Abstract
Nine Pseudomonas aeruginosa strains showing very high levels of resistance to various aminoglycosides have been isolated from clinical specimens in seven separate Japanese hospitals in five prefectures since 1997. These strains harbor the newly identified 16S rRNA methylase gene (rmtA). When an rmtA gene probe was hybridized with genomic DNAs of the nine strains digested with EcoRI, two distinct patterns were observed. The 11.1- and 15.8-kb regions containing the rmtA genes of strains AR-2 and AR-11, respectively, were sequenced and compared. In strain AR-2, a transposase gene-like sequence (sequence 1) and a probable tRNA ribosyltransferase gene (orfA) were located upstream of rmtA, and a Na+/H+ antiporter gene-like sequence (sequence 2) was identified downstream of rmtA. This 6.2-kbp insert (the rmtA locus) was flanked by 262-bp κγ elements. Part of the orfQ gene adjacent to an inverted repeat was found outside of the rmtA locus. In strain AR-11, the rmtA gene and sequence 2 were found, but the 5′ end of the orfA gene was truncated and replaced with IS6100. An orfQ-orfI region was present on each side of the rmtA gene in strain AR-11. The G+C content of the rmtA gene was about 55%, and since the newly identified rmtA gene may well be mediated by some mobile genetic elements such as Tn5041, further dissemination of the rmtA gene could become an actual clinical problem in the near future.
Pseudomonas aeruginosa is an important opportunistic pathogen that is capable of causing chronic and severe invasive diseases in critically ill and immunocompromised patients. Aminoglycosides are clinically effective agents for treating infections caused by P. aeruginosa as well as other gram-negative bacilli. However, multidrug resistance is rapidly emerging in P. aeruginosa, whose spectrum of resistance often includes aminoglycosides as well as broad-spectrum β-lactams and fluoroquinolones (15). The most frequently encountered molecular mechanism for aminoglycoside resistance in P. aeruginosa is the production of aminoglycoside-modifying enzymes such as plasmid-dependent acetyltransferase (AAC), adenylyltransferase (AAD), and phosphotransferase (APH) (6, 17, 23). Among these, production of AAC(6′)-II and AAD(2")-I is the most common mechanism for resistance to aminoglycosides in P. aeruginosa (1), although ribosomal mutations also play some part in aminoglycoside resistance (20). Arbekacin, one of the semisynthetic aminoglycosides belonging to the kanamycin group, is very efficacious for treatment of infections caused by both gram-positive and gram-negative bacteria, and since 1990 it has been approved, for chemotherapy of methicillin-resistant Staphylococcus aureus (MRSA) infections only, by the Japanese health insurance system. Unlike the other aminoglycosides, arbekacin is not inactivated by most of the modifying enzymes listed above. Only the bifunctional modifying enzyme composed of aminoglycoside-6′-N-acetyltransferase and 2"-O-phosphotransferase activity [AAC(6′)/APH(2")] is able to inactivate arbekacin. However, such enzymes have not been found in gram-negative bacilli to date.
We recently reported a P. aeruginosa strain that was highly resistant to most aminoglycosides, including arbekacin. This strain harbors a novel aminoglycoside resistance gene named rmtA, which encodes a new 16S rRNA methylase (29). Production of 16S rRNA methylase had been reported among aminoglycoside-producing actinomycetes, including Micromonospora spp. and Streptomyces spp., but this novel aminoglycoside resistance mechanism had not been identified in clinical pathogens before, although a similar putative 16S rRNA methylase, ArmA, was found quite recently in Klebsiella pneumoniae in Europe (11). In the present study, we investigated the genetic environments of the rmtA genes harbored by two different P. aeruginosa strains isolated in separate Japanese hospitals.
(Some of the findings presented in this manuscript have been reported at the 102nd General Meeting of the American Society for Microbiology [abstr. A-28, 2002] by Y. Doi and at its 103rd General Meeting [abstr. A-105, 2003] by K. Yamane.)
MATERIALS AND METHODS
Screening of 16S rRNA methylase producers.
