Abstract
Klebsiella pneumoniae strain KG525, which showed high-level resistance to broad-spectrum cephalosporins, was isolated from the neonatal intensive care unit (NICU) of a Japanese hospital in March 2002. The ceftazidime resistance of strain KG525 was transferable to Escherichia coli CSH-2 by conjugation. Cloning and sequence analysis revealed that production of a novel extended-spectrum class A β-lactamase (pI 7.0), designated GES-3, which had two amino acid substitutions of M62T and E104K on the basis of the sequence of GES-1, was responsible for resistance in strain KG525 and its transconjugant. The blaGES-3 gene was located as the first gene cassette in a class 1 integron that also contained an aacA1-orfG fused gene cassette and one unique cassette that has not been described in other class 1 integrons and ended with a truncated 3′ conserved segment by insertion of IS26. Another five ceftazidime-resistant K. pneumoniae strains, strains KG914, KG1116, KG545, KG502, and KG827, which were isolated from different neonates during a 1-year period in the same NICU where strain KG525 had been isolated, were also positive for GES-type β-lactamase genes by PCR. Pulsed-field gel electrophoresis and enterobacterial repetitive intergenic consensus-PCR analyses displayed genetic relatedness among the six K. pneumoniae strains. Southern hybridization analysis with a GES-type β-lactamase gene-specific probe showed that the locations of blaGES were multiple and diverse among the six strains. These findings suggest that within the NICU setting genetically related K. pneumoniae strains carrying the blaGES gene were ambushed with genetic rearrangements that caused the multiplication and translocation of the blaGES gene.
Resistance to β-lactam antibiotics mainly depends on the production of β-lactamases. To date, a large variety of β-lactamases which were classified by their amino acid sequences and functional substrate specificity profiles in various gram-negative bacilli such as Pseudomonas spp. and members of the family Enterobacteriaceae have been documented (6). Since the late 1980s, extended-spectrum β-lactamases (ESBLs) derived from TEM- and SHV-type penicillinases capable of hydrolyzing the oxymino-cephalosporins have been spreading globally, mainly in the Enterobacteriaceae, including Klebsiella pneumoniae and Escherichia coli (5, 23, 29). Moreover, various non-TEM-, non-SHV-type class A β-lactamases exhibiting extended-spectrum activities, including CTX-M-type (13, 31, 38, 39, 41), SFO-type (18), VEB-type (12, 20, 25), and GES-type (10, 11, 19, 24, 28, 37) β-lactamases, have also been reported in various gram-negative bacilli. Among the GES-type β-lactamases, GES-1, which was found to be produced by K. pneumoniae ORI-1, identified from a child transferred from French Guiana to France in 1998, was the first report of the GES-type class A β-lactamase (24); and GES-1-producing K. pneumoniae strains have caused nosocomial infections in Portugal (9). IBC-1 was identified in an Enterobacter cloacae clinical isolate from Greece in 1999 (11), and IBC-1-producing E. cloacae has also been reported to cause nosocomial infections in a neonatal intensive care unit (NICU) (17). GES-2, which displayed more extended-spectrum activity against imipenem compared with that of GES-1, was reported in Pseudomonas aeruginosa from South Africa (28) in 2000, and GES-2 producers also caused a nosocomial infection (27). All three genes, blaGES-1, blaGES-2, and blaIBC-1, were found to be located as a gene cassette within similar class 1 integrons.
Recently, six clinical isolates of K. pneumoniae showing high-level resistance to various broad-spectrum cephalosporins, including ceftazidime, were identified from the NICU of a Japanese hospital, and conventional PCR analyses for TEM-derived ESBLs and CTX-M enzymes failed to specify their genetic determinants. In the present study, therefore, we characterized the molecular mechanism underlying the multiple-cephalosporin resistance among these six strains, as well as the organizations of their genetic environments.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Six K. pneumoniae clinical strains had been isolated from neonates over 1 year, from September 2001 to August 2002, and were stored in the clinical microbiology laboratory of the hospital until this study. Biochemical phenotypic identification of these strains was carried out by the analytical profile index procedure (API 20E system; bioMerieux, Marcy l'Etoile, France). A preliminary double-disk synergy test was carried out with disks containing ceftazidime and amoxicillin-clavulanate. Bacteria were grown in Luria-Bertani (LB) broth supplemented with appropriate antibiotics, unless specified otherwise.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or Reference |
|---|---|---|
| K. pneumoniae KG914, KG1116, KG525, KG545, KG502, KG827 | Clinical isolates from neonatal specimens | This study |
| E. coli | ||
| CSH-2 | metB F− nalidixic acidr rifampinr | T. Sawai, Chiba University |
| XL1-Blue | supE44 recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 lac [F−proAB+ lac1qZΔM15::Tn10(Tetr)] | Stratagene |
| BL21(DE3)pLysS | F−ompT hsdSB (rB−mB−) gal dcm (DE3) pLysS (Camr) | Invitrogen |
| Plasmids | ||
| pKGC525 | A natural plasmid carrying blaGES-3 of K. pneumoniae KG525 | This study |
| pKGB525 | A recombinant plasmid carrying a 6.7-kb BamHI fragment containing blaGES-3 of K. pneumoniae KG525 | This study |
| pKGM525 | A recombinant plasmid carrying a 11.6-kb BamHI fragment containing blaGES-3 of K. pneumoniae KG525 | This study |
| pTAGES3 | A recombinant plasmid carrying a PCR fragment with the entire blaGES-3 sequence and its promoter region cloned into the pCR2.1 vector | This study |
| pGES3 | A recombinant plasmid carrying EcoR1 fragment from pTAGES3 | This study |
| pIBC1 | A recombinant plamid carrying blaIBC-1 constructed from pGES3 | This study |
| pBCSK+ | A cloning vector; chloramphenicolr | Stratagene |
| pCR2.1 | A cloning vector; ampicillinr kanamycinr | Invitrogen |
| pET29a(+) | An expression vector; kanamycinr | Novagen |
| pET-GES3 | A recombinant plasmid carrying PCR-amplified blaGES-3 gene ligated to pET29a(+) | This study |
Antibiotic susceptibility testing.
