Abstract
Over the last decades, Candida spp have been responsible for an increasing number of infections, especially in patients requiring intensive care. Knowledge of local epidemiology and analysis of the spread of these pathogens is important in understanding and controlling their transmission. The aim of this study was to evaluate the genetic diversity of 31 Candida albicans and 17 Candida glabrata isolates recovered from intensive care unit patients from the tertiary hospital in Krakow between 2011-2012. The strains were typed by random amplified polymorphic DNA (RAPD) polymerase chain reaction using five primers (CD16AS, HP1247, ERIC-2, OPE-3 and OPE-18). The results of the present investigation revealed a high degree of genetic diversity among the isolates. No clonal relationship was found among the C. albicans strains, whereas two C. glabrata isolates were identical. The source of Candida infection appeared to be mostly endogenous; however, the presence of two clonal C. glabrata strains suggested the possibility of cross-transmission of these pathogens. Our study confirmed the high discriminatory power of the RAPD technique in the molecular typing of Candida clinical isolates. This method may be applied to the evaluation of transmission routes of pathogenic fungi on a local level.
Keywords: Candida albicans, Candida glabrata, genotyping, epidemiology, RAPD PCR
Over the last few decades, the incidence of fungal infections in humans has increased, especially in patients requiring intensive care. The Extended Prevalence of Infection in Intensive Care study demonstrated that Candi-da spp accounted for 18.5% of microorganisms isolated from intensive care unit (ICU) patients in both Western and Eastern Europe (Vincent et al. 2009). Invasive candidiasis and candidaemia, caused most commonly by Candida albicans and Candida glabrata, are of growing concern in the ICU (Morace & Borghi 2010, Pfaller et al. 2011, Rodrigues et al. 2013). Polish surveillance involving 94 units in 20 hospitals (including surgery, intensive care, neonatology, haematology and others) indicated that the highest degree of candidaemia was found in the ICU (30.8%) (Nawrot et al. 2013).
Furthermore, C. albicans and C. glabrata bloodstream infections have been associated with a high mortality rate (Weinberger et al. 2005). Risk factors for invasive fungal infection in ICU patients include the use of broad-spectrum antibacterial agents and immunosuppressive agents, parenteral nutrition, central venous catheters and mechanical ventilations. Immune status, comorbid diseases and the age of patients also contribute to the incidence of fungal diseases (Lass-Flörl 2009, Arendrup 2010). Although the most cases of Candida infections appear to originate from an endogenous source, there are cases of nosocomial transmission (Vrioni & Matsiota-Bernard 2001, Ben Abdeljelil et al. 2011). Identification of relatedness between Candida species is crucial for control measures to reduce the incidence of nosocomial candidiasis, but little is known of the epidemiology of these species in southern Poland. For this purpose, it is important to perform genotyping analysis that would allow evaluation of the spread of these pathogens. The most commonly used molecular typing methods are restriction endonuclease analysis, microsatellite analysis, multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, multilocus sequence typing and random amplified polymorphic DNA polymerase chain reaction (RAPD PCR) (Krawczyk et al. 2009, Abbes et al. 2010, Karaman et al. 2013). The latter method was used in this study to evaluate the level of genetic diversity of C. albicans and C. glabrata strains isolated from ICU patients from the tertiary hospital in Krakow, Poland over a two-year period.
SUBJECTS, MATERIALS AND METHODS
Patients and isolates - A total of 136 and 145 yeast strains were isolated from patients hospitalised in the ICU of the Rydygier’s Specialized Hospital in Krakow in 2011 and 2012, respectively. The species distribution is shown in Table I. The predominant species were C. albicans (162 strains) and C. glabrata (45 strains), obtained from endotracheal aspirates (ETA) and urine. Among the most frequently isolated species, subjected to further molecular studies, were 48 strains, including 31 C. albicans and 17 C. glabrata isolates cultured at concentrations of ≥ 104 colony-forming unit (cfu)/mL. Of the study group patients, 20 were female, 22 were male and the age range was 19-94 (mean 65) years. In 2011, there were 13 recovered C. albicans isolates and seven recovered C. glabrata isolates, whereas in 2012 there were 18 C. albicans and 10 C. glabrata isolated strains, respectively. Twenty nine strains of C. albicans and 17 strains of C. glabrata were isolated from ETA, while two C. albicans isolates were taken from urine. All strains were obtained once per patient, except for C. albicans CA11 and CA12 strains, which were isolated from the ETA and urine of the same patient. From five of the patients tested, both C. albicans and C. glabrata were cultured (Table II). Antifungal prophylaxis was not administered to the patients included in this study. This research was approved by the Ethical Committee of Jagiellonian University Medical College (protocol KBET/129/B/2011).