In October 2001, a total of 903 nonrepetitive clinical strains of P. aeruginosa were collected from 278 medical institutions located in 22 prefectures across Japan. Potential producers of rmtA were first screened for a lack of susceptibility to gentamicin, amikacin, and arbekacin (MICs, ≥32 μg/ml). Our bacterial stock of 210 P. aeruginosa strains isolated clinically since 1997 was also subjected to a screening test for the rmtA gene. Strains that formed colonies on aminoglycoside-containing Mueller-Hinton agar plates were subjected to PCR analyses to check whether or not they harbored the rmtA gene. Primers used for amplification of the rmtA gene were RMTA-F (5′-CTA GCG TCC ATC CTT TCC TC-3′) and RMTA-R (5′-TTT GCT TCC ATG CCC TTG CC-3′), which amplify a 635-bp DNA fragment within the rmtA gene. Template DNAs used were prepared by boiling the bacterial suspension at 100°C for 10 min. Cycling parameters consisted of an initial cycle at 94°C for 5 min; 30 cycles of 94°C for 30 s, annealing at 60°C for 30 s, and extension at 74°C for 2 min; and a final 5-min incubation at 74°C. Detection of AAC(6′)/APH(2") was carried out as described by Ida et al. (13). Clinical isolates and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain(s) or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa AR-2, AR-3, AR-11, AR-15, AR-26, AR-101, AR-105, AR-112, and AR-118 | Clinical isolates carrying the rmtA gene | This study |
| E. coli XL1-Blue | supE44 recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 lac [F−proAB+lacIqZΔM15::Tn10(Tetr)] | Stratagene |
| Plasmids | ||
| pBCSK+ | Cloning vector; chloramphenicol resistant | Stratagene |
| pBCRMTH2 | Recombinant plasmid carrying a 6.8-kb HindIII fragment containing the rmtA gene of P. aeruginosa strain AR-2 | This study |
| pBCRMTE2 | Recombinant plasmid carrying a 10.3-kb EcoRI fragment containing the rmtA gene of P. aeruginosa strain AR-2 | This study |
| pBCRMTE11 | Recombinant plasmid carrying a 15.8-kb EcoRI fragment containing the rmtA gene of P. aeruginosa strain AR-11 | This study |
Antibiotics and susceptibility testing.
Antibiotics were obtained from the following sources: amikacin, Bristol Pharmaceuticals K. K., Tokyo, Japan; arbekacin, kanamycin, and streptomycin, Meiji Seika Kaisha Ltd., Tokyo, Japan; chloramphenicol, Sankyo Co., Ltd., Tokyo, Japan; gentamicin and sisomicin, Schering-Plough K. K., Osaka, Japan; hygromycin B, Sigma-Aldrich Japan K. K., Tokyo, Japan; isepamicin, Asahi Kasei Corporation, Tokyo, Japan; neomycin, Nippon Kayaku Co., Ltd., Tokyo, Japan; rifampin, Daiichi Pharmaceutical Co., Ltd., Tokyo, Japan; tobramycin, Shionogi Pharmaceutical Co., Ltd., Osaka, Japan. MICs were determined by the agar dilution method according to the protocol recommended by the National Committee for Clinical Laboratory Standards in document M7-A5 (19).
PFGE analysis.
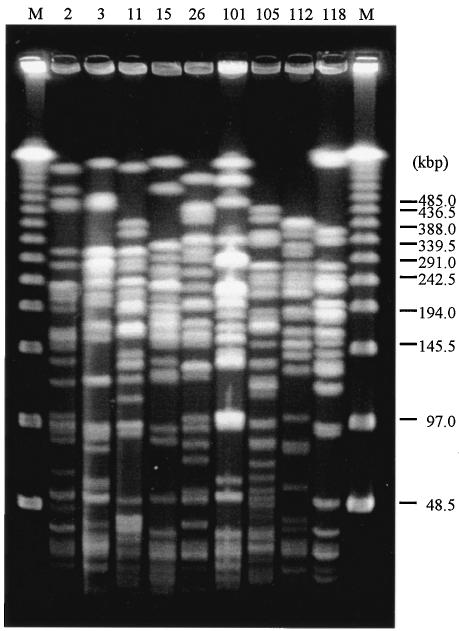
SpeI (New England Biolabs, Beverly, Mass.)-digested genomic DNAs of P. aeruginosa isolates were subjected to pulsed-field gel electrophoresis (PFGE) analysis by using a CHEF-DRII system (Bio-Rad Laboratories, Hercules, Calif.) under conditions described elsewhere (5). The pulses were increased linearly from 4 to 8 s for 10 h, after which the phase was 8 to 50 s for 12 h in this study. Banding patterns of the strains were compared visually; distinct patterns were defined by more than three fragment differences, in accordance with the criteria proposed by Tenover et al. (27).