The following antibiotics were obtained from the indicated sources: ampicillin, amoxicillin, and cefminox, Meiji Seika Kaisha, Ltd., Tokyo, Japan; piperacillin, Toyama Chemical Co., Ltd., Toyama, Japan; cephaloridine and moxalactam, Shionogi & Co., Ltd., Osaka, Japan; cefmetazole and chloramphenicol, Sankyo Co., Ltd., Tokyo, Japan; cefotaxime and cefpirome, Aventis Pharma, Ltd., Tokyo, Japan; ceftazidime and clavulanic acid, GlaxoSmithKline K. K., Tokyo, Japan; sulbactam, Pfizer Pharmaceutical Inc., Tokyo, Japan; tazobactam, Taiho Pharmaceutical Co., Ltd., Tokyo, Japan; cefepime, Bristol Pharmaceuticals K. K., Tokyo, Japan; aztreonam, Eizai Co., Ltd., Tokyo, Japan; imipenem, Banyu Pharmaceutical Co., Ltd., Tokyo Japan; and rifampin, Daiichi Pharamaceutical Co., Ltd., Tokyo, Japan. The MICs of the β-lactams were determined by the agar dilution method, according to the recommendations of National Committee for Clinical Laboratory Standards document M7-A5 (21). E. coli ATCC 25922 and ATCC 35218 were purchased from the American Type Culture Collection (ATCC) and served as control strains in the antimicrobial susceptibility testing.
PCR amplification.
To amplify the broad-spectrum β-lactamase genes from the six clinical strains, PCR analyses were performed with sets of primers specific for various β-lactamase genes found in Japan—including the TEM-derived extended-spectrum β-lactamase (39); CMY-2-, MOX-1-, and DHA-1-type βlactamases (8, 40, 41); and CTX-M-1-, CTX-M-2-, CTX-M-9-, IMP-1-, IMP-2-, and VIM-2-type β-lactamases (13, 26, 30, 31, 33, 39)—under the conditions described elsewhere (33). Detection of the SHV-type β-lactamase gene was not performed because most clinical K. pneumoniae strains carry the LEN-1 and/or SHV-1 β-lactamase gene on their chromosomes (1, 7). In order to detect the GES-type β-lactamase gene, an 827-bp internal fragment of the gene was amplified with primers GES-A (5′-CTT CAT TCA CGC ACT ATT AC-3′) and GES-B (5′-TAA CTT GAC CGA CAG AGG-3′) under the conditions described above.
Conjugal transfer of β-lactam resistance.
Conjugal transfer of the ceftazidime resistance of K. pneumoniae KG525 to a recipient E. coli strain, strain CSH-2 (F− metB, resistant to nalidixic acid and rifampin), was performed by the filter mating method. Transconjugants were selected on LB agar plates containing ceftazidime (2 μg/ml), rifampin (100 μg/ml), and nalidixic acid (50 μg/ml).
Cloning experiment and DNA sequencing.
Basic recombinant DNA techniques were performed as described by Sambrook et al. (32). Total DNA of K. pneumoniae KG525 was extracted and digested with BamHI. The resultant fragments were ligated into cloning vector pBCSK+ (Stratagene, La Jolla, Calif.) restricted with the same enzyme. Transformants were selected on LB agar plates containing chloramphenicol (30 μg/ml) and ampicillin (50 μg/ml) or ceftazidime (2 μg/ml). The nucleotide sequence of the cloned fragment was determined with BigDye terminator cycle Sequencing Ready Reaction kits and an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, Calif.) by using custom sequencing primers.
Site-directed mutagenesis for comparison of GES-3 with IBC-1.
PCR-based site-directed mutagenesis of the blaGES-3 gene was performed with the LA PCR In Vitro Mutagenesis kit (TAKARA Bio Inc., Ohtsu, Japan). In brief, the entire blaGES-3 gene and its promoter region were amplified by PCR and cloned into plasmid pCR2.1 with the TA cloning kit (Invitrogen, NV, Leek, The Netherlands). One plasmid, pTAGES3, was selected after it was confirmed that it contained no amplification error and was then digested with EcoRI. The resultant fragment was recloned into pBCSK+. The resultant plasmid, pGES3, with an insert carrying the blaGES-3 gene and its promoter region was used to introduce a single nucleotide mutation (C to T) at nucleotide position 167, which leads to an amino acid substitution (T to M) at position 62 in GES-3, resulting in the conversion of the gene product from GES-3 to IBC-1 expressed under the same promoter.
Pulsed-field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensus (ERIC)-PCR analyses.
Total DNA was prepared from six K. pneumoniae strains (34) and digested overnight with XbaI (New England Biolabs, Beverly, Mass.). The digested DNA was electrophoresed with a CHEF-DRII Drive Module (Bio-Rad Laboratories, Hercules, Calif.) under the following conditions: pulses ranging from 10 to 40 s at 6 V/cm for 20 h at 16°C. Six K. pneumoniae strains were also typed with the primer ERIC-2 (5′-AAG TAA GTG ACT GGG GTG AGC G-3′). The PCR was carried out under the conditions described elsewhere (36).
Southern hybridization.
Large plasmids were prepared from six K. pneumoniae strains by the procedure described by Kado and Liu (16). The chromosomal DNA was extracted from each isolate by the method of Stauffer et al. (35). Both plasmid and chromosomal DNA preparations were separately subjected to Southern hybridization experiments. The 827-bp DNA probes were amplified by a PCR with primers 5′-CTT CAT TCA CGC ACT ATT AC-3′ and 5′-TAA CTT GAC CGA CAG AGG-3′. The PCR amplicons were labeled with digoxigenin (DIG) by a random priming labeling method with the PCR DIG detection system, as recommended by the manufacturer (Roche Diagnostics, Tokyo, Japan). Southern hybridization was performed by the protocol of the manufacturer (Roche Diagnostics).