TABLE I. Yeast species isolated from the intensive care unit patients of the Rydygier’s Specialized Hospital in Krakow, Poland, between 2011-2012.
| Yeast species | Year of isolation
|
|
|---|---|---|
| 2011 | 2012 | |
|
| ||
| n (%) | n (%) | |
| Candida albicans | 76 (55.9) | 86 (59.3) |
| Candida glabrata | 23 (16.9) | 22 (15.2) |
| Candida tropicalis | 11 (8.1) | 8 (5.5) |
| Candida krusei | 5 (3.7) | 8 (5.5) |
| Candida kefyr | 4 (2.9) | 6 (4.1) |
| Candida dubliniensis | 3 (2.2) | 2 (1.4) |
| Candida parapsilosis | 3 (2.2) | 6 (4.1) |
| Othera | 11 (8.1) | 7 (4.8) |
|
| ||
| Total | 136 (100) | 145 (100) |
a: Candida lusitaniae, Candida lipolytica, Candida guilliermondii, Candida inconspicua, Saccharomyces cerevisiae, Geotrichum sp.
TABLE II. Description of 31 Candida albicans and 17 Candida glabrata isolates comprising gender and age of patients, date of isolation, clinical sample, antifungal agents minimal inhibitory concentration (MIC) values and results of random amplified polymorphic DNA polymerase chain reaction (RAPD) analysis.
| Isolate code | Patient code | Gender | Age | Date of isolation | Clinical sample | MIC values (mg/L)
|
RAPD profile | RAPD cluster | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | FLC | 5FC | VRC | ||||||||
| CA1 | SE | F | 78 | 12 Jan 2011 | ETA | 1 | 4 | ≤ 1 | ≤ 0.12 | A | A |
| CA2 | DM | M | 25 | 16 Mar 2011 | ETA | 1 | ≤ 1 | ≤ 1 | ≤ 0.12 | U1 | U |
| CA3 | WM | M | 80 | 30 May 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | R | R |
| CA4 | BL | M | 53 | 07 Jun 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | D | D |
| CA5 | PA | M | 50 | 18 Aug 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | N | N |
| CA6 | LP | M | 28 | 05 Sep 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | K1 | K |
| CA7 | KL | M | 28 | 21 Sep 2011 | Urine | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | O1 | O |
| CA8 | PB | F | 43 | 22 Sep 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | O2 | O |
| CA9 | SR | M | 38 | 05 Oct 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | K2 | K |
| CA10 | SL | M | 90 | 07 Oct 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | L | L |
| CA11 | BM | F | 66 | 15 Nov 2011 | ETA | 1 | ≤ 1 | ≤ 1 | ≤ 0.12 | P | P |
| CA12 | BM | F | 66 | 16 Nov 2011 | Urine | 1 | ≤ 1 | ≤ 1 | ≤ 0.12 | M1 | M |
| CA13 | LF | M | 90 | 29 Dec 2011 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | M2 | M |
| CA14 | WH | M | 34 | 03 Jan 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | C | C |
| CA15 | BuM | F | 53 | 12 Jan 2012 | ETA | 1 | ≤ 1 | ≤ 1 | ≤ 0.12 | E1 | E |
| CA16 | PZ | M | 87 | 09 Mar 2012 | ETA | 1 | ≤ 1 | ≤ 1 | ≤ 0.12 | T1 | T |
| CA17 | ZW | M | 83 | 04 Apr 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | E2 | E |
| CA18 | KT | M | 61 | 04 Apr 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | F | F |
| CA19 | NA | F | 61 | 16 Apr 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | E4 | E |