Southern hybridization analysis of the rmtA gene.
Total DNAs of all strains were digested with EcoRI (New England Biolabs), electrophoresed through 1.0% agarose gels, transferred to nylon membranes (Bio-Rad Laboratories) by the method of Southern (25), and then hybridized with digoxigenin-labeled rmtA gene fragments by use of the PCR DIG detection system (Roche Diagnostics, Tokyo, Japan).
Cloning of the rmtA gene.
Basic recombinant-DNA techniques were carried out as described by Sambrook et al. (21). EcoRI and HindIII (New England Biolabs) were used for digestion of genomic DNA. The resultant fragments were ligated into the plasmid vector pBCSK+ (Stratagene, La Jolla, Calif.), and electrocompetent Escherichia coli XL1-Blue (Stratagene) was transformed with these recombinant plasmids. Transformants were selected on Luria-Bertani agar plates supplemented with 4 μg of arbekacin/ml and 30 μg of chloramphenicol/ml.
DNA sequencing.
DNA sequences were determined as described by Sanger et al. (22) with BigDye Terminator Cycle Sequencing Ready Reaction kits and a model 3100 DNA sequence analyzer (Applied Biosystems, Foster City, Calif.). The sequences of the cloned fragments were determined with custom sequencing primers. Nucleotide sequence alignment was performed with GENETYX-MAC (version 10.1.1; Software Development Co., Ltd., Tokyo, Japan). The nucleotide sequence was analyzed by the FASTA service of the DNA Data Bank of Japan (DDBJ) homology search system.
Nucleotide sequence accession numbers.
The nucleotide sequence data determined in this study will appear in the DDBJ database under nucleotide accession numbers AB083212 and AB120321.
RESULTS
Bacterial strains.
Among 903 strains collected in October 2001, the MICs of arbekacin, gentamicin, and amikacin for 23 strains (2.5%) were greater than 32 μg/ml. Of these, four strains (AR-101, AR-105, AR-112, and AR-118), accounting for 0.4% of all isolates, were found to be positive for rmtA by PCR analysis. From our bacterial collection of 210 P. aeruginosa strains, 5 strains (AR-2, AR-3, AR-11, AR-15, and AR-26) were PCR positive for rmtA. AAC(6′)/APH(2") was not detected in any of these nine strains by PCR analysis. Strains AR-2 and AR-3 were isolated from a hospital, as were strains AR-101 and AR-105. These nine rmtA-positive strains have been isolated from seven separate medical institutions in five prefectures in Eastern and Central Japan since 1997.
Susceptibility to antimicrobial agents.
MICs of representative aminoglycosides for these nine strains carrying the rmtA gene are shown in Table 2. All the strains were highly resistant to 4,6-disubstituted deoxystreptamines such as kanamycin, amikacin, tobramycin, and arbekacin, which belong to the kanamycin group, as well as to gentamicin, isepamicin, and sisomicin, belonging to the gentamicin group. In contrast, levels of resistance to neomycin, streptomycin, and hygromycin B varied. Strain AR-11 showed a multidrug-resistant profile to ceftazidime, imipenem, and ciprofloxacin as well as to most aminoglycosides.
TABLE 2.
Results of antibiotic susceptibility testing
| Aminoglycoside | MIC (μg/ml) for the following P. aeruginosa strain:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AR-2 | AR-3 | AR-11 | AR-15 | AR-26 | AR-101 | AR-105 | AR-112 | AR-118 | |
| Kanamycin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Amikacin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Tobramycin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Arbekacin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Gentamicin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Sisomicin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Isepacin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Neomycin | >1,024 | >1,024 | >1,024 | 128 | >1,024 | 1,024 | 512 | >1,024 | 1,024 |
| Hygromycin B | >1,024 | 1,024 | 256 | 128 | 512 | 128 | 128 | 256 | 512 |
| Streptomycin | 128 | 128 | 128 | >1,024 | 512 | 64 | 128 | 128 | 32 |
PFGE profiles.