Purification of GES-3 β-lactamase.
To overproduce GES-3 β-lactamase in E. coli, the blaGES-3 gene was amplified by using two primers, primer GES-F (5′-CAT ATG CGC TTC ATT CAC GCA CTA TTA CTG-3′), which was designed to add an NdeI linker (underlined), and primer GES-R (5′-GTC GAC CTA TTT GTC CGT GCT CAG GAT GAG-3′), which was designed to add an SalI linker (underlined), and DNA polymerase (Expand High Fidelity PCR System; Roche Diagnostics), according to the instructions of the manufacturers. The resulting products were cloned into plasmid pCR2.1 with the TA cloning kit (Invitrogen, NV) and subjected to confirmatory sequencing. One plasmid with no amplification error was selected and was partially double digested with NdeI and SalI and then subcloned into pET-29a(+) (Novagen, Madison, Wis.), which had been digested with the same enzymes. The expression vector constructed, named pET-GES3, was introduced into E. coli BL21(DE3) pLysS (Novagen). E. coli BL21(DE3) pLysS carrying plasmid pET-GES3 was cultured in 1 liter of LB broth containing kanamycin (50 μg/ml). Isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) was added when the culture reached an A600 of 0.6, and the culture was incubated for an additional 2 h. The cells were harvested by centrifugation and were suspended in 5 ml of 20 mM bis-Tris buffer (pH 6.5). The suspension was passed through a French pressure cell twice and was then centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was used for subsequent chromatographic purification. Size-exclusion chromatography was performed on a HiLoad 16/60 Superdex 200 prep-grade column (Pharmacia Biotech, Uppsala, Sweden) preequilibrated with 20 mM bis-Tris buffer (pH 6.5). Fractions containing β-lactamase activity were collected and applied to an anion-exchange Hitrap Q HP column with the same buffer. β-Lactamase activity was recovered in the flowthrough and was dialyzed against 20 mM Tris-HCl buffer (pH 8.0) overnight at 4°C. This partially purified enzyme was loaded onto a Hitrap Q HP column (Pharmacia Biotech) preequilibrated with the same buffer and eluted with a linear gradient of NaCl. Fractions presenting high levels of activity were pooled and dialyzed against 50 mM phosphate buffer (pH 7.0).
Isoelectric focusing (IEF).
Fifty milliliters of the bacterial culture was centrifuged, and the cell pellet was suspended in 5 ml of distilled water. A crude periplasmic preparation containing β-lactamase was obtained by freezing-thawing the bacterial suspension three times, followed by ultracentrifugation (40,000 × g) for 1 h. The supernatant was condensed to 1/10 volume with an Ultrafree-15 Centrifugal Filter Device (Millipore Corporation, Bedford, Mass.). To determine the isoelectric point (pI), 5 μl of the condensed supernatant containing β-lactamase was loaded onto an Ampholine PAG plate (pH 3.5 to 9.5; Pharmacia Biotech) with a Multiphor II electrophoresis system (Pharmacia Biotech). The pI of the β-lactamase was measured by staining the gel with a 0.05% solution of nitrocefin. Purified GES-3 β-lactamase was also electrophoresed on the Ampholine PAG plate and stained with Coomassie blue.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession number AB113580.
RESULTS
Characteristics of six K. pneumoniae clinical isolates.
The susceptibilities of the six isolates to β-lactams are presented in Table 2. All isolates were resistant to piperacillin, ceftazidime, and aztreonam. The MICs of cefotaxime, cefmetazole, cefepime, and imipenem for the isolates were variable. Despite the addition of clavulanic acid, these isolates kept their high-level resistance to ceftazidime (MICs, ≥128 μg/ml). This observation was consistent with the negative results of the double-disk synergy test with two disks containing ceftazidime and amoxicillin-clavulanate, respectively. Metallo-β-lactamase production was not detected by using a thiol compound (2). PCR analyses performed preliminarily to detect broad-spectrum β-lactamase genes including TEM derivatives, CTX-M-1, CTX-M-2, CTX-M-9, MOX-1 (CMY-9), CMY-2, DHA-1, IMP-1, IMP-2, and VIM-2, all of which had already been identified in Japan, failed to give positive results.
TABLE 2.
MICs for six K. pneumoniae clinical isolates from a NICU
| K. pneumoniae strain | Date of isolation (mo/day/yr) | Site of isolation | MIC (μg/ml)a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX + CLA | PIP | PIP + TZB | CAZ | CAZ + CLA | CTX | ATM | CMZ | FEP | IPM | GEN | AMK | LVX | CIP | |||
| KG914 | 9/14/01 | Bronchial secretion | >128 | >128 | 128 | >1,024 | 512 | 64 | 64 | 16 | 32 | 0.13 | 1 | 32 | <0.06 | <0.06 |
| KG1116 | 11/16/01 | Bronchial secretion | >128 | >128 | 128 | >1,024 | 128 | 64 | 64 | 16 | 32 | 0.13 | 0.5 | 32 | <0.06 | <0.06 |
| KG525 | 3/4/02 | Stool | >128 | 128 | 128 | >1,024 | 256 | 64 | 64 | 16 | 16 | 0.13 | 2 | 64 | <0.06 | <0.06 |
| KG545 | 3/7/02 | Nasal mucosa | >128 | >128 | >128 | >1,024 | 1,024 | 128 | 128 | 128 | 64 | 0.5 | 2 | 64 | 0.25 | 0.25 |
| KG502 | 5/2/02 | Pus | >128 | 128 | 64 | 1,024 | 512 | 16 | 32 | >128 | 8 | 8 | 2 | 32 | 0.13 | <0.06 |
| KG827 | 8/27/02 | Bronchial secretion | >128 | 128 | 32 | >1,024 | 256 | 16 | 64 | >128 | 32 | 0.5 | 2 | 32 | 2 | 1 |
Abbreviations: AMX, amoxicillin; CLA, clavulanic acid; PIP, piperacillin; TZB, tazobactam; CAZ, ceftazidime; CTX, cefotaxime; ATM, aztreonam; CMZ, cefmetazole; FEP, cefepime; IPM, imipenem; GEN, gentamicin; AMK, amikacin; LVX, levofloxacin; CIP, ciprofloxacin.
