strain E759. Thus, those changes are not associated with the smeDEF-overproducing phenotype.
TABLE 1.
Bacterial strains
| Strain | Description | Reference or source |
|---|---|---|
| E. coli M15(pPS6) | E. coli strain that expresses His-tagged SmeT | 12 |
| S. maltophilia | ||
| D457 | Bronchial aspirate isolate (January 1992) | 1, 3 |
| D457R | Single-step spontaneous SmeDEF-overproducing mutant derived from D457 | 1, 3 |
| C048 | Bronchial aspirate isolate (November 1990) | 2 |
| C357 | Urinary isolate (March 1991) | 2 |
| D388 | Urinary isolate (December 1991) | 2 |
| E301 | Urinary isolate (October 1992) | 2 |
| E539 | Infected wound isolate (January 1993) | 2 |
| E729 | Urinary isolate (March 1993) | 2 |
| E759 | Sputum isolate (March 1993) | 2 |
| E923 | Sputum isolate (June 1993) | 2 |
| E999 | Respiratory secretion isolate (July 1993) | 2 |
| F227 | Blood culture isolate (November 1993) | 2 |
| F375 | Blood culture isolate (January 1994) | 2 |
| F861 | Sputum isolate (August 1994) | 2 |
| G51 | Blood culture isolate (November 1994) | 2 |
FIG. 1.
Schematic representation of the intergenic smeD-smeT region (A) and the smeT gene (B). MDR strains are marked with an asterisk. Single base substitutions are represented in black. Continuous, dashed, and dashed-dotted lines indicate different base substitutions in a given position. Base insertions are highlighted in red. Base deletions are highlighted in blue. The diamonds in panel B represent mutations producing a nonconservative amino acid change that can be involved in the MDR phenotype (see text and Table 2).
Most nucleotide changes in smeT did not render changes in its amino acid sequence. Furthermore, most amino acid changes in the smeDEF-overproducing strains were also present in some wild-type strains (Table 2). Six amino acid changes (underlined in Table 2) were exclusively found in the MDR strains and thus might be responsible for the MDR phenotype. Two MDR strains had one change each: clinical isolate E729 had a Thr197Pro substitution and in vitro mutant D457R had a Leu166Gln change. Strain E923 had two amino acid changes: the Leu166Gln substitution observed in D457R and another Arg123Lys change. Finally, strain C357 had four changes: Arg123Lys (found in E923), Leu144Pro, Arg148Gln, and Ala204Glu. SmeDEF-nonoverproducing strain E999 contained a similar substitution at position 148, the change being in this case Arg148Lys. Only four amino acid changes (boldface in Table 2) were nonconservative: Leu144Pro, Leu166Gln, Thr197Pro, and Ala204Glu. All of the changes were clustered in the carboxylic region of the SmeT protein, suggesting a relevant role of the carboxy terminus of SmeT in its function.
TABLE 2.
Amino acid changes in SmeT proteins of clinical S. maltophilia isolates
| Strain | Amino acid changea
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V30 | V63 | D80 | M93 | N121 | R123 | M128 | A136 | L144 | R148 | L166 | E182 | T197 | A204 | |
| D457 | ||||||||||||||
| D457Rb | Q | |||||||||||||
| F861 | L | D | ||||||||||||
| F375b | L | D | ||||||||||||
| E729b | L | D | D | P | ||||||||||
| C048 | ||||||||||||||
| D388 | ||||||||||||||
| E539 | ||||||||||||||
| E759 | E | |||||||||||||
| E923b | E | D | K | Q | ||||||||||
| E999 | D | L | G | K | D | |||||||||
| G51 | L | |||||||||||||
| F227 | T | E | ||||||||||||
| E301 | D | D | ||||||||||||
| C357b | D | K | P | Q | E | |||||||||
The changes that are found only in smeDEF-overproducing strains are underlined. Nonhomologous changes are highlighted in bold.
Strains that overexpress smeDEF. Data are from reference 2.
The sequences of smeT and the intergenic smeT-smeD region were exactly the same as those of strains F861 (wild type) and F375 (smeDEF overproducer). Thus, smeDEF overproduction in F375 is the consequence of mutations in other loci, and factors other than SmeT must be involved in the regulation of smeDEF. A similar situation was described in nalC Pseudomonas aeruginosa mutants that overexpress the mexAB-oprM multidrug efflux pump but that do not have mutations in the mexR gene encoding its local transcriptional repressor (13). Increasing evidence supports the idea that the regulation of MDR pump expression is complex (6). One of the clearest examples of this is the regulation of acrAB expression in Escherichia coli, in which several transcription factors are involved (10, 11). So, it seems that MDR pumps expression needs to be finely tuned, probably in response to different environmental inputs.