The results of the PFGE analysis are shown in Fig. 1. The SpeI-digested patterns of the total DNAs of nine strains harboring the rmtA gene were apparently different from each other. This finding suggests not a clonal expansion of an rmtA-carrying strain but plasmid-mediated transmission of the rmtA gene among clinical strains with different genetic backgrounds by the help of some movable genetic elements such as a transposon and transferable plasmids.
FIG. 1.
PFGE fingerprinting of total DNAs from P. aeruginosa isolates digested with SpeI. M, PFGE molecular weight marker. The number above each lane indicates the AR strain number shown in Table 1.
Southern hybridization.
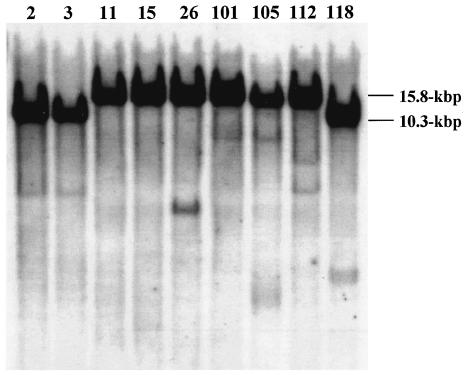
DNA fragments digested with EcoRI showed two hybridization patterns. The rmtA probe hybridized with a 10.3-kbp EcoRI fragment for strains AR-2, AR-3, and AR-118 and with a 15.8-kbp fragment for strains AR-11, AR-15, AR-26, AR-101, AR-105, and AR-112 (Fig. 2).
FIG. 2.
Southern hybridization patterns of EcoRI-digested genomic DNAs. The number above each lane represents the AR strain number shown in Table 1. The nine strains tested appeared to be divided into two groups by the sizes of EcoRI-digested fragments (10.3 and 15.8 kbp, respectively).
Genetic environments harboring rmtA genes.
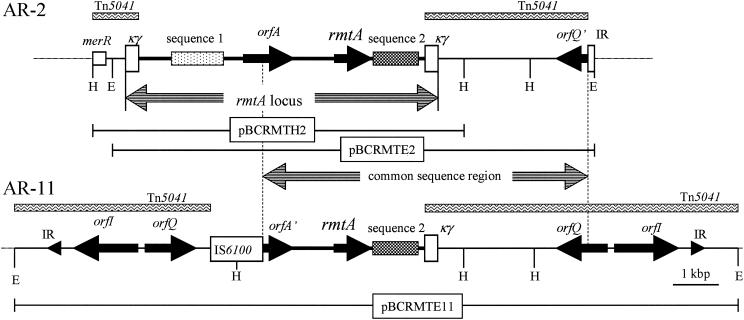
A 6.8-kbp HindIII fragment and a 10.3-kbp EcoRI fragment containing the rmtA gene of AR-2 were cloned into the plasmid vector pBCSK+. The 6.8- and 10.3-kbp fragments were inserted into pBCRMTH2 and pBCRMTE2, respectively. The schematic structure of the 11.1-kbp sequenced region cloned from strain AR-2 is shown in Fig. 3. The rmtA gene was located within a 6.2-kbp genetic locus (the rmtA locus) flanked by a 262-bp sequence named the κγ element that was previously found in Tn5041 and predicted to be a relic of mobile genetic elements (Fig. 3). The elements of the 6.2-kbp rmtA locus, comprising rmtA, orfA, and two additional specific sequences, were located in the following order: transposase gene-like sequence (sequence 1), probable tRNA ribosyltransferase gene (orfA), rmtA, and Na+/H+ antiporter gene-like sequence (sequence 2) (Fig. 3). The 5′ end of the HindIII fragment flanked merR of the mer operon found in Tn5041. However, the 3′ end of the EcoRI fragment was located within a 17-bp sequence which was completely identical to a part of the terminal inverted repeat of Tn1721. This 17-bp sequence was within orfQ, located upstream of orfI in Tn5041. The G+C content of the 6.2-kbp rmtA locus was about 55%. The 15.8-kbp EcoRI fragment of AR-11 containing the rmtA gene was also cloned into the plasmid vector pBCSK+, and the resultant recombinant plasmid was designated pBCRMTE11. In the 15.8-kbp EcoRI fragment, a 5′-truncated orfA (orfA′), rmtA, and sequence 2 were found between IS6100 and a κγ element, and the sequence was completely identical to that of the corresponding region of the 6.2-kbp rmtA locus cloned from strain AR-2. The orfQ and orfI sequences of Tn5041 were present both upstream of IS6100 and downstream of a κγ element in the 15.8-kbp EcoRI fragment cloned from strain AR-11. In the sequenced areas, the fragments harboring the rmtA gene appeared to be inserted between the κγ sequences found in Tn5041 (Fig. 3).