Transfer and cloning of β-lactamase genes.
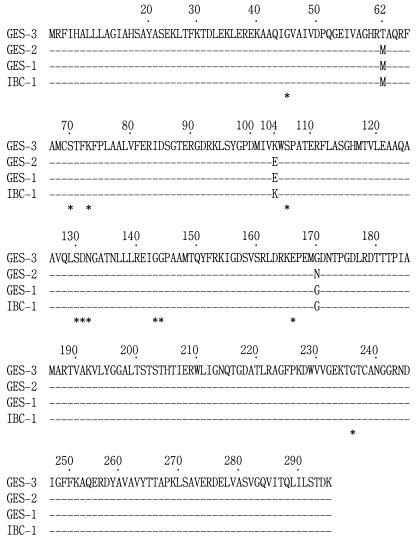
The ceftazidime resistance determinant of representative strain K. pneumoniae KG525 was successfully transferred to a recipient strain, E. coli CSH-2; and this finding indicated that the genetic determinant was located on a transferable plasmid. Two ceftazidimeresistant E. coli clones, each of which harbored a plasmid containing BamHI fragment inserts of approximately 6.7 and 11.6 kb, respectively, were obtained as a result of the cloning experiment. These two recombinant plasmids contained the same 864-bp open reading frame (ORF) encoding a putative β-lactamase which had conserved structural features of the active site of Ambler class A β-lactamases. The deduced amino acid sequence of the β-lactamase showed an amino acid substitution of M62T (a point mutation of T to C at nucleotide position 167) compared with the amino acid sequences of GES-1 (24), GES-2, and IBC-1, as well as an additional E104K substitution in comparison with the amino acid sequences of GES-1 and GES-2 (Fig. 1). Moreover, an N170G substitution was found in GES-3 compared with the amino acid sequence of GES-2, although the G residue at amino acid position 170 was conserved in IBC-1 and GES-1, as well as in GES-3. Therefore, we named this novel class A β-lactamase GES-3, although GES-1 is based on “Guiana extended spectrum” (24).
FIG. 1.
Comparison of the amino acid sequence of GES-3 with those of the GES-1, GES-2, and IBC-1 β-lactamases. Only the substituted amino acid residues are indicated by the single-letter amino acid code. Dashes represent positions where no amino acid substitution was observed among the four enzymes. The amino acid residues conserved among class A β-lactamases are indicated with asterisks.
Antibiotic susceptibilities.
The MICs of the β-lactams for parent strain K. pneumoniae KG525, transconjugant E. coli CSH-2(pKGC525), and transformant E. coli XL1-Blue(pKGB525) are listed in Table 3. Parental strain K. pneumoniae KG525 was resistant to most β-lactams except the cephamycins and carbapenems. The tranconjugant and transformant were resistant to ceftazidime, and the MICs of the other β-lactams were lower for the tranconjugant and the transformant than for the parent strain. The changes in the MICs of cefotaxime and ceftazidime for parent strain KG525 were apparently observed by the addition of β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam, while decreases in the MICs of amoxicillin, ampicillin, and piperacillin, as well as cefotaxime and ceftazidime, were observed for the E. coli tranconjugant and transformant in the presence of the inhibitors.
TABLE 3.
MICs of antimicrobial agents for the parental strain, transconjugant, and transformant
| β-Lactama | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| K. pneumoniae KG525 | E. coli CSH-2(pKGC525)b | E. coli CSH-2 | E. coli XL1-Blue(pKGB525)c | E. coli XL1-Blue(pBCSK+) | |
| Ampicillin | >128 | >128 | 4 | >128 | 4 |
| Ampicillin + sulbactam | >128 | 2 | 2 | 2 | 2 |
| Amoxicillin | >128 | >128 | 8 | >128 | 4 |
| Amoxicillin + clavulanate | >128 | 32 | 4 | 32 | 4 |
| Piperacillin | 128 | 16 | 1 | 16 | 1 |
| Piperacillin + tazobactam | 128 | 0.5 | 1 | 0.5 | 0.5 |
| Cefotaxime | 64 | 2 | 0.13 | 2 | 0.13 |
| Cefotaxime + clavulanate | 8 | 0.06 | 0.06 | 0.06 | 0.06 |
| Cefotaxime + sulbactam | 32 | 0.06 | 0.06 | 0.06 | 0.06 |
| Cefotaxime + tazobactam | 64 | 0.06 | 0.06 | 0.06 | 0.06 |
| Ceftazidime | >1,024 | 128 | 0.13 | 128 | 0.13 |
| Ceftazidime + clavulanate | 256 | 4 | 0.13 | 4 | 0.06 |
| Ceftazidime + sulbactam | >128 | 0.25 | 0.13 | 0.5 | 0.13 |
| Ceftazidime + tazobactam | >128 | 0.5 | 0.13 | 0.5 | 0.13 |
| Cephaloridine | >128 | 16 | 2 | 16 | 2 |
| Cefminox | 8 | 0.5 | 0.5 | 1 | 0.5 |
| Moxalactam | 4 | 0.25 | 0.13 | 0.5 | 0.13 |
| Cefpirome | >128 | 1 | 0.06 | 2 | 0.06 |
| Cefepime | 16 | 0.13 | 0.06 | 0.25 | 0.06 |
| Aztreonam | 64 | 4 | 0.06 | 4 | 0.06 |
| Imipenem | 0.25 | 0.5 | 0.25 | 0.13 | 0.13 |
| Gentamicin | 2 | 0.13 | 0.13 | <0.06 | <0.06 |
| Amikacin | 64 | 2 | 0.25 | 4 | 0.25 |
| Levofloxacin | <0.06 | 0.13 | 0.13 | <0.06 | <0.06 |
| Ciprofloxacin | <0.06 | <0.06 | <0.06 | <0.06 | <0.06 |
Clavulanate, tazobactam, and sulbactam were used at a fixed concentration of 4 μg/ml each.