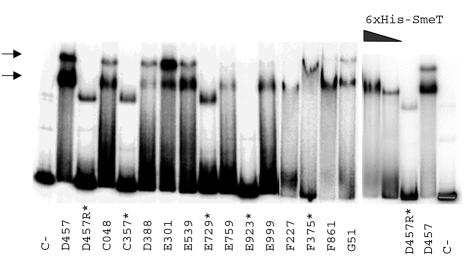
Using whole-cell extracts and a purified His-tagged SmeT protein, we showed that the wild-type SmeT protein binds to the intergenic smeT-smeD region, whereas the Leu166Gln SmeT mutant was incapable of such binding (12). Although the intergenic smeT-smeD region is polymorphic in our clinical isolates, the region between the transcriptional origins of smeT and smeD is conserved. Since this region comprises the operator sequences of smeT and smeD and, most probably, the binding sites of SmeT, we analyzed wild-type strain S. maltophilia D457, one of our clinical isolates, for the presence of cellular factors capable of binding to the intergenic smeT-smeD region. As shown in Fig. 2, all isolates that did not overproduce smeDEF had a protein(s) that was able to bind to the promoter region of the operon in the same way as the wild-type strain D457 does. The retarded complex was of the same size as that obtained by using a His-tagged recombinant SmeT protein. On the other hand, the MDR strains with mutations in SmeT presented the same pattern as D457R, an MDR mutant obtained in the laboratory. This indicates that mutant SmeT proteins of these strains are unable to bind to the intergenic smeT-smeD region. We did find, however, that cellular extracts from MDR strain F375 were able to bind to the intergenic smeT-smeD region. These data agree with the fact that SmeT in this strain did not have any relevant amino acid change. Since in all cases mutations map out of the HTH motif, an explanation for the lack of SmeT activity might be a reduced stability of the protein. By using an anti-SmeT antibody obtained in our laboratory, it has been determined by Western blotting (3) that the amount of SmeT was variable among these clinical isolates; however, none of the MDR mutants presented lower SmeT levels than wild-type strains (data not shown), indicating that the quantity of SmeT is not the limiting factor in the MDR phenotype.
FIG. 2.
Extracts obtained from wild-type S. maltophilia strains are capable of retarding the intergenic smeT-smeD region. Two retarding complexes (marked with arrows) were detected in the case of extracts from the wild-type strains. These complexes were not detected when extracts from MDR strains, (marked with an asterisk) were assayed. A band smaller than that corresponding to the SmeT-DNA complex was observed in the band shifts by using cellular extracts from the smeDEF-overproducing mutants. With the available data, it difficult to know whether this band is nonspecific or whether it is the consequence of the binding of a cellular factor required for smeDEF transcription. Cellular extracts from E. coli M15(pPS6), which contains a His-tagged SmeT protein able to bind to and retard the intergenic smeT-smeD region (12), were used as controls. The band shift obtained with this recombinant SmeT protein is dose dependent and is of the same size as one of the bands obtained with extracts from S. maltophilia D457. 6×His, six-His tag.
Few data comparing sequences from different S. maltophilia strains are available; however, a high degree of diversity similar to that observed in this work was previously reported for beta-lactamases (5) and the topoisomerase II and IV quinolone resistance-determining regions (14). Bacterial diversity and evolution can be driven by mutation (8) and recombination (7, 9). Data from our work and others indicate that mutation might have a relevant role in the evolution of antibiotic resistance in S. maltophilia. However, comparison of the DNA sequences comprising smeT and the intergenic smeT-smeD region in strains E759, E923, and E999 has shown that they have a mosaic structure, with some completely conserved regions and other divergent regions, with very clear boundaries between them. Previous work in our laboratory has shown the presence of genes in the genome of S. maltophilia that originated from gram-positive organisms (4). Together, these data indicate that recombination should also have a relevant role in the evolution of S. maltophilia.
Acknowledgments
We thank Fernando Rojo for fruitful discussions and suggestions on this work.
The research in our laboratory is aided by grants QLRT-2000-1339, QLRT-2000-00873, BIO2001-1081, CAM08.2/0020-1/2001, and GEN2001/4689/C05.
REFERENCES
- 1.Alonso, A., and J. L. Martínez. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, A., and J. L. Martinez. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, A., and J. L. Martinez. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44:3079-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, A., P. Sanchez, and J. L. Martinez. 2000. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob. Agents Chemother. 44:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence, J. G. 1999. Gene transfer, speciation, and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2:519-523. [DOI] [PubMed] [Google Scholar]
- 8.Martinez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 10.Rahmati, S., S. Yang, A. L. Davidson, and E. L. Zechiedrich. 2002. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 43:677-685. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez, P., A. Alonso, and J. L. Martinez. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 46:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valdezate, S., A. Vindel, A. Echeita, F. Baquero, and R. Canton. 2002. Topoisomerase II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob. Agents Chemother. 46:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]