| CA20 | ZJ | F | 68 | 08 Oct 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | J1 | J |
| CA21 | OJ | M | 69 | 08 Oct 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | T2 | T |
| CA22 | PP | M | 19 | 10 Oct 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | E3 | E |
| CA23 | NN | F | 41 | 12 Oct 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | J2 | J |
| CA24 | OA | F | 45 | 17 Oct 2012 | ETA | ≤ 0.25 | ≤ 1 | ≤ 1 | ≤ 0.12 | G | G |
| CA25 | MR | M | 86 | 12 Nov 2012 | ETA | ≤ 0.25 | 4 | ≤ 1 | ≤ 0.12 | U2 | U |
| CA26 | TP | M | 57 | 26 Nov 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | H1 | H |
| CA27 | ZaJ | F | 79 | 28 Nov 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | H2 | H |
| CA28 | ZS | F | 94 | 28 Nov 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | S | S |
| CA29 | KM | F | 76 | 18 Dec 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | I1 | I |
| CA30 | CM | F | 69 | 24 Dec 2012 | ETA | ≤ 0.25 | ≤ 1 | ≤ 1 | ≤ 0.12 | B | B |
| CA31 | KH | F | 62 | 28 Dec 2012 | ETA | 0.5 | ≤ 1 | ≤ 1 | ≤ 0.12 | I2 | I |
| CG1 | SZ | F | 80 | 28 Apr 2011 | ETA | ≤ 0.25 | 8 | ≤ 1 | ≤ 0.12 | a | a |
| CG2 | WM | M | 80 | 30 May 2011 | ETA | 0.5 | 4 | ≤ 1 | 0.25 | f1 | f |
| CG3 | PJ | M | 80 | 15 Jun 2011 | ETA | 1 | 8 | ≤ 1 | ≤ 0.12 | g1 | g |
| CG4 | WJ | F | 47 | 29 Jun 2011 | ETA | 0.5 | 8 | ≤ 1 | ≤ 0.12 | h | h |
| CG5 | CA | M | 57 | 16 Sep 2011 | ETA | 1 | 8 | ≤ 1 | ≤ 0.12 | g2 | g |
| CG6 | SL | M | 90 | 07 Oct 2011 | ETA | 1 | 4 | ≤ 1 | ≤ 0.12 | i | i |
| CG7 | SJ | M | 70 | 28 Oct 2011 | ETA | 0.5 | 32 | ≤ 1 | ≤ 0.12 | f2 | f |
| CG8 | WE | F | 37 | 31 Jan 2012 | ETA | 1 | 8 | ≤ 1 | 1 | e2 | e |
| CG9 | PZ | M | 87 | 04 Apr 2012 | ETA | 0.5 | 4 | ≤ 1 | 0.25 | d1 | d |
| CG10 | LG | F | 72 | 18 Apr 2012 | ETA | 1 | 4 | ≤ 1 | ≤ 0.12 | d1 | d |
| CG11 | JJ | M | 57 | 07 Aug 2012 | ETA | 0.5 | 4 | ≤ 1 | ≤ 0.12 | e1 | e |
| CG12 | BG | F | 77 | 08 Aug 2012 | ETA | 0.5 | 8 | ≤ 1 | ≤ 0.12 | c | c |
| CG13 | ZJ | F | 68 | 04 Oct 2012 | ETA | 0.5 | 2 | ≤ 1 | ≤ 0.12 | d3 | d |
| CG14 | OJ | M | 69 | 05 Oct 2012 | ETA | 1 | 2 | ≤ 1 | ≤ 0.12 | d2 | d |
| CG15 | GG | F | 78 | 25 Oct 2012 | ETA | ≤ 0.25 | 2 | ≤ 1 | ≤ 0.12 | d4 | d |
| CG16 | GH | F | 87 | 18 Dec 2012 | ETA | 0.5 | 2 | ≤ 1 | ≤ 0.12 | j | j |
| CG17 | CM | F | 69 | 24 Dec 2012 | ETA | 0.5 | 8 | ≤ 1 | ≤ 0.12 | b | b |
AMB: amphotericin B; CA1-CA31: C. albicans strains; CG1-CG17: C. glabrata strains; ETA: endotracheal aspirate; F: female; FLC: fluconazole; M: male; VRC: voriconazole; 5FC: 5-fluorocytosine.
Identification and antifungal susceptibility testing of Candida strains - All analysed clinical specimens, from ETA (46) and urine (2), were cultured by quantitative technique on Sabouraud dextrose agar (bioMérieux, Poland) and incubated at 37ºC for 48 h. Fungal concentrations between 104 cfu/mL and 106 cfu/mL for ETA and 105 cfu/mL for urine were observed after 48 h of incubation (Müller et al. 1986).