pKGC525 is a resident plasmid found in K. pneumoniae strain KG525, and it carries the blaGES-3 gene.
pKGB525 is a recombinant plasmid that carries a 6.7-kb BamHI insert that mediates the blaGES-3 gene.
Genetic environment of blaGES-3.
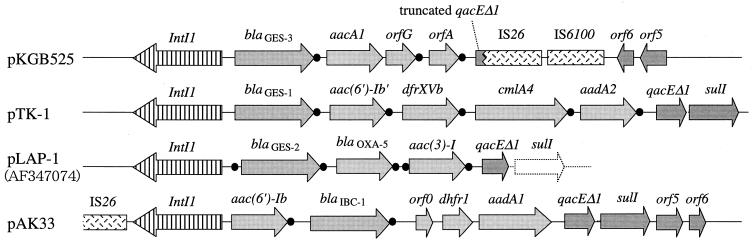
Two distinct BamHI fragments carrying the blaGES-3 gene were cloned; their sizes were approximately 6.7 and 11.6 kb, respectively, and pKGB525 had the 6.7-kb fragment. Sequencing analysis of the entire insert on pKGB525 revealed that the blaGES-3 gene was located as a gene cassette within a class 1 integron structure, as was observed in the other GES-type β-lactamase genes, blaGES-1, blaGES-2, and blaIBC-1 (Fig. 2). The 59-base element downstream of the blaGES-3 gene was made up of 110 bp and was different from that of the truncated 59-base element of blaGES-1 on pTK1 (GenBank accession number AF156486), but shared it 99% nucleotide identity with those of blaGES-2 on pLAP-1 (GenBank accession number AF326355) and blaIBC-1 on pHT9-2 (GenBank accession number AF208529).
FIG. 2.
Schematic comparison of the class 1 integron on pKGB525 with those on pTK1 (GenBank accession number AF156486), pLAP-1 (GenBank accession number AF326355), and pAK33 (34). Filled circles indicate the positions of GTTRRRY (core site) or the 59-base elements around the gene cassettes. pKGM525, which carries the 11.6-kb BamHI insert, was also sequenced; and the nucleotide sequence from intI1 to IS26 was the same as that found in pKGB525.
The second gene cassette adjacent to the blaGES-3 gene was a fused aacA1-orfG gene cassette. The results of the disk diffusion test indicated that the presence of an aacA1 component, which encodes aminoglycoside-6′-N-acetyltransferase, conferred kanamycin resistance to the transformant E. coli XL1-Blue(pKGB525) (data not shown). The nucleotide sequence of this fused gene cassette shared 100% identity with that in a class 1 integron on plasmid pCMXR1 (GenBank accession number AB061794). The function of the product encoded by orfG has not been characterized in detail. The third gene cassette is 327 bp and was named orfA. The orfA gene was suggested to be a gene cassette by recognition of the features typical of these elements: (i) the presence at the cassette boundaries of 7-bp core site sequences that completely fit the consensus sequence and (ii) the presence of a 59-base element of 78 bp downstream of the orfA gene. However, no remarkable similarity between the hypothetical protein encoded by orfA and any other known protein sequences was detected in a search performed with the BLAST program. The 3′ conserved segment of this integron showed a characteristic organization. The qacEΔ1 gene was truncated at nucleotide position 114 by the insertion sequence IS26. In the region downstream of IS26, an IS6100 element and two ORFs of unknown function, i.e., orf5 and orf6, were found. The nucleotide sequence of the 1.4-kb region containing IS6100, orf5, and orf6 was identical to that seen downstream of the sulI gene of the class 1 integron in the chromosomal multidrug resistance locus of Salmonella enterica subsp. enterica serovar Typhimurium (GenBank accession number AF261825). Sequencing analysis of pKGM525 carrying the second blaGES-3 gene on an 11.6-kb fragment was also done, and the blaGES-3 gene was also found in a class 1 integron structure with the same gene cassette configuration as in pKGB525. The nucleotide sequence of the region from intI1 to IS26 was the same as that found in pKGB525 carrying a 6.7-kb BamHI fragment.
Construction of IBC-1 by site-directed mutagenesis.
Only one amino acid substitution, M62T, was found between the sequences of GES-3 and IBC-1 (Fig. 1). Therefore, to examine whether this amino acid substitution affects the MICs of βlactams for the E. coli clones producing each enzyme, we constructed plasmid pIBC1, which encodes the IBC-1 enzyme under the same promoter as that for GES-3, by site-directed mutagenesis of the blaGES-3 gene within the parental plasmid, pGES3. However, this single substitution did not markedly influence the MICs for the E. coli clones (data not shown).
PCR detection and genotypic comparison.