FIG. 3.
Comparison of the genetic organizations of AR-2 and AR-11. Double-headed striped arrows indicate the position of the rmtA locus and that of the region common to both sequenced areas. Inserts of pBCRMTH2, pBCRMTE2, and pBCRMTE11 are indicated by horizontal lines. Rectangles filled with wavy lines, sequences similar to part of Tn5041. Solid arrowheads in the 15.8-kbp EcoRI fragment, terminal inverted repeats. mer, the mercury resistance operon, includes merR. Sequence 1, transposase gene-like sequence; sequence 2, Na+/H+ antiporter-like sequence; orfA, probable tRNA ribosyltransferase gene; orfQ′, part of orfQ; orfA′, part of orfA; IR, probable inverted repeat. Restriction sites: H, HindIII; E, EcoRI. Sequences 1 and 2 encode no complete proteins due to several frameshifts and deletions.
DISCUSSION
Aminoglycoside-producing actinomycetes such as Micromonospora spp. and Streptomyces spp. protect their 30S ribosome through methylation of its 16S rRNA at the aminoglycoside-binding A site (10, 30). For example, Kgm, which was isolated from Micromonospora purpurea (28), methylates G1405, and Kam, which was isolated from Streptomyces tenjimariensis (24), methylates A1408 (2). The 16S rRNA methylases had been thought to exist among aminoglycoside-producing environmental actinomycetes such as Micromonospora spp. or Streptomyces spp (7). However, we recently reported a novel 16S rRNA methylase, RmtA, that was identified in a P. aeruginosa clinical strain, AR-2 (29). This strain demonstrated an extraordinarily high level of aminoglycoside resistance to various 4,6-disubstituted deoxystreptamines, including semisynthetic arbekacin, as well as to gentamicin and kanamycin. In the present study, we investigated the genetic environments mediating the rmtA genes found in two different strains of P. aeruginosa. The G+C content of the rmtA gene was 55%, and those of 16S rRNA methylase genes found in aminoglycoside-producing actinomycetes were 64 to 72%. These observations suggested that the rmtA gene might have been acquired by P. aeruginosa from some environmental bacteria such as aminoglycoside-producing actinomycetes, although the armA gene, with a 30% G+C content, was speculated to have originated from unknown bacteria other than actinomycetes. At any rate, lateral gene transfer across bacterial genera would become much more important for acquisition of new antibiotic resistance profiles hereafter.
Although the PFGE patterns of the nine RmtA-producing strains in this study were highly divergent, Southern hybridization showed only two hybridization patterns when genomic DNAs were digested with EcoRI. This finding indicated that the rmtA gene might be mediated by some mobile genetic elements sharing similar genetic environments and spreading among genetically unrelated strains in geographically separate hospitals. This speculation would be supported by the finding that even strains AR-2 and AR-3, isolated at the same hospital, showed different PFGE patterns. Strains AR-101 and AR-105 also demonstrated quite different PFGE profiles despite being isolated at the same hospital. Furthermore, the arbakacin resistance profile of AR-2 was transferable to another P. aeruginosa strain by conjugation (29). This suggested that rmtA was mediated by some transferable plasmids in strain AR-2, but we failed to visualize the plasmid either by the method of Kado and Liu (14) or by cesium chloride-ethidium bromide density gradient ultracentrifugation (21). This is possibly due to the instability or the very low copy number of the plasmid which mediates the rmtA gene.