Preliminary fungal strain identification was based on colony morphology on CHROMAgar Candida (bioMérieux), while the identification to the species level was confirmed by the Vitek 2 Compact automatic system (bioMérieux). Susceptibility of strains to amphotericin B, fluconazole, 5-fluorocytosine and voriconazole was determined by using the Vitek 2 Compact with AST-YS01 cards and by interpreting results according to the Clinical and Laboratory Standards Institute criteria (CLSI 2008).
DNA extraction - Fungal genomic DNA was extracted with a Genomic Mini AX YEAST isolation kit (A&A Biotechnology, Poland) according to the manufacturer’s instructions. Evaluation of the DNA concentration and purity was performed by spectrophotometry at 260 nm and the ratio of the absorbance at 260 nm and 280 nm, respectively. The total genomic DNA was stored at -20ºC for further molecular studies.
RAPD reaction - Five arbitrary primers were used for RAPD analysis of C. albicans and C. glabrata strains: CD16AS (5’-CTCTTGAAACTGGGGAGACTTGA-3’), HP1247 (5’-AAGAGCCCGT-3’), ERIC-2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’) (Trojanowska et al. 2010), OPE-3 (5’-CCAGATGCAC-3’) and OPE-18 (5’-GGACTGCAGA-3’) (Marol & Yücesoy 2008) (Blirt, Poland). The RAPD reactions were performed with 25 ng of Candida genomic DNA in a final volume of 25 μL containing 100 pmol of primer (Blirt, Poland), 100 μM of each nucleotide (Fermentas, Lithuania), 0.75 mM MgCl2, 0.625 U of GoTaq DNA Polymerase in 5 μL and 5x colourless GoTaq Flexi Buffer (pH 8.5) provided by the manufacturer (Promega, USA). Amplification was carried out in a thermocycler T personal (Biometra, Germany) with an initial denaturation at 95ºC for 2 min followed by 40 cycles of denaturation at 95ºC for 1 min, annealing at 47ºC for 1 min, elongation at 72ºC for 2 min and a final extension at 72ºC for 5 min. The DNA fragments were then separated by 2% agarose gel electrophoresis and visualised by ethidium bromide staining (Sigma-Aldrich, Germany). The PCR product size was determined by comparison with a molecular weight standard (O’Gene Ruler 100 bp DNA Ladder Plus; Thermo Scientific, USA).
The analyses of the genetic relatedness of the Candi-da isolates were performed by Bio 1D++ software (Vilber-Lormat, France) via unweighted pair-group method with arithmetic averages using the Dice coefficient. When the similarity value was 100%, the strains were determined to be identical, while isolates with values ranging between 80-99% were considered related and those with values under 80% were considered unrelated. The experiments were carried out in duplicate and there were no differences in the RAPD profiles.
RESULTS
The minimal inhibitory concentration (MIC) values for amphotericin B, fluconazole, 5-fluorocytosine and voriconazole for all tested isolates are shown in the Table II. Forty-seven Candida strains were susceptible to the antifungal agents tested in this study, except for one C. glabrata strain (CG7) that had intermediate susceptibility to fluconazole (MIC 32 mg/L).
Genetic diversity of 31 C. albicans and 17 C. glabrata isolates were examined in this study. C. albicans isolates displayed from one to nine; two-nine, four-nine, four-10 and three-13 bands for OPE-18, HP1247, OPE-3, CD16AS and ERIC-2 primers, respectively. The RAPD patterns obtained for C. glabrata were composed of one-five, one-eight, two-10, six-11 and three-12 DNA fragments for HP1247, CD16AS, ERIC-2, OPE-3 and OPE-18 oligonucleotides, respectively. The results of the RAPD reactions obtained with five primers were used for the construction of dendrograms.