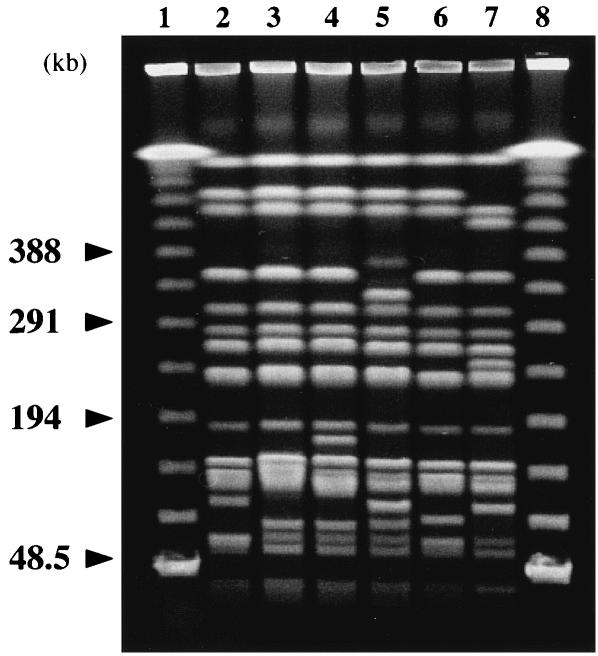
The remaining five nonrepetitive ceftazidime-resistant K. pneumoniae strains, strains KG914, KG1116, KG545, KG502, and KG827, were all found to be blaGES positive by PCR. The results of PFGE analysis of all six isolates are shown in Fig. 3. Their fingerprinting patterns were very similar but in some cases were distinct. We examined the fingerprinting patterns from 48.5 to 194 kb in detail under other conditions (data not shown). Overall, there were from three to seven band differences among the six strains examined. The ERIC-PCR patterns amplified with the ERIC-2 primer were indistinguishable from one another (data not shown). Taken together with the fact that these isolates were collected over a 1-year period, we speculate that they were genetically related and had probably spread via nosocomial transmission of an endemic clone.
FIG. 3.
PFGE analysis of K. pneumoniae isolates. Lanes: 1 and 8, PFGE marker; 2, K. pneumoniae KG914; 3, K. pneumoniae KG1116; 4, K. pneumoniae KG525; 5, K. pneumoniae KG545; 6, K. pneumoniae KG502; 7, K. pneumoniae KG827.
Plasmid profiles and Southern hybridization.
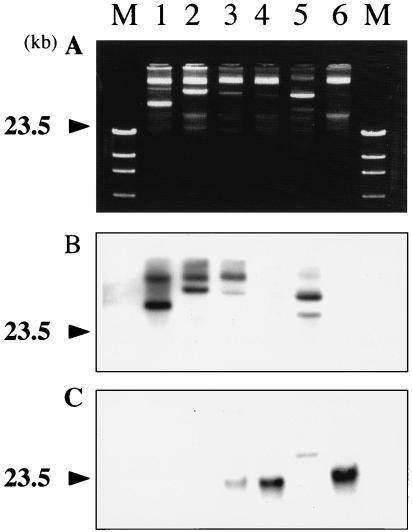
The plasmid profiles of the six blaGES-positive strains showed the presence of a large plasmid of approximately similar size in five of the six strains (Fig. 4A), while some of them possessed additional plasmids which were smaller and more diverse in size. Hybridization analyses with the probe specific for the GES-type βlactamase genes, including blaIBC-1, revealed that the location of this gene varied among the strains tested. Hybridization signals for large plasmids were detected for strains KG914, KG1116, and KG502 (Fig. 4B). Hybridization signals for both plasmids and chromosomal positions were observed for KG525. One of the hybridized plasmids from each of KG914, KG1116, and KG525 were similar in size. A single hybridization signal corresponding to the chromosomal position was detected for each of the strains KG545 and KG827 (Fig. 4C).
FIG. 4.
Plasmid profiles and Southern hybridization analysis. (A) Plasmid profiles of each strain prepared by the method of Kado and Liu (16); (B) hybridization to large plasmids harbored by each strain; (C) hybridization to the chromosomal position of each strain. The photograph of the results of gel electrophoresis of chromosomal DNAs prepared by the method of Stauffer et al. (35) was omitted. The large plasmids and chromosomal DNA were separately extracted by using freshly prepared reagents to avoid cross contamination of nicked or physicochemically amputated DNA fragments. For strains KG545 and KG827, the blaGES gene was suggested to be encoded by the chromosome. In strain KG525, the blaGES gene was suggested to be encoded by both the plasmid and the chromosome. Lanes: M, HindIII-digested DNA marker; 1, K. pneumoniae KG914; 2, K. pneumoniae KG1116; 3, K. pneumoniae KG525; 4, K. pneumoniae KG545; 5, K. pneumoniae KG502; 6, K. pneumoniae KG827.
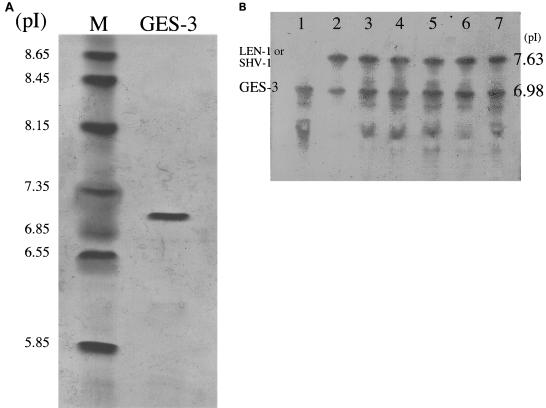
pIs of β-lactamases.
The pI value of the purified GES-3 enzyme was determined to be 7.0 (Fig. 5A). IEF of crude extracts from six GES-type β-lactamase-producing clinical strains revealed two bands with pIs of 7.6 and 7.0 (Fig. 5B). The band with pI 7.0 was also detected in a GES-3-producing E. coli transformant which harbored the blaGES-3 gene of K. pneumoniae strain KG525. The band with a pI of 7.6 corresponds to the chromosomally encoded LEN-1 (1) or SHV-type penicillinase of K. pneumoniae.
FIG. 5.