Tn5041 was previously identified in a strain of a Pseudomonas species as a mercury resistance transposon (3, 16). Tn5041 carries a 4-kbp insert of unknown origin between orfQ and the mer operon, and several nonfunctional pseudogenes and possible mobile elements such as the κγ element locate in this region. The 262-bp κγ element, containing 38 bp of imperfect inverted repeats starting with the sequence GGGG and terminating internally with the sequence TAAG, falls into the inverted repeats of Tn3 family (4). Transposons belonging to the Tn3 family usually contain transposase and resolvase genes and some additional genes encoding resistance to antimicrobial agents or heavy metals such as mercury between the terminal inverted repeats. The 6.2-kbp rmtA locus found in this study was flanked by an insertion element-like κγ element. Moreover, the rmtA locus had a transposase gene-like sequence (sequence 1) whose 5′ part showed 80.2% identity with part of the transposase gene derived from Pseudomonas putida (accession number AF109307); the 3′ part of sequence 1 had 67.2% identity with part of the transposase gene derived from Pseudomonas pseudoalcaligenes (accession number AF028594), but this sequence had no apparent initiation and stop codons. Thus, the 6.2-kb rmtA locus itself is unlikely to be an active transposon, although the nucleotide sequences outside of the two κγ elements were completely identical to the corresponding regions of Tn5041. The Na+/H+ antiporter gene-like sequences (sequence 2) found in strains AR-11 and AR-2 were completely identical, although they seemed nonfunctional. Multicopy expression of the intact transposase-like gene and the Na+/H+ antiporter-like gene might disturb systematic bacterial cell growth, so these genes might have been inactivated during replication and translocation of the rmtA locus.
To examine whether strains other than AR-2 and AR-11 also carry part of the sequence found in Tn5041, Southern hybridization analysis was performed using a Tn5041-specific DNA probe containing a sequence between the right-hand κγ element and the orfQ gene, which is conserved in both strains AR-2 and AR-11. The DNA probes and the rmtA gene probe hybridized to the same fragments in all nine strains (data not shown). This finding strongly suggests the probable implication of some mobile genetic elements such as Tn5041 in the dissemination of the rmtA gene among strains of P. aeruginosa.
The 5′ end of the rmtA locus was replaced by IS6100 in strain AR-11. IS6100 was originally discovered in Mycobacterium fortuitum (accession number X53635) (18) and was subsequently found in several gram-negative and -positive bacteria (9, 26). It has been reported that transposition of IS6100 stimulates genetic rearrangement (12). Thus, it may be possible to speculate that the region containing orfQ and orfI found upstream of IS6100 might be duplicated during IS6100-mediated recombination in strain AR-11. The outside sequences of both inverted repeats had no DNA homology to the genomic DNA of P. aeruginosa PAO-1. This finding suggests that the 15.8-kb EcoRI fragment of strain AR-11 might be carried by a much longer mobile genetic element, since the arbekacin-resistant profile of AR-11 was not transferred to another P. aeruginosa strain by conjugation, and no apparent plasmid was detected in this strain by the method of Kado and Liu (14). Additionally, rmtA gene probes hybridized to the position of chromosomal DNA (data not shown). These findings strongly suggested that the rmtA gene and its adjacent regions might be integrated into the chromosomal DNA in strain AR-11.
P. aeruginosa strains harboring the rmtA gene have already been found in several separate clinical settings in Japan, and a gene encoding the same kind of 16S rRNA methylase, called armA, has also been identified in members of the family Enterobacteriaceae, such as Citrobacter freundii (accession number NC004464) and K. pneumoniae (11) (accession number AY220558), in Europe. ArmA shares 29% identity with RmtA at the amino acid sequence level. Moreover, a new plasmid-mediated 16S rRNA methylase, RmtB, that shares 82% identity with RmtA at the amino acid sequence level, has also been identified in Serratia marcescens in Japan (8) (accession number AB103506). From our preliminary study on a bacterial stock, the presence of these genes was also suggested in several strains of K. pneumoniae, E. coli, and Acinetobacter species isolated in Japan. Thus, further dissemination of these genetic determinants to various pathogenic gram-negative bacilli could become a serious concern in the near future.