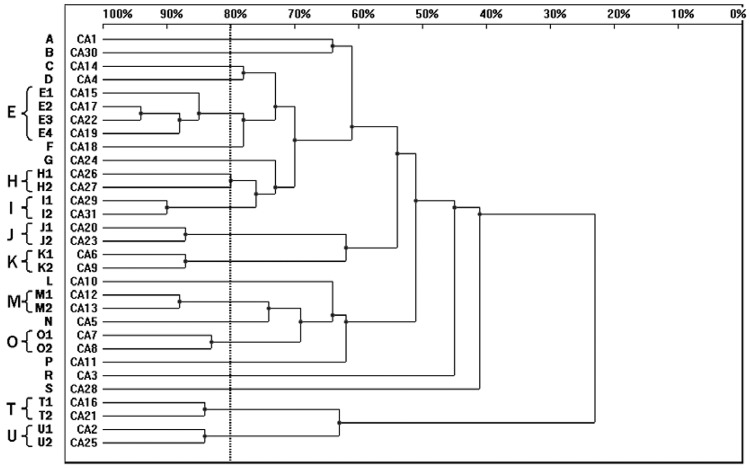
Among 31 C. albicans strains, 31 RAPD profiles were distinguished. Twenty C. albicans isolates were allocated into nine clusters (E, H-K, M, O, T, U) with homology levels higher than 80%, whereas the remaining 11 genotypes (A-D, F, G, L, N, P, R, S) were represented by single isolates only. Four isolates belonged to cluster E, while clusters H-K, M, O, T and U consisted of two strains each (Fig. 1).
Fig. 1. : dendrogram presenting the genetic relatedness of 31 Candida albicans strains from intensive care unit patients generated by random amplified polymorphic DNA polymerase chain reaction, using CD16AS, HP1247, ERIC-2, OPE-3 and OPE-18 primers. The vertical line divides the strains according to the level of genetic similarity into related and unrelated.
The strains CA11 and CA12, cultured from ETA and urine of the same patient, were unrelated with homology levels under 80%. Two pairs of isolates, CA17-CA22 (cluster E) and CA29-CA31 (cluster I) showed the highest degree of similarity with 94% and 90% homology, respectively. None of the C. albicans isolates revealed the same RAPD profile when results from all primers were combined to generate the dendrogram (Fig. 1, Table II).
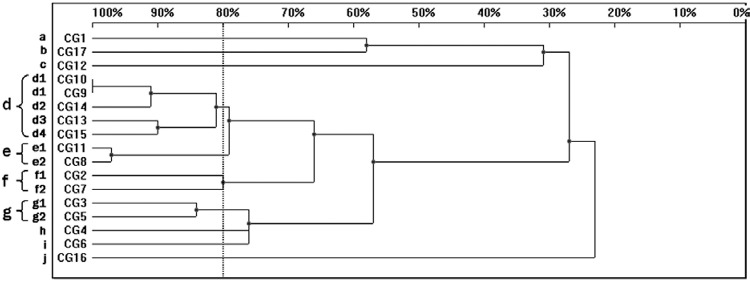
Sixteen patterns were observed in the RAPD analysis of the 17 C. glabrata strains. When 80% homology was used as a cut-off point, the C. glabrata strains were allocated into four genetic similarity groups (d-g) and six unique profiles were distinguished (a-c, h-j) (Fig. 2). The cluster d comprised of five isolates that presented four different patterns, whereas the remaining four clusters, e-g, contained two isolates each. Among the C. glabrata strains, only two isolates, CG9 and CG10 (d1), were identical. Both of them were collected in April 2012 from two patients hospitalised in the ICU. Despite 100% homology, they presented different MIC values to the antifungal agents amphotericin B (MIC 0.5 and 1 mg/L) and voriconazole (MIC 0.25 and ≤ 0.12 mg/L) (Table II). The analysis of the dendrogram revealed three groups of isolates: CG8 and CG11 (cluster e), CG9, CG10 and CG14 (cluster d), CG13 and CG15 (cluster d), with 97%, 91% and 90% homology, respectively.
Fig. 2. : dendrogram presenting the genetic similarity among 17 Candida glabrata strains generated by random amplified polymorphic DNA polymerase chain reaction, using CD16AS, HP1247, ERIC-2, OPE-3 and OPE-18 primers. The vertical line divides the strains according to the level of genetic similarity into related and unrelated.
The discriminatory power (DP) of the RAPD method was calculated for each species according to the Simpson index (Hunter & Gaston 1988). When all tested oligonucleotides were combined, the DP value was 1 for C. albicans and 0.99 for C. glabrata.