IEF. (A) IEF and staining with Coomassie blue. Lanes: M, pI marker; GES-3, purified GES-3 enzyme. (B) IEF and staining with nitrocefin. Lanes: 1, GES-3-producing E. coli transformant that harbors pKGB525 carrying blaGES-3; 2, K. pneumoniae KG914; 3, K. pneumoniae KG1116; 4, K. pneumoniae KG525; 5, K. pneumoniae KG545; 6, K. pneumoniae KG502; 7, K. pneumoniae KG827. The bands of pI 7.63 (pI 7.6 in the text) are the chromosomally encoded LEN-1 or SHV-1 β-lactamase of K. pneumoniae, and the bands at pI 6.98 (pI 7.0 in the text) are GES-3. Several β-lactamases with activities at pIs lower than 7.0 were speculated to be partially unfolded GES-3 β-lactamase, because these bands were also found in the IEF gels of an E. coli clone harboring only the blaGES-3 gene (data not shown).
DISCUSSION
Considerable differences in the levels of resistance to various cephalosporins were observed among the E. coli clones producing GES-1, GES-2, and IBC-1, although the level of production of each enzyme may differ in individual clones. For instance, the MIC of ceftazidime for an E. coli clone producing GES-1 was 128 μg/ml (24), while that for an E. coli clone producing GES-2 was 8 μg/ml (28). The single amino acid substitution in the Ω loop observed between GES-1 and GES-2, G170N, may well contribute to the difference in the substrate specificities of these enzymes. On the other hand, the MIC of ceftazidime for an E. coli clone producing IBC-1 was >256 μg/ml (11). The only amino acid substitution observed between GES-1 and IBC-1, E104K, might well also be attributed to a higher level of resistance of IBC-1 than that of GES-1 to ceftazidime, cefotaxime, and aztreonam (11, 19, 24). The MIC of ceftazidime for an E. coli clone producing GES-3 was 128 μg/ml, and GES-3 has a single M62T substitution compared with the sequence of IBC-1, which also confers high-level resistance to ceftazidime in an E. coli clone (MIC, >256 μg/ml). We investigated whether this one amino acid substitution observed between GES-3 and IBC-1 affects the MICs for E. coli clones expressing each enzyme. However, this substitution did not result in significant changes in the MICs for the E. coli clones. This finding suggests that the amino acid substitution at position 62 may not play a crucial role in the extended substrate specificity of GES-3 against ceftazidime and that those at positions 104 and 170 would be crucial for extended-spectrum enzyme activity.
In the present study, we also isolated a novel GES-type class A enzyme, GES-3, from K. pneumoniae strains which caused neonatal nosocomial infections in 2002 in Japan. Sequence analysis of the genetic environments of the blaGES-3 genes on pKGB525 carrying a 6.7-kb insert and pKGM525 carrying a 11.6-kb insert revealed that the blaGES-3 genes were located as gene cassettes in class 1 integrons, as observed in other GES-type β-lactamase genes, including blaIBC-1 (Fig. 2). Integrons are very sophisticated site-specific recombination systems that capture various gene cassettes, including antibiotic resistance genes, between their 5′ and 3′ conserved segments (14, 15, 22). The gene cassettes for the GES-type enzymes with a very close phylogenetic relationship might have originated as a single clone and then disseminated worldwide with the help of class 1 integrons possessing very similar genetic organizations. These integrons are mediated by self-transmissible plasmids with a wide host range. Since very similar GES enzymes have so far been found in French Guiana, Greece, South Africa, Portugal, and Japan, these GES-type β-lactamase-producing strains might have been scattered globally by the recent extensive international travel or dissemination of humans, foods, and animals.
In the present study, we analyzed genetic relatedness using PFGE and ERIC-PCR of all six GES-type β-lactamase-producing K. pneumoniae strains isolated in a NICU over a 1-year period. Since the fingerprinting patterns obtained by PFGE and ERIC-PCR were very similar, these isolates were suggested to belong to the same genetic lineage that caused the nosocomial spread. The minor differences in the fingerprinting patterns obtained by PFGE might be due to the occurrence of genetic rearrangements over the course of the nosocomial spread. Interestingly, the results of Southern hybridization suggested that the locations of the blaGES genes were multiple and diverse among the six strains studied. By consideration of the results of PFGE, ERIC-PCR, and Southern hybridization, it can be speculated that an endemic strain containing the blaGES genes might have spread within the NICU setting over the 1-year period and might have undergone genetic rearrangements, including translocation and multiplication of the blaGES gene.
The presence of multiple blaGES-3 genes in strain KG525 is probably the result of these genetic rearrangements. Translocation and multiplication of the blaGES gene might be facilitated by mediation of a site-specific recombination system of an integron or a transposon. A similar example of multiple locations of the same antibiotic resistance gene has been reported by Yagi et al. (38). In their study, a single clinical E. coli isolate was found to carry three distinguishable Toho-1-like β-lactamase genes, which were later identified as blaCTX-M-2, by their restriction digestion patterns on the chromosome. These multiple locations of the same β-lactamase gene would be beneficial to bacteria, since they increase the chance of amino acid substitutions necessary for extension of the substrate profiles of β-lactamases as well as the multicopy effect of gene expression. Bradford et al. (3) reported that point mutations leading to ESBLs (ESBLs TEM-1 to TEM-10 and TEM-12) occurred on the plasmids of a single K. pneumoniae clinical isolate. A notable finding presented in that report was the distinct hydrolyzing activity between TEM-10 and TEM-12. TEM-10 had hydrolyzing activity against ceftazidime, while TEM-12 also hydrolyzed cefotaxime and aztreonam, in addition to ceftazidime. A variety of susceptibility profiles for cephamycins were also observed among the six K. pneumoniae strains in the present study. For instance, strains KG914, KG1116, and KG525 were susceptible to cefmetazole (MICs, 16 μg/ml), whereas strains KG545, KG502, and KG827 were resistant to this agent (MICs, ≥128 μg/ml). In particular, strain KG502 showed high-level resistance to other cephamycins, such as cefoxitin (MIC, >128 μg/ml), cefminox (MIC, >128 μg/ml), and moxalactam (MIC, 128 μg/ml). An evolutionary event similar to that observed in the TEM enzymes (3) might have occurred in these K. pneumoniae strains to give them further resistance to a broad range of antibiotics. The MIC of imipenem for strain KG502 was 8 μg/ml, and this might be due to the hyperproduction of some β-lactamase with an extended substrate specificity as well as the loss of some outer membrane protein, as reported by Bradford et al. (4). Further molecular characterization of the cephamycin resistance observed in strain KG502 will be undertaken in the next study.