In Japanese clinical settings, various aminoglycosides have been used in the treatment of bacterial infections, since these agents still have very high efficacies against both gram-positive and gram-negative bacteria. Arbekacin is a semisynthetic aminoglycoside belonging to the kanamycin-group. It has been approved, for MRSA infection only, since 1990, and it is still very efficacious for MRSA infection. Under such clinical circumstances, arbekacin has been preferentially used in many clinical settings, although arbekacin-resistant strains which produce the bifunctional enzyme AAC(6′)/APH(2") have emerged in MRSA. No such bifunctional enzymes, however, have been found in gram-negative bacilli to date. Thus, acquisition of 16S rRNA methylase would give gram-negative bacteria a great advantage in coping with clinical environments where huge amounts of semisynthetic aminoglycosides, including arbekacin, are consumed. Hence, one should recall again that bacteria can survive and proliferate in clinical environments, given their natural hereditary capacity to overcome the hazards of any environment.
Acknowledgments
This work was supported by grants H12-Shinkou-19, H12-Shinkou-20, H15-Shinkou-9, and H15-Shinkou-10 from the Ministry of Health, Labor, and Welfare of Japan. The travel costs for presenting part of the data reported in this paper at the 103rd General Meeting of the American Society for Microbiology was supported by a grant from The Japan Antibiotics Research Association, sponsored by the Pfizer Infectious Diseases Research Fund 2003.
REFERENCES
- 1.Aminoglycoside Resistance Study Groups. 1994. Resistance to aminoglycosides in Pseudomonas. Trends Microbiol. 2:347-353. [PubMed] [Google Scholar]
- 2.Beauclerk, A. A. D., and E. Cundliffe. 1987. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J. Mol. Biol. 193:661-671. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanova, E. S., S. Z. Mindlin, E. S. Kalyaeva, and V. G. Nikiforov. 1988. The diversity of mercury reductases among mercury-resistant bacteria. FEBS Lett. 234:280-282. [DOI] [PubMed] [Google Scholar]
- 4.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz. 2002. Mobile DNA II. ASM Press, Washington, D.C.
- 5.D'Agata, E., L. Venkataraman, P. DeGirolami, and M. Samore. 1997. Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J. Clin. Microbiol. 35:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, J., and G. D. Wright. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5:234-240. [DOI] [PubMed] [Google Scholar]
- 7.Davies, J. D. 1983. Resistance to aminoglycosides: mechanisms and frequency. Rev. Infect. Dis. 5:S261-S267. [Google Scholar]
- 8.Doi, Y., K. Yokoyama, K. Yamane, J., Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa,. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois, V., C. Arpin, P. Noury, and C. Quentin. 2002. Clinical strain of Pseudomons aeruginosa carrying a blaTEM-21 gene located on a chromosomal interrupted TnA type transposon. Antimicrob. Agents Chemother. 46:3624-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouemy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1347-1371. [DOI] [PubMed] [Google Scholar]
- 11.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunos, G., B. Smith, and P. Dyson. 1999. Genetic instability associated with insertion of IS6100 into one end of the Streptomyces lividans chromosome. Microbiology 145:2203-2208. [DOI] [PubMed] [Google Scholar]
- 13.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlowsky, J. A., D. C. Draghi, M. E. Jones, C. Thornsberry, I. R. Friedland, and D. F. Sahm. 2003. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob. Agents Chemother. 47:1681-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kholodii, G. Y., O. V. Yurieva, Z. M. Gorlenko, S. Z. Mindlin, I. A. Bass, O. L. Lomovskaya, A. V. Kopteva, and V. G. Nikiforov. 1997. Tn5041: a chimeric mercury resistance transposon closely related to the toluene degradative transposon Tn4651. Microbiology 143:2549-2556. [DOI] [PubMed] [Google Scholar]
- 17.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, C., J. Timm, J. Rauzier, R. Gomez-Lus, J. Davies, and B. Gicquel. 1990. Transposition of an antibiotic resistance element in mycobacteria. Nature 345:739-743. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1988. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Recht, M. I., and J. D. Puglisi. 2001. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob. Agents Chemother. 45:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzames. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skeggs, P. A., J. Thompson, and E. Cundliffe. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics in clones of Streptomyces lividans carrying DNA from Streptomyces tenjimariensis. Mol. Gen. Genet. 200:415-421. [DOI] [PubMed] [Google Scholar]
- 25.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 26.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48:117-129. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J., P. A. Skeggs, and E. Cundliffe. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol. Gen. Genet. 201:168-173. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa,. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]
- 30.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 17:6437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]