DISCUSSION
C. albicans is an important cause of nosocomial infections; however, in the last decade, non-albicans species were responsible for an increasing number of infections (Bacelo et al. 2010, Nawrot et al. 2013). Yeasts are members of the normal microflora of the skin, mucous membranes and gastrointestinal tract in healthy individuals. However, as opportunistic pathogens, they may also cause endogenous infections in immunocompromised patients. Exogenous Candida infections are observed less frequently (Bacelo et al. 2010, Bonfim-Mendonça et al. 2013). Knowing the source of infection and detection of possible routes of transmission can help with the prevention of clonal spread of pathogenic fungi. RAPD is one of the most frequently used genotyping methods for epidemiological investigations of Candida infections worldwide (Lian et al. 2004, Marol & Yücesoy 2008, Muthig et al. 2010, Karaman et al. 2013). This technique is easy to perform as well as rapid, cost-effective and suitable for the typing of a large number of strains (Noumi et al. 2009). RAPD may result in low interlaboratory reproducibility, nevertheless this technique can be applied for the investigation of Candida epidemiology on a local level (Saghrouni et al. 2013).
In our study, we performed RAPD reactions to evaluate the genetic relatedness of C. albicans and C. glabrata isolates from southern Poland. We noted a high DP of the RAPD technique. This fact has also been supported by other investigators who reported that RAPD was able to clearly distinguish the genomic variability within the Candida isolates (Robert et al. 1995, Pinto et al. 2004).
In the molecular typing of C. albicans, there were no strains that generated identical RAPD profiles, which may indicate an endogenous source of infection. These findings were in accordance with results obtained by da Costa et al. (2012) who studied the genetic homology of 15 C. albicans strains using five primers. Similar results were found by Marol and Yücesoy (2008) who used four primers for typing 42 C. albicans strains recovered from ICU patients. Furthermore, four C. albicans isolates cultured from various specimens from the same patients were determined to be different (Marol & Yücesoy 2008). In our study, the strains CA11 and CA12, isolated from the respiratory tract and urinary system of a single patient, were also unrelated. However, in some studies, C. albicans strains cultured from two sources from the same patient were genotypically identical. Brillowska-Dabrowska et al. (2010) typed 34 C. albicans strains that revealed the same RAPD patterns in blood samples and colonisation specimens collected from a single patient. Bonfim-Mendonça et al. (2013) also observed that C. albicans strains recovered from different sites from a single patient were identical. Such results indicate the migration of yeasts from colonisation sites and confirm that previous colonisation is an important factor predisposing for systemic infection.
Molecular typing of 17 C. glabrata isolates revealed 16 different genotypes. We observed two strains generating identical RAPD profiles. Another study analysing the diversity of 19 C. glabrata isolates originating from oral and vaginal specimens from Tunisia hospitals, revealed an even higher degree of genetic diversity among strains (Noumi et al. 2009). Much lower genotypic variability of C. glabrata isolates, with two primers tested, was shown in a recent study from Krasnodębska-Szponder et al. (2005). The investigation performed by Szweda et al. (2013) revealed 26 genotypes among 54 C. glabrata isolates, but it is noteworthy that the authors used only one primer in the study. Additionally, Ergon and Gülay (2005) observed a lower degree of genetic diversity among C. glabrata strains. The authors suggested that it is possible that primers used in the study were not appropriate for the typing of non-albicans species (Ergon & Gülay 2005). Some authors highlighted that the strains classified as identical by one primer are not always classified as belonging to the same cluster when analysed by another primer (Bonfim-Mendonça et al. 2013). Therefore, the data obtained with a number of oligonucleotides should be combined to generate higher DP (Soll 2000).
In the present study, we noticed a few groups of C. albicans and C. glabrata strains with similarity values between 80-99%, which represented related, but not identical clinical isolates. These findings may suggest microevolutionary changes of a single strain occurring during adaptation to environmental conditions (Soll 2000, Bonfim-Mendonça et al. 2013). Furthermore, there was the possibility that the unique strains presented in the dendrograms belonged to other species, such as Candida africana or Candida bracarensis, which are closely related to C. albicans and C. glabrata. However, these species can be distinguished only with molecular methods, including sequencing of the D1/D2 region of 26S rDNA (Taverna et al. 2013).
Footnotes
Financial support: Jagiellonian University Medical College (with funds from maintenance of the research potential of the Department of Pharmaceutical Microbiology JU MC)
REFERENCES
- Abbes S, Amouri I, Sellami H, Sellami A, Makni F, Ayadi A. A review of molecular techniques to type Candida glabrata isolates. Mycoses. 2010;53:463–467. doi: 10.1111/j.1439-0507.2009.01753.x. [DOI] [PubMed] [Google Scholar]
- Arendrup MC. Epidemiology of invasive candidiasis. Curr Opin Crit Care. 2010;16:445–452. doi: 10.1097/MCC.0b013e32833e84d2. [DOI] [PubMed] [Google Scholar]
- Bacelo KL, Costa KR, Ferreira JC, Candido RC. Biotype stability of Candida albicans isolates after culture storage determined by randomly amplified polymorphic DNA and phenotypical methods. Mycoses. 2010;53:468–474. doi: 10.1111/j.1439-0507.2009.01741.x. [DOI] [PubMed] [Google Scholar]
- Ben Abdeljelil J, Saghrouni F, Emira N, Valentin-Gomez E, Chatti N, Boukadida J, Ben Saïd M, Agudo LDC. Molecular typing of Candida albicans isolates from patients and health care workers in a neonatal intensive care unit. J Appl Microbiol. 2011;111:1235–1249. doi: 10.1111/j.1365-2672.2011.05121.x. [DOI] [PubMed] [Google Scholar]
- Bonfim-Mendonça PS, Fiorini A, Shinobu-Mesquita CS, Baeza LC, Fernandez MA, Svidzinski TI. Molecular typing of Candida albicans isolates from hospitalized patients. Rev Inst Med Trop Sao Paulo. 2013;55:385–391. doi: 10.1590/S0036-46652013000600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillowska-Dabrowska A, Bergmann O, Jensen IM, Jarløv JO, Arendrup MC. Typing of Candida isolates from patients with invasive infection and concomitant colonization. Scand J Infect Dis. 2010;42:109–113. doi: 10.3109/00365540903348336. [DOI] [PubMed] [Google Scholar]
- CLSI - Clinical and Laboratory Standards Institute Reference method for broth dilution antifungal susceptibility testing of yeasts - approved standard. 3rd. CLSI M27-A3; Wayne: 2008. 13 [Google Scholar]
- Costa KR, Ferreira JC, Lavrador MA, Baruffi MD, Candido RC. Virulence attributes and genetic variability of oral Candida albicans and Candida tropicalis isolates. Mycoses. 2012;55:e97–e105. doi: 10.1111/j.1439-0507.2011.02125.x. [DOI] [PubMed] [Google Scholar]
- Ergon MC, Gülay Z. Molecular epidemiology of Candida species isolated from urine at an intensive care unit. Mycoses. 2005;48:126–131. doi: 10.1111/j.1439-0507.2004.01086.x. [DOI] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman M, Firinci F, Karaman O, Uzuner N, Bahar IH. Long-term oropharyngeal colonization by C. albicans in children with cystic fibrosis. Yeast. 2013;30:429–436. doi: 10.1002/yea.2977. [DOI] [PubMed] [Google Scholar]
- Krasnodębska-Szponder B, Szymaniak L, Wojciechowska-Koszko I, Mnichowska M, Michalska-Krzanowska G, Giedrys-Kalemba S. Species and genetic diversity of Candida genus fungi isolated from respiratory tract of intensive care unit patients. Med Mycol. 2005;12:93–98. [Google Scholar]
- Krawczyk B, Leibner-Ciszak J, Mielech A, Nowak M, Kur J. PCR melting profile (PCR MP) - a new tool for differentiation of Candida albicans strains. 177BMC Infect Dis. 2009;9 doi: 10.1186/1471-2334-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass-Flörl C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses. 2009;52:197–205. doi: 10.1111/j.1439-0507.2009.01691.x. [DOI] [PubMed] [Google Scholar]
- Lian C, Zhao J, Zhang Z, Liu W. Genotype of Candida species associated with different conditions of vulvovaginal candidosis. Mycoses. 2004;47:495–502. doi: 10.1111/j.1439-0507.2004.01049.x. [DOI] [PubMed] [Google Scholar]
- Marol S, Yücesoy M. Molecular epidemiology of Candida species isolated from clinical specimens of intensive care unit patients. Mycoses. 2008;51:40–49. doi: 10.1111/j.1439-0507.2007.01435.x. [DOI] [PubMed] [Google Scholar]
- Morace G, Borghi E. Fungal infections in ICU patients: epidemiology and the role of diagnostics. Minerva Anestesiol. 2010;76:950–956. [PubMed] [Google Scholar]
- Müller J, Kappe R, Jaeger R, Kubitza D. Pathogen identification in indigenous deep mycoses. La Roche; Basel: 1986. 32 [Google Scholar]
- Muthig M, Hebestreit A, Ziegler U, Seidler M, Müller FM. Persistence of Candida species in the respiratory tract of cystic fibrosis patients. Med Mycol. 2010;48:56–63. doi: 10.3109/13693780802716532. [DOI] [PubMed] [Google Scholar]
- Nawrot U, Pajączkowska M, Fleischer M, Przondo-Mordarska H, Samet A, Piasecka-Pazik D, Komarnicka J, Sulik-Tyszka B, Swoboda-Kopeć E, Cieślik J, Mikucka A, Gospodarek E, Ozorowski T, Mól A, Tryniszewska E, Kłosowska W, Krawczyk M, Golec K, Szymaniak L, Giedrys-Kalemba S, Bilska I, Prawda-Zołotar J, Juszczyk-Grudzińska M, Wróblewska M, Burdynowski K. Candidaemia in Polish hospitals - a multicentre survey. Mycoses. 2013;56:576–581. doi: 10.1111/myc.12077. [DOI] [PubMed] [Google Scholar]
- Noumi E, Snoussi M, Saghrouni F, Ben Said M, Del Castillo L, Valentin E, Bakhrouf A. Molecular typing of clinical Candida strains using random amplified polymorphic DNA and contour-clamped homogenous electric fields electrophoresis. J Appl Microbiol. 2009;107:1991–2000. doi: 10.1111/j.1365-2672.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M. Candida bloodstream infections: comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008-2009. Antimicrob Agents Chemother. 2011;55:561–566. doi: 10.1128/AAC.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PM, Resende MA, Koga-Ito CY, Tendler M. Genetic variability analysis among clinical Candida spp isolates using random amplified polymorphic DNA. Mem Inst Oswaldo Cruz. 2004;99:147–152. doi: 10.1590/s0074-02762004000200006. [DOI] [PubMed] [Google Scholar]
- Robert F, Lebreton F, Bougnoux ME, Paugam A, Wassermann D, Schlotterer M, Tourte-Schaefer C, Dupouy-Camet J. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J Clin Microbiol. 1995;33:2366–2371. doi: 10.1128/jcm.33.9.2366-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CF, Silva S, Henriques M. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2013;33:673–688. doi: 10.1007/s10096-013-2009-3. [DOI] [PubMed] [Google Scholar]
- Saghrouni F, Ben Abdeljelil J, Boukadida J, Ben Said M. Molecular methods for strain typing of Candida albicans: a review. J Appl Microbiol. 2013;114:1559–1574. doi: 10.1111/jam.12132. [DOI] [PubMed] [Google Scholar]
- Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–370. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweda P, Gucwa K, Naumiuk L, Romanowska E, Dzierzanowska-Fangrat K. Evaluation of possibilities of genotyping of Candida glabrata clinical isolates with RAPD-PCR method and microsatellite analysis. Scientific Journal of Microbiology. 2013;2:194–200. [Google Scholar]
- Taverna CG, Bosco-Borgeat ME, Murisengo OA, Davel G, Boité MC, Cupolillo E, Canteros CE. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem Inst Oswaldo Cruz. 2013;108:178–185. doi: 10.1590/0074-0276108022013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowska D, Zwolinska-Wcislo M, Tokarczyk M, Kosowski K, Mach T, Budak A. The role of Candida in inflammatory bowel disease. Estimation of transmission of C. albicans fungi in gastrointestinal tract based on genetic affinity between strains. Med Sci Monit. 2010;16:CR451–CR457. [PubMed] [Google Scholar]
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K, EPIC II Group of Investigators International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- Vrioni G, Matsiota-Bernard P. Molecular typing of Candida isolates from patients hospitalized in an intensive care unit. J Infect. 2001;42:50–56. doi: 10.1053/jinf.2000.0778. [DOI] [PubMed] [Google Scholar]
- Weinberger M, Leibovici L, Perez S, Samra Z, Ostfeld I, Levi I, Bash E, Turner D, Goldschmied-Reouven A, Regev-Yochay G, Pitlik SD, Keller N. Characteristics of candidaemia with Candida-albicans compared with non-albicans Candida species and predictors of mortality. J Hosp Infect. 2005;61:146–154. doi: 10.1016/j.jhin.2005.02.009. [DOI] [PubMed] [Google Scholar]