Acknowledgments
We are grateful to Leonidas S. Tzouvelekis, Laboratory of Bacteriology, Hellenic Pasteur Institute, Athens, Greece, for kindly providing E. cloacae HT9-producing IBC-1 and E. coli DΗ5α containing pHT9-2 and to Kumiko Kai for technical help.
This work was supported by grants H12-Shinko-19 and H12-Shinko-20 from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Arakawa, Y., M. Ohta, N. Kido, Y. Fujii, T. Komatsu, and N. Kato. 1986. Close evolutionary relationship between the chromosomally encoded βlactamase gene of Klebsiella pneumoniae and the TEM β-lactamase gene mediated by R-plasmids. FEBS Lett. 207:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamaseproducing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burwen, D. R., S. N. Banerjee, R. P. Gaynes, et al. 1994. Ceftazidime resistance among selected nosocomial gram-negative bacilli in the United States. J. Infect. Dis. 170:1622-1625. [DOI] [PubMed] [Google Scholar]
- 6.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaves, J., M. G. Ladona, C. Segura, A. Coira, R. Reig, and C. Ampurdanes. 2001. SHV-1 β-lactamase is mainly a chromosomally encoded species-specific enzyme in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi, Y., N. Shibata, K. Shibayama, K. Kamachi, H. Kurokawa, K. Yokoyama, T. Yagi, and Y. Arakawa. 2002. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 46:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte, A., F. Boavida, F. Grosso, M. Correia, L. M. Lito, J. M. Cristino, and M. J. Salgado. 2003. Outbreak of GES-1 β-lactamase-producing multidrug-resistant Klebsiella pneumoniae in a university hospital in Lisbon, Portugal. Antimicrob. Agents Chemother. 47:1481-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 15.Hall, R. M., and C. M. Collis. 1995. Site-specific insertion of genes into integrons: role of 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 16.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kartali, G., E. Tzelepi, S. Pournaras, C. Kontopoulou, F. Kontos, D. Sofianou, A. N. Maniatis, and A. Tsakris. 2002. Outbreak of infections caused by Enterobacter cloacae producing the integron-associated β-lactamase IBC-1 in a neonatal intensive care unit of a Greek hospital. Antimicrob. Agents Chemother. 46:1577-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto, Y., and M. Inoue. 1999. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavroidi, A., E. Tzelepi, A. Tsakris, V. Miriagou, D. Sofianou, and L. S. Tzouvelekis. 2001. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J. Antimicrob. Chemother. 48:627-630. [DOI] [PubMed] [Google Scholar]
- 20.Naas, T., F. Benaoudia, S. Massuard, and P. Nordmann. 2000. Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46:703-711. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Paulsen, I. T., T. G. Littlejohn, P. Radstrom, L. Sundstrom, O. Skold, G. Swedberg, and R. A. Skurray. 1993. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit, A., D. L. Sirot, C. M. Chanal, J. L. Sirot, R. Labia, G. Gerbaud, and R. A. Cluzel. 1988. Novel plasmid-mediated β-lactamase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime than to other broad-spectrum cephalosporins. Antimicrob. Agents Chemother. 32:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., G. F. Weldhagen, C. De Champs, and P. Nordmann. 2002. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum β-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 49:561-565. [DOI] [PubMed] [Google Scholar]
- 28.Poirel, L., G. F. Weldhagen, T. Naas, C. De Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, Z. Y., P. Y. Liu, Y. J. Lau, Y. H. Lin, and B. S. Hu. 1996. Epidemiological typing of isolates from an outbreak of infection with multidrug-resistant Enterobacter cloacae by repetitive extragenic palindromic unit b1-primed PCR and pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stauffer, G. V., M. D. Plamann, and L. T. Stauffer. 1981. Construction and expression of hybrid plasmids containing the Escherichia coli glyA genes. Gene 14:63-72. [DOI] [PubMed] [Google Scholar]
- 36.van der Zee, A., N. Steer, E. Thijssen, J. Nelson, A. van't Veen, and A. Buiting. 2003. Use of multienzyme multiplex PCR amplified fragment length polymorphism typing in analysis of outbreaks of multiresistant Klebsiella pneumoniae in an intensive care unit. J. Clin. Microbiol. 42:798-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vourli, S., L. S. Tzouvelekis, E. Tzelepi, E. Lebessi, N. J. Legakis, and V. Miriagou. 2003. Characterization of In111, a class 1 integron that carries the extended-spectrum β-lactamase gene blaIBC-1. FEMS Microbiol. Lett. 225:149-153. [DOI] [PubMed] [Google Scholar]
- 38.Yagi, T., H. Kurokawa, K. Senda, S. Ichiyama, H. Ito, S. Ohsuka, K. Shibayama, K. Shimokata, N. Kato, M. Ohta, and Y. Arakawa. 1997. Nosocomial spread of cephem-resistant Escherichia coli strains carrying multiple Toho-1-like β-lactamase genes. Antimicrob. Agents Chemother. 41:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagi, T., H. Kurokawa, N. Shibata, K. Shibayama, and Y. Arakawa. 2000. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS. Microbiol. Lett. 184:53-56. [DOI] [PubMed] [Google Scholar]
- 40.Yan, J. J., W. C. Ko, Y. C. Jung, C. L. Chuang, and J. J. Wu. 2002. Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 β-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40:3121